Abstract
Membrane type 1 matrix metalloproteinase (MT1-MMP) plays a critical role in cancer cell biology by proteolytically remodeling the extracellular matrix. Utilizing fluorescence resonance energy transfer (FRET) imaging, we have developed a novel biosensor, with its sensing element anchoring at the extracellular surface of cell membrane, to visualize MT1-MMP activity dynamically in live cells with subcellular resolution. Epidermal growth factor (EGF) induced significant FRET changes in cancer cells expressing MT1-MMP, but not in MT1-MMP-deficient cells. EGF-induced FRET changes in MT1-MMP-deficient cells could be restored after reconstituting with wild-type MT1-MMP, but not MMP-2, MMP-9, or inactive MT1-MMP mutants. Deletion of the transmembrane domain in the biosensor or treatment with tissue inhibitor of metalloproteinase-2, a cell-impermeable MT1-MMP inhibitor, abolished the EGF-induced FRET response, indicating that MT1-MMP acts at the cell surface to generate FRET changes. In response to EGF, active MT1-MMP was directed to the leading edge of migrating cells along micropatterned fibronectin stripes, in tandem with the local accumulation of the EGF receptor, via a process dependent upon an intact cytoskeletal network. Hence, the MT1-MMP biosensor provides a powerful tool for characterizing the molecular processes underlying the spatiotemporal regulation of this critical class of enzymes.
Extracellular matrix macromolecules present cancer cells with a structural barrier that serves to limit their unregulated growth and movement (1, 2). In turn, cancer cells have been postulated to use matrix metalloproteinases (MMPs),2 a class of zinc-dependent proteolytic enzymes, as a means to dissolve these extracellular matrix barriers during neoplastic progression (1-3). Although the human MMP family is comprised of 16 secreted and seven membrane-tethered enzymes, increasing evidence suggests that a subclass of the membrane-anchored proteinases, termed the membrane type (MT) MMPs, plays dominant roles in controlling cancer cell behavior (1, 4-8). The MT-MMPs are expressed either as type I transmembrane proteins (i.e. MT1, 2, 3, and 5 MMPs) or in a glycosylphosphatidyl-inositol-anchored format (i.e. MT4- and MT6-MMP) (1). Among these enzymes, MT1-MMP is considered to be the family member most closely linked to neoplastic cell behavior (6, 8). Indeed, recent studies have demonstrated that MT1-MMP plays a direct and essential role in allowing tumor cells to degrade and invade multiple connective tissue barriers through a mechanism independent of MMP-2, an effector protease that operates downstream of MT1-MMP (4, 7).
Synthesized as a catalytically inactive proenzyme, the MT1-MMP precursor undergoes proteolytic processing in the transGolgi complex wherein the prodomain is removed by members of the proprotein convertase family (9). Subsequently, the mature enzyme traffics to the plasma membrane via a process controlled by Rab8 and the microtubular apparatus (6, 10). Little is known with regard to the signaling molecules that mobilize MT1-MMP to the cell surface, but EGF, an extracellular ligand of EGF receptor family members (EGFRs; also known as ERBB/HERs), has been reported to induce cancer invasion by modulating MT1-MMP expression (11, 12). Once delivered to the cell surface, the short cytoplasmic tail of MT1-MMP regulates its internalization and turnover at the plasma membrane, possibly through its interactions with adaptor protein 2, which resides in clathrin-coated pits (13, 14). Migrating cells are then thought to localize MT1-MMP to various cellular domains by complex processes involving caveolar proteins, the actin network, and extracellular binding partners (5, 10, 15-17). Src kinase has also been shown to phosphorylate the cytoplasmic tail of MT1-MMP and regulate its functions (18). Despite these insights, however, few of these principles have been evaluated directly because techniques have not yet been developed for monitoring MT1-MMP activity with subcellular resolution in live cells.
Recent advances in fluorescent probes have enabled the study of protein activity and distribution in live cells (19, 20). Along these lines, we previously developed a biosensor based on fluorescence resonance energy transfer (FRET) to monitor Src activity in real time at the single cell level (21). We report here the development and characterization of a new, FRET-based MT1-MMP biosensor (GenBank™ accession number EU545473) anchored at the plasma membrane wherein a MT1-MMP-sensitive substrate has been concatenated between the high efficiency FRET pair, ECFP and YPet (22). Utilizing this biosensor, MT1-MMP activity and distribution have been resolved in live cells with high spatiotemporal resolution. Further, new mechanistic insights have been developed into the means by which the local activity of MT1-MMP is coordinated at the surface of migrating cancer cells.
EXPERIMENTAL PROCEDURES
Protein Expression, in Vitro Spectroscopy, and Cleavage Assays—Chimeric proteins were expressed with N-terminal His6 tags in Escherichia coli and purified by nickel chelation chromatography as described (21). Emission ratios of ECFP/YPet (476 nm/526 nm) were measured by a fluorescence plate reader (TECAN, Sapphire II) before and after adding the recombinant catalytic domain of human MT1-, MT2-, or MT3-MMP (2 μg/ml; Calbiochem) or of human MMP-2 or MMP-9 (6 μg/ml; Calbiochem) into the proteolysis assay buffer (50 mm HEPES, 10 mm CaCl2, 0.5 mm MgCl2, 50 μm ZnCl2, and 0.01% Brij-35, pH 6.8) at 37 °C (23). The samples were separated by 10% SDS-PAGE gels followed by Coomassie Blue staining. After destaining (50% v/v methanol in water with 10% acetic acid), the protein was visualized, and the image was recorded by digital camera (Olympus).
Gene Construction and DNA Plasmids—The substrate peptide sequence CPKESCNLFVLKD was derived from the MT1-MMP cleavage site identified in proMMP-2 (24) and used for the MT1-MMP FRET biosensor. The YPet cDNA was amplified by PCR with a sense primer containing a BglII site and a reverse primer containing a SacI site. ECFP was amplified by PCR with a sense primer containing a SacI site and the sequence of the MT1-MMP substrate peptide and a reverse primer containing a PstI site, a stop codon, and a HindIII site. The PCR products were fused together and cloned into pRSETb (Invitrogen) using BglII/HindIII sites for bacterial expression and into pDisplay (Invitrogen) using BglII/PstI sites for mammalian cell expression (see Fig. 1A). The pDisplay vector contains an N-terminal murine Ig κ-chain leader sequence, which directs the biosensor protein to the secretory pathway, and a C-terminal transmembrane domain of the platelet-derived growth factor receptor β (i.e. PDGFRβ)), which targets the biosensor protein to the plasma membrane. The cytosolic MT1-MMP biosensor for mammalian cell expression was constructed by PCR amplification of the fused full-length gene encoding the MT1-MMP biosensor and subcloned into pcDNA3.1 (Invitrogen) with BamHI and EcoRI sites. A mutant MT1-MMP biosensor was constructed by replacing the sequence encoding NL with IV in the sense primer used for YPet amplification.
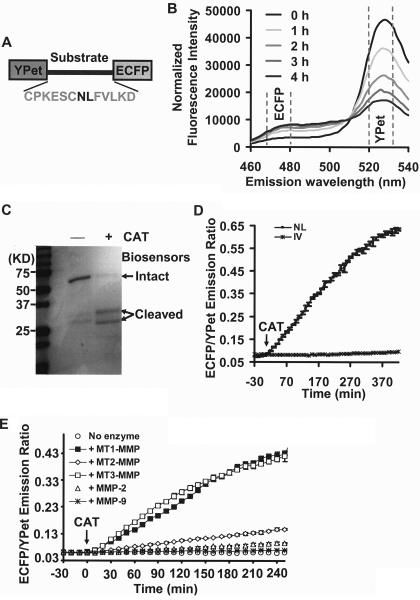
FIGURE 1.
In vitro characterization of the MT1-MMP biosensor. A, domain structure of MT1-MMP biosensor with a YPet and ECFP at its N and C termini connected by a substrate peptide for MT1-MMP. B, emission spectra of the MT1-MMP biosensor (excited at 437 nm) before and after incubation with the catalytic domain of MT1-MMP (CAT) for various periods of time as indicated. The broken lines highlight the regions encompassing emission maxima of ECFP and YPet. C, PAGE gel showing the sizes of MT1-MMP biosensor with or without the incubation of CAT following a 15-h incubation period. D, the time courses of ECFP/YPet emission ratio (means ± S.D.) of wild-type (▪) and mutant (NL to IV mutation; *) biosensor before and after the incubation with CAT. E, the time courses of ECFP/YPet emission ratio (mean ± S.D.) of wild-type biosensor before and after the incubation with the catalytic domains of MT1-MMP, MT2-MMP, MT3-MMP, and active MMP-2 and MMP-9, as indicated.
The mCherry-conjugated MT1-MMP was constructed by PCR amplification of the cDNA encoding MT1-MMP with a sense primer containing a HindIII site and a reverse primer containing a gene sequence encoding GGS as a linker and an EcoRI site. The PCR product of cDNA encoding mCherry was fused at the C-terminal of MT1-MMP with GGS and cloned into pcDNA3.1 with HindIII/XhoI sites. The GFP-fused EGFR construct was a gift from Dr. Sorkin at the Department of Pharmacology, University of Colorado (25). The constructs for MT1-MMP and various other plasmids: control vector (PCR3.1 Uni; Invitrogen); human MT1-MMP; catalytically inactive human MT1-MMP (E/A mutant, Glu240 → Ala); transmembrane-deleted human MT1-MMP (MT1ΔTM); cytoplasmic tail-deleted human MT1-MMP (MT1ΔCT); and human MMP-2, MMP-9, and their convertase-activable forms (MMP-2RXKR and MMP-9RXKR) have been previously published (26).
Micropatterning and Migration Assay—Glass coverslips (Fisher) were cleaned with a solution containing H2SO4 and H2O2 prior to the silanization in 2% dimethyl dichlorosilane (Aldrich) in dichlorobenzene for 10 s. The treated slips were rinsed with acetone, ethanol, and water, blown dry, and oxidized by UV-generated ozone (UVO Cleaner; Jelight, Irvine, CA) for 1 min (27). These treated coverslips were stored at 4 °C before usage. The polydimethylsiloxane microchannel mold was created by soft lithography. Negative photo-resist Epon SU8 2015 was coated on a silicon wafer, which was then exposed to UV light through a transparency mask with parallel lines and spacing before being developed. Polydimethylsiloxane prepared by mixing two liquid components (Sylgard 184 kit; Dow Corning) was poured onto the developed wafer and cured. After solidification, the polydimethylsiloxane mold with microgrooves was peeled off and sealed on treated coverslips to create microfluidic channels. Fibronectin solution (40 μg/ml) was perfused through the channel driven by pressure gradient to coat the coverslips with defined parallel lines (10 μm in width with 30-μm spacing). The coverslips were then backfilled with pluronic acid (F127) (BASF Corporation; 0.5% in phosphate-buffered) to prevent the cell adhesion outside of the patterned areas.
After co-transfection with wild-type biosensor and MT1-MMP for 36-48 h, HeLa cells were detached with 4 mm EDTA (pH 7.4) in phosphate-buffered and seeded on fibronectin-coated glass-bottomed dishes or fibronectin stripes for 3 h before EGF stimulation.
Microscopy, Image Acquisition, and Analysis—Cells expressing various exogenous proteins were starved in 0.5% fetal bovine serum for 36-48 h before EGF (50 ng/ml) treatment. During the imaging process, the cells were maintained in serum-free CO2-independent medium (Invitrogen) at 37 °C. The images were collected with a Zeiss axiovert inverted microscope equipped with a cooled charge-coupled device camera (Cascade 512B; Photometrics) using MetaFluor 6.2 software (Universal Imaging). The parameters of dichroic mirrors, excitation and emission filters for FRET, and different fluorescence proteins are shown in supplemental Table S1. The fluorescence intensity of nontransfected cells were quantified as the background signal and subtracted from the ECFP and YPet (FRET) signals on transfected cells. There is minimal cross-talk between ECFP and YPet channels (i.e. the direct excitation of YPet with ECFP excitation wavelength and the ECFP emission bleed-through into YPet channel) with our filter settings (data not shown). Hence, the pixel-by-pixel ratio images of ECFP/YPet were directly calculated based on the background-subtracted fluorescence intensity images of ECFP and YPet by the MetaFluor software to represent the FRET efficiency and activation levels of biosensor. Emission ratios of ECFP/YPet were averaged on chosen regions of interest to allow the quantification and statistical analysis by Excel (Microsoft) and Matlab (The MathWorks).
Statistical Analysis—For statistical analysis, we used the Bonferroni multiple comparison test of means at 95% confidence interval, which is provided by the multcompare function in the MATLAB statistics toolbox.
RESULTS
In Vitro Characterization of the MT1-MMP Biosensor—To generate a sensitive biosensor for detecting MT1-MMP activity, a substrate peptide (31CPKESCNLFVLKD43) derived from the MT1-MMP cleavage site in the propeptide sequence of MMP-2 (24) was flanked by a fluorescence protein pair, ECFP and YPet (22) for FRET (Fig. 1A). In this construct, we reasoned that active MT1-MMP would cleave the biosensor substrate peptide and thus separate ECFP and YPet, resulting in a change in FRET that could be tracked by an increase in the emission ratio of ECFP/YPet. Indeed, following incubation of the engineered biosensor with the catalytic domain of MT1-MMP (CAT), a significant decrease in the YPet spectrum max (526 nm) was detected along with a concomitant increase in the ECFP spectrum max (476 nm), indicative of FRET attenuation (Fig. 1B). Analysis by gel electrophoresis further revealed that the biosensor can be cleaved by CAT (Fig. 1C). If, however, the critical cleavage site NL was mutated to IV (24), the FRET response of the biosensor was abolished (Fig. 1D). Taken together, these results indicate that MT1-MMP hydrolyzes the biosensor at the designed cleavage site, which leads to the expected FRET change in vitro. The specificity of biosensor was further examined by comparing the catalytic domains of MT1-MMP with other MMP family membranes. MT1-MMP displayed a much stronger activity toward the biosensor in comparison with MT2-MMP, MMP-2, and MMP-9, although MT3-MMP can also efficiently induce FRET change of the biosensor (Fig. 1E). Gel electrophoresis results on biosensor cleavage are consistent with the FRET signals (data not shown). These results indicate that the biosensor is sensitive to the catalytic domains of both MT1- and MT3-MMP in vitro, but not to MT2-MMP, MMP-2, and MMP-9.
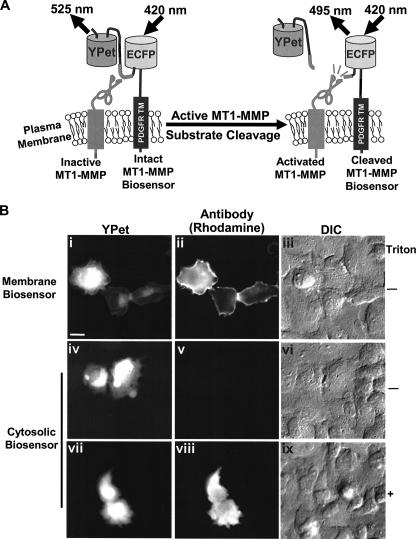
Targeting the MT1-MMP Biosensor to the Extracellular Face of the Cell Plasma Membrane—Because the catalytic domain of MT1-MMP is directed toward the extracellular face of the plasma membrane (1), the cDNA encoding the MT1-MMP biosensor was subcloned into pDisplay vector which contains an N-terminal murine Ig κ-chain leader sequence for secretory pathway targeting and a C-terminal transmembrane domain of PDGFRβ for plasma membrane targeting. Because both MT1-MMP and PDGFRβ have been demonstrated previously to colocalize at the cell surface (28), the functional domain of the biosensor protrudes outward from the surface of plasma membrane in proximity to the MT1-MMP catalytic domain (Fig. 2A). Following transfection of HeLa cells with the biosensor, the full-length construct was oriented properly on the cell surface as evidenced by the strong staining observed with an ECFP/YPet-reactive antibody in nonpermeabilized cells (Fig. 2B, panels i-iii). By contrast, when cells were transfected with a biosensor construct lacking PDGFR_TM, ECFP/YPet could only be immunodetected after the membrane was permeabilized (Fig. 2B, panels iv-ix). These results confirmed that the MT1-MMP biosensor is correctly targeted to the cell surface. Of note, a portion of the membrane-targeted biosensor was localized to perinuclear regions, possibly reflecting biosensors trapped in an endo/exocytosis recycling route before being transported to the plasma membrane (29).
FIGURE 2.
Design strategy and membrane localization of the MT1-MMP biosensor in mammalian cells. A, design strategy of the membrane-tethered MT1-MMP biosensor. The biosensor is fused to the transmembrane domain of PDGFR to position its sensing element on the extracellular surface of the plasma membrane and accessible to MT1-MMP. Active MT1-MMP can cleave the substrate peptide to separate ECFP and YPet, which leads to the observed FRET decrease. B, extracellular surface localization of the membrane-targeted MT1-MMP biosensors. HeLa cells were transfected with the membrane-targeted (panels i-iii) or cytosolic (panels iv-ix) biosensor, and stained with GFP-reactive antibodies with (panels i-vi) or without (panels vii-ix) permeabilization of the plasma membrane. panels i, iv, and vii, the fluorescence intensity image of biosensor; panels ii, v, and viii, the immunostained image of biosensors with ECFP/YPet-reactive antibody and rhodamine-conjugated secondary antibody; panels iii, vi, and ix, the differential interference contrast image of HeLa cells. Scale bar, 20 μm.
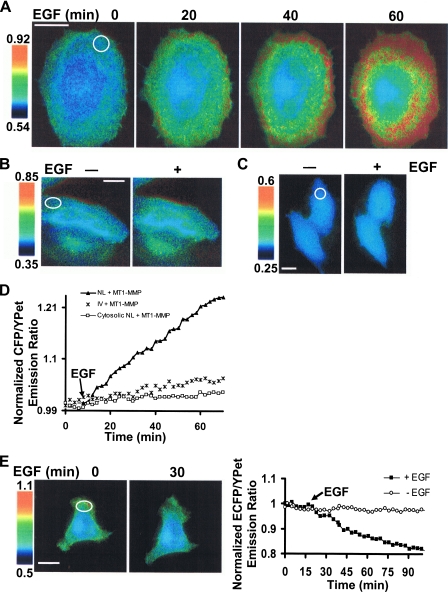
Functional Characterization of the MT1-MMP Biosensor in Live Cells—To begin characterizing the utility of the MT1-MMP biosensor, HeLa cells (a cell type that expresses minimal levels of endogenous MT1-MMP) (30) were co-transfected with wild-type MT1-MMP and the membrane-targeted biosensor. In the presence of EGF, a significant FRET change was induced over a 60-min monitoring period (Fig. 3A and supplemental Movie S1). By contrast, the FRET response was ablated when the NL → IV cleavage-resistant (24) biosensor was coexpressed with MT1-MMP (Fig. 3B). As expected, deletion of PDGFR_TM in the biosensor probe, a modification that precludes its trafficking to the cell surface, also abolished the EGF-induced FRET response (Fig. 3C), supporting the conclusion that EGF-induced MT1-MMP activation occurs preferentially at the cell surface. Quantification of FRET responses further confirmed that EGF-induced biosensor cleavage occurs only with the wild-type membrane-anchored construct but not in the cleavage-resistant mutant or cytosol-directed biosensors (Fig. 3D). Finally, in a fashion consistent with the extracellular proteolysis of the biosensor, tissue inhibitor of metalloproteinase-2 (TIMP-2), an endogenous, ∼25-kDa inhibitor of MT1-MMP(7), reversed the EGF-induced FRET change in the MT1-MMP-transfected HeLa cells (Fig. 3E). The observed decrease in the ECFP/YPet emission ratio that occurred in the presence of TIMP-2 is most likely attributed to an enhanced turnover rate of the biosensor as well as MT1-MMP at the plasma membrane upon EGF stimulation (see “Discussion”).
FIGURE 3.
FRET response of the MT1-MMP biosensor in HeLa cells. A, ECFP/YPet emission ratio images of a cell co-transfected with the MT1-MMP biosensor and MT1-MMP before and after EGF as a function of time. B and C, ECFP/YPet emission ratio images of mutant (IV) (B) and cytosol-directed MT1-MMP biosensor (C) in HeLa cells before and after EGF stimulation for 60 min. D, representative time courses of normalized ECFP/YPet emission ratio in HeLa cells transfected with MT1-MMP and different MT1-MMP biosensors: the wild-type membrane-targeted (▴), mutant membrane-targeted (IV; *), or cytosol-directed (□). The emission ratios were averaged over the indicated regions of interest at the cell periphery (white circles) as shown in A-C. E, HeLa cells expressing the MT1-MMP biosensor and MT1-MMP were treated with 2.5 μg/ml TIMP-2 for 10 min before EGF stimulation. ECFP/YPet emission ratio images before and after EGF are shown on the left. Representative time course of normalized ECFP/YPet emission ratio averaged over the indicated region of interest (white circle on the left) is shown on the right together with a control time course in which cells were not exposed to EGF. In all the panels, the scale bars represent the ECFP/YPet emission ratio, with cold and hot colors representing low and high activities of MT1-MMP, respectively. Because the FRET change in the same cell before and after stimulation is studied, the dynamic ranges of scale bars are set to be similar with the absolute ratio values varying dependent on the different biosensor used.
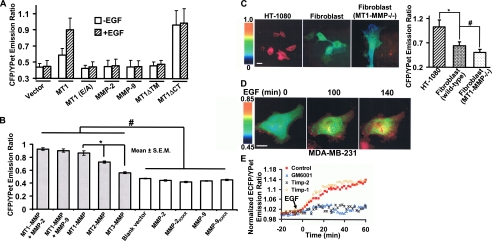
To further delineate the structural characteristics that underlie the MT1-MMP-dependent hydrolysis of the biosensor, EGF-treated HeLa cells were co-transfected with MT1-MMP constructs harboring either an inactivating point mutation in the catalytic domain (14) (i.e. MT1 (E/A)) or a transmembrane-deletion mutant that results in the secretion of a truncated, but active, form of the enzyme (6) (i.e. MT1ΔTM). In either case, EGF was unable to induce a significant FRET response in HeLa cells (Fig. 4A and supplemental Fig. S1). Similarly, HeLa cells transfected with either MMP-2 or MMP-9 did not generate a significant FRET response following EGF treatment (Fig. 4A and supplemental Fig. S1). By contrast, when the cytosolic tail of MT1-MMP was deleted (MT1ΔCT), the membrane-tethered mutant generated a high ECFP/YPet ratio with or without EGF stimulation (Fig. 4A and supplemental Fig. S1). Western blot analysis of cell lysates further revealed that EGF triggered biosensor cleavage only in cells transfected with MT1-MMP, as evidenced by the generation of a ∼35-kDa fragment of the biosensor (supplemental Fig. S2), most likely reflecting the ECFP-PDGFR_TM fusion protein that remains tethered to the plasma membrane after cleavage (Fig. 2A). Interestingly, MT1ΔCT resulted in a very low level of both intact and cleaved biosensors (supplemental Fig. S2), possibly representing a sustained high level of cleavage and degradation of the biosensor. Serum-supplemented medium also induced a FRET change in biosensor-expressing cells transfected with wild-type MT1-MMP, but not MT1 (E/A) or the control vector (supplemental Fig. S3). Further examination revealed that in the presence of serum, the biosensor is sensitive to MT1-MMP, but not to wild-type or constitutively active forms of MMP-2 and MMP-9 (Fig. 4B). The expression of MMP-2 or MMP-9 with MT1-MMP did not cause significant increase in FRET changes when comparing with cells expressing MT1-MMP only. Interestingly, although the catalytic domain of MT3-MMP displayed a substantial activity toward the biosensor in vitro (Fig. 1E), in live cells, the biosensor is most sensitive to MT1-MMP as compared with MT2- or MT3-MMP (Fig. 4B). Hence, in live cells, the biosensor preferably monitors the activity of MT1-MMP, although MT2- and MT3-MMP may also contribute to the signal depending on their respective levels of activities.
FIGURE 4.
The MT1-MMP biosensor monitors MT1-MMP activity. A, bar graph representing the ECFP/YPet emission ratio of HeLa cells transfected with the MT1-MMP biosensor together with different MT1-MMP mutants (MT1, wild-type MT1-MMP; MT1 (E/A), catalytically inactive MT1-MMP; MT1ΔTM, tail-deleted MT1-MMP; MT1ΔCT, transmembrane-domain-deleted MT1-MMP (13)) or other types of MMP (MMP-2 and MMP-9), before (open bars) and after (shaded bars) EGF treatment for 3 h. B, bar graph representing the ECFP/YPet emission ratio of HeLa cells transfected with the biosensor together with a control vector or different MMPs in the presence of 10% fetal bovine serum. The * and # signs represent statistically significant differences between the indicated groups. C, the ECFP/YPet emission ratio images of MT1-MMP biosensor in HT-1080 cells, wild-type or MT1-MMP-/- fibroblasts (left side). The bar graph on the right side represents the ECFP/YPet emission ratio (means ± S.D.) averaged over the whole bodies of cells. The * and # signs represent statistically significant differences between the indicated groups. D, ECFP/YPet emission ratio images of MT1-MMP biosensor before and after EGF (50 ng/ml) stimulation in MDA-MB-231 cells. E, representative time courses of normalized ECFP/YPet emission ratio of MT1-MMP biosensor averaged over the whole bodies of MDA-MB-231 cells treated with TIMP-2, TIMP-1, GM6001, or solvent control (0.1% Me2SO).
To monitor the ability of the biosensor to detect endogenous MT1-MMP activity, FRET imaging was assessed in (i) HT-1080 cells that are known to express high levels of the proteinase or (ii) mouse dermal fibroblasts isolated from either MT1MMP+/+ or MT1-MMP-/- mice (7). Among these cells, the ECFP/YPet emission ratio was highest in HT-1080 cells, modest in wild-type fibroblasts, and lowest in MT1-MMP-/- fibro-blasts (Fig. 4C). Western blot analyses confirmed that the expression of MT1-MMP in MT1-MMP-/- fibroblasts is ablated (supplemental Fig. S4). Although the wild-type fibro-blasts generate only a modest signal (consistent with their low level of MT1-MMP activity (7)), the addition of the synthetic MMP inhibitor, GM6001, caused strong FRET changes while exerting only small effects on MT1-MMP-/- cells (supplemental Fig. S5), presumably because of their MT3-MMP activity (31). These results reinforced the note that MT1-MMP plays a more dominant role than MT3-MMP in biosensor cleavage. Because both MT1-MMP and EGFR are abundant in the MDA-MB-231 breast cancer cell line (32, 33), we further examined whether the biosensor can monitor the dynamic activation of endogenous MT1-MMP in response to EGF stimulation. Indeed, a significant FRET change was observed in MDA-MB-231 cells upon EGF stimulation with the highest level of activity found at the cell periphery (Fig. 4D). Time course analyses further revealed that this EGF-induced FRET change was inhibited by TIMP-2 and GM6001 (a general, synthetic MMP inhibitor (34)), but not TIMP-1 (an inhibitor that preferentially targets secreted MMPs (7)) (Fig. 4E). Together, these results suggest that the biosensor can preferably report MT1-MMP activity in live cells.
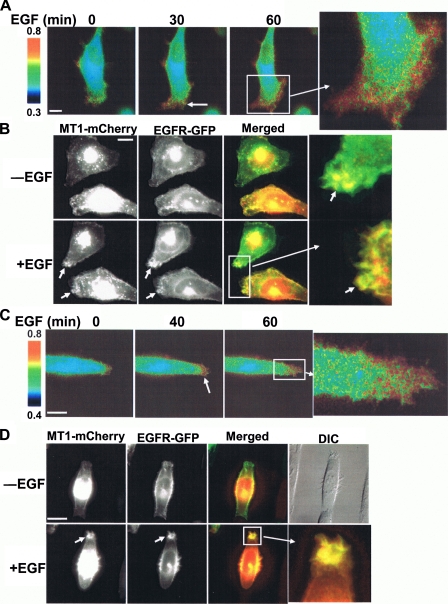
EGF-induced Mobilization of MT1-MMP Activity to the Migrating Cell Front—To further characterize the spatiotemporal effect of EGF on MT1-MMP activity, HeLa cells were co-transfected with both the biosensor and MT1-MMP and then seeded atop fibronectin-coated surfaces to initiate a motile response. Following EGF treatment, there was a significant increase of directional FRET signals with intensified activity pattern appearing at the migrating front relative to the lower activity monitored at the rear of the cell (Fig. 5A, supplemental Fig. S6, and supplemental Movie S2), although no clear patterns in ECFP fluorescence intensity can be observed at the migrating front after EGF stimulation (supplemental Fig. S7A). Histograms of ECFP intensity versus the ratiometric signal (ECFP/YPet) in MT1-MMP-transfected HeLa cells cultured in the absence or presence of EGF indicate that there is no significant correlation between the fluorescence intensity and FRET ratio (supplemental Fig. S7B), with an overall shift toward higher ratio of pixels inside cells after EGF stimulation (supplemental Fig. S7C). Hence, the directional FRET signals detected in this system should represent MT1-MMP spatial activity, but not artifacts caused by local differences in fluorescence intensity. Given these results, EGFP and mCherry (a version of red fluorescence protein (25)) were then fused to EGFR and MT1-MMP, respectively, to study their dynamic localization in migrating HeLa cells. Interestingly, EGFP-EGFR and mCherry-MT1-MMP co-localized at the migrating front of HeLa cells in response to EGF stimulation (Fig. 5B and supplemental Movie S3). To further characterize this directional MT1-MMP activity, we constrained HeLa cell movement by growing the cells on 10-μm-wide parallel stripes coated with fibronectin (supplemental Fig. S8A). As shown in Fig. 5 (C and D), EGF induced a high MT1-MMP activity (supplemental Movie S4) and a concomitant clustering of MT1-MMP and EGFR at the leading edge of the cell migrating along the fibronectin stripes (supplemental Movie S5). The dynamic regions flanking the nucleus wherein MT1-MMP and EGFR are co-localized likely represent transient filopodia protrusions probing the surrounding environment of the fibronectin-coated stripes (Fig. 5D). Again, no clear patterns in ECFP fluorescence intensity can be observed at the migrating front after EGF stimulation (supplemental Fig. S8B). Hence, EGF preferentially mobilizes both MT1-MMP activity and EGFR to the leading edge of live cells in response to migratory signals.
FIGURE 5.
EGF-induced polarization of MT1-MMP activity and co-localization of MT1-MMP and EGFR at the leading edge of migrating cells. A, HeLa cells co-transfected with MT1-MMP biosensor and MT1-MMP were plated on 2 μg/cm2 fibronectin for 3 h before EGF stimulation as indicated. ECFP/YPet emission ratio images were shown to represent the distribution of MT1-MMP activity. B, MT1-MMP fused with mCherry and EGFR fused with EGFP were co-transfected into HeLa cells, plated on fibronectin, and stimulated with EGF. The images of MT1-MMP-mCherry and EGFR-EGFP were displayed and overlaid to demonstrate their co-localization at the migrating front. C, HeLa cells co-transfected with MT1-MMP biosensor and MT1-MMP were constrained on micropatterned parallel stripes (10 μm in width) coated with fibronectin before stimulation with EGF. ECFP/YPet emission ratio images are shown to represent the distribution of MT1-MMP activity. D, HeLa cells were co-transfected with MT1-MMP-mCherry and EGFR-EGFP, plated on fibronectin-coated stripes, and stimulated with EGF. The images of MT1-MMP-mCherry and EGFR-EGFP were displayed and overlaid to demonstrate their co-localization at the leading edge along the stripes. The differential interference contrast image of cells is shown in the upper right corner.
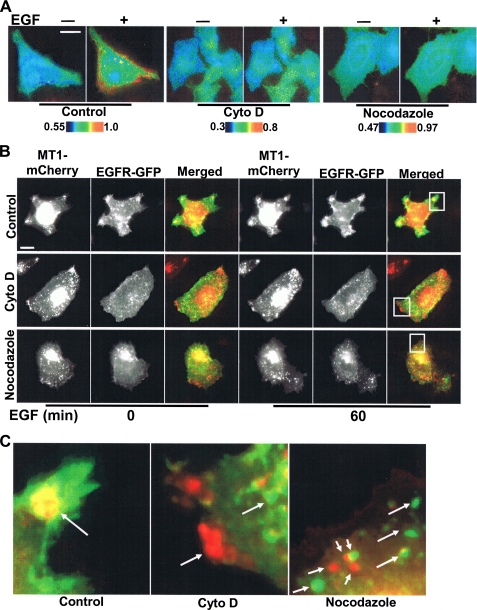
The EGF-induced MT1-MMP Activation Depends on Cytoskeletal Integrity—An intact cytoskeletal network may be important for regulating the proteolytic activity of MT1-MMP (35). As such, we examined the role of the cytoskeleton in modulating EGF-induced MT1-MMP activity. Following disruption of either actin filaments with cytochalasin D (Cyto D) or microtubules with nocodazole (data not shown), MT1-MMP activity and directional responses were blunted in response to EGF (Fig. 6A and supplemental Fig. S9). Further, Cyto D as well as nocodazole prevented the EGF-induced co-localization of MT1-MMP and EGFR at the cell periphery (Fig. 6, B and C). Taken together, cytoskeletal integrity plays a critical role in controlling the EGF-dependent localization and activity of MT1-MMP, possibly by mediating interactions between EGFR and MT1-MMP.
FIGURE 6.
EGF-induced MT1-MMP activation is dependent on cytoskeletal integrity. A, HeLa cells cotransfected with the MT1-MMP biosensor and MT1-MMP were treated with Cyto D or nocodazole for 1 h before EGF stimulation. The color images represent the ECFP/YPet emission ratio of MT1-MMP biosensor before and after EGF stimulation for 60 min. B, HeLa cells co-transfected with EGFR-GFP and MT1-MMP-mCherry were treated with Cyto D or nocodazole for 1 h before EGF stimulation. The images of MT1-MMP-mCherry and EGFR-EGFP before and after EGF stimulation for 60 min were displayed and overlaid to assess the co-localization of MT1-MMP and EGFR. C, enlarged images of selected regions in B showing the overlay of MT1-MMP-mCherry and EGFR-EGFP after EGF stimulation.
DISCUSSION
We have developed a sensitive FRET biosensor for detecting MT1-MMP activity with high spatiotemporal resolution in live cells. Although other membrane type MMPs, e.g. MT2- and MT3-MMP, may also contribute the FRET signals to some extent, the biosensor preferentially monitors MT1-MMP activity. With this biosensor, EGF was observed to induce a directional and cytoskeleton-dependent mobilization of MT1-MMP activity that concentrated at the leading edge of migrating cells. Given the importance of MT1-MMP in cancer invasion and metastasis (1), this biosensor provides not only a means to study the subcellular trafficking of MT1-MMP activity in live cell systems but also a convenient reporter system for high throughput screening of new MT1-MMP inhibitors.
Recently, a MMP-2 FRET biosensor utilizing CFP and YFP was described. Although this biosensor can detect the activity of MMP-2 in cell-free systems, a high level of recombinant MMP-2 was introduced into culture medium to induce detectable signals in live cells (36). Using a new construct that employs ECFP and YPet as the FRET pair for live cell imaging, we have established that this MT1-MMP biosensor can monitor MT1-MMP activity at the intact cell surface both in vitro and in vivo. The use of this FRET pair was based largely on its superior dynamic range at 37 °C relative to other FRET pairs. Indeed, MT1-MMP biosensors employing ECFP/EYFP, ECFP/CpVenus (37), or CyPet/YPet (22) because the FRET pair did not provide significant FRET changes upon EGF stimulation.3 Using this ECFP/YPet-based biosensor, we demonstrated that EGF did not induce FRET changes with the cytosol-directed biosensor (Fig. 3C), thus suggesting that the EGF-induced hydrolysis by active MT1-MMP does not occur intracellularly. Further, no intracellular signals were generated when the membrane-targeted biosensor was co-expressed with a soluble form of active MT1-MMP (i.e. MT1ΔTM) (Fig. 4A), and TIMP-2, a cell-impermeable MT1-MMP inhibitor, decreased the FRET ratio of the membrane-targeted biosensor (Fig. 3E). Together, these results suggest that the enhanced cleavage of membrane-targeted biosensor induced by EGF stimulation occurs primarily at the cell surface. The quantification of biosensor and MT1-MMP further revealed that the surface level of MT1-MMP, but not the biosensor, is moderately enhanced upon EGF stimulation (data not shown). Hence, the elevated expression of MT1-MMP at the cell surface upon EGF stimulation may also contribute to the reported FRET signals. Although the reversed response of the biosensor upon EGF stimulation in the presence of TIMP-2 is surprising, we reason that there is a basal level of MT1-MMP activity at the cell surface even in the absence of EGF. As such, the consequent cleavage of a fraction of the biosensor pool at the cell surface would result in a relatively high ECFP/YPet ratio. Following EGF stimulation, the turnover rate of the biosensor would be predicted to increase, resulting in a decrease in the fraction of cleaved biosensor at the cell surface as the pool of intact biosensors increases in the absence of MT1-MMP activity. In fact, TIMP-2 incubation in the absence of EGF caused a minor and slow reduction of ECFP/YPet ratio, most probably reflecting a basal level of turnover (Fig. 3E).
MT-serine proteases are expressed in HeLa (e.g. MT-SP1) (38) as well as MDA-MB-231 cells (e.g. KIAA 1363) (39). In our experiments, both TIMP-2 (a specific inhibitor for membrane type MMPs) and GM6001 (a general, synthetic inhibitor of MMPs) blocked the EGF-induced FRET change of the biosensors (Figs. 3E and 4E). Further, the FRET signals of the biosensor in HeLa cells (which express abundant MT-SP1, but are deficient in MT1-MMP) are low in the presence of either EGF or 10% fetal bovine serum (Fig. 4, A and B, and supplemental Figs. S1 and S3). These results suggest that MT-serine proteases may not significantly contribute to the FRET signals of the biosensors in live cell assays.
During random cell migration, a subtle spatial gradient of MT1-MMP activity was detected at the cell periphery prior to EGF stimulation. Because integrins are known to regulate MT1-MMP function (4, 40) and have been shown to co-localize with MT1-MMP at the leading edge of migrating cells (41), the detected activity likely reflects zones of integrin-dependent recruitment of proteolytic activity. Further, integrin engagement can also lead to the activation of EGFRs in the absence of EGF (42). Following EGF stimulation, however, MT1-MMP activity was mobilized and concentrated at the leading edge, possibly reflecting a synergistic effect of EGFR and integrins on MT1-MMP function (43). Given that either EGFR or integrin ligation can cause the recruitment and activation of Src kinase (44, 45), Src may mediate the effects of EGF on MT1-MMP through FAK/endophilin A2 (46) and caveolin (47). Recent evidence also indicates that Src can directly phosphorylate MT1-MMP at its cytoplasmic tail and regulate its functions (18), but it should be noted that MT1-MMP activity was readily detected at the cell surface even when its cytosolic tail was deleted. Presently, the detailed molecular mechanisms by which Src or related signaling molecules regulate the EGF-induced MT1-MMP activity remain to be characterized.
Following the ligation of cell surface receptors and integrins, the cytoskeleton plays an important role in both mediating signal transduction cascades (48) and regulating MT1-MMP function (6, 14, 35). Microtubules participate in intracellular trafficking and help control the level of surface receptors (49), whereas actin filaments are important for the proper localization of EGFR and MT1-MMP (35, 50). Our findings indicate that cytoskeletal integrity is critical for both the co-localization of MT1-MMP and EGFR at the cell periphery as well as the directional regulation of surface MT1-MMP activity in response to EGF (Fig. 6). Although earlier studies have reported that high doses of Cyto D can enhance MT1-MMP levels at the plasma membrane of HT-1080 cells (51), we find that at a 10-fold lower dose, Cyto D disrupted actin filaments and inhibited EGF-initiated effects on MT1-MMP activity (data not shown). Further studies have demonstrated that the extraction of cholesterol and disruption of lipid rafts by methyl-β-cyclodextran also inhibits EGF-induced MT1-MMP activity.3 As such, lipid rafts may also provide a docking platform that serves as a mediator for the regulation of MT1-MMP activity. Interestingly, neither Cyto D, nocodazole nor methyl-β-cyclodextran blocked the EGF-induced tyrosine phosphorylation of EGFR (data not shown) (52), thus indicating that these inhibitors alter the function of MT1MMP at a level downstream of EGFR.
The ability to monitor the spatiotemporal activity of a critical membrane-anchored proteinase in live cells provides a new and powerful tool for characterizing the manner in which cells integrate motile responses with the localized remodeling of the surrounding extracellular matrix. Interestingly, recent studies have demonstrated that cell adhesion, cytoskeletal organization, and mobility are distinct within the confines of the three-dimensional extracellular matrix versus two-dimensional environment (53, 54). As such, the new techniques described herein should prove useful for eventually extending these analyses into the more complex third dimension (55, 56).
Supplementary Material
Acknowledgments
We are very grateful to Dr. Roger Y. Tsien for mCherry cDNA, Dr. Patrick S. Daugherty for YPet cDNA, and Drs. Ning Wang and Stephen A. Boppart for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA71699 (to S. J. W.) and HL77448 (Y.-J. S.). This work was also supported by grants from Wallace H. Coulter Foundation and the Beckman Laser Institute, Inc. (to Y. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Movies S1-S5, Table S1, and Figs. S1-S9.
Footnotes
The abbreviations used are: MMP, matrix metalloproteinase; MT1-MMP, membrane type 1 MMP; FRET, fluorescence resonance energy transfer; TIMP-2, tissue inhibitor of metalloproteinase-2; EGF, epidermal growth factor; EGFR, EGF receptor; PDGFR, platelet-derived growth factor receptor; GFP, green fluorescent protein; EGFP, enhanced GFP; Cyto D, cytochalasin D.
M. Ouyang and Y. Wang, unpublished observation.
References
- 1.Seiki, M. (2003) Cancer Lett. 194 1-11 [DOI] [PubMed] [Google Scholar]
- 2.Visse, R., and Nagase, H. (2003) Circ. Res. 92 827-839 [DOI] [PubMed] [Google Scholar]
- 3.Deryugina, E. I., and Quigley, J. P. (2006) Cancer Metastasis Rev. 25 9-34 [DOI] [PubMed] [Google Scholar]
- 4.Deryugina, E. I., Ratnikov, B., Monosov, E., Postnova, T. I., DiScipio, R., Smith, J. W., and Strongin, A. Y. (2001) Exp. Cell Res. 263 209-223 [DOI] [PubMed] [Google Scholar]
- 5.Itoh, Y., Takamura, A., Ito, N., Maru, Y., Sato, H., Suenaga, N., Aoki, T., and Seiki, M. (2001) EMBO J. 20 4782-4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remacle, A. G., Rozanov, D. V., Baciu, P. C., Chekanov, A. V., Golubkov, V. S., and Strongin, A. Y. (2005) J. Cell Sci. 118 4975-4984 [DOI] [PubMed] [Google Scholar]
- 7.Sabeh, F., Ota, I., Holmbeck, K., Birkedal-Hansen, H., Soloway, P., Balbin, M., Lopez-Otin, C., Shapiro, S., Inada, M., Krane, S., Allen, E., Chung, D., and Weiss, S. J. (2004) J. Cell Biol. 167 769-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato, H., Takino, T., Okada, Y., Cao, J., Shinagawa, A., Yamamoto, E., and Seiki, M. (1994) Nature 370 61-65 [DOI] [PubMed] [Google Scholar]
- 9.Pei, D., and Weiss, S. J. (1995) Nature 375 244-247 [DOI] [PubMed] [Google Scholar]
- 10.Bravo-Cordero, J. J., Marrero-Diaz, R., Megias, D., Genis, L., Garcia-Grande, A., Garcia, M. A., Arroyo, A. G., and Montoya, M. C. (2007) EMBO J. 26 1499-1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato, T., Iwai, M., Sakai, T., Sato, H., Seiki, M., Mori, Y., and Ito, A. (1999) Br. J. Cancer 80 1137-1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Meter, T. E., Broaddus, W. C., Rooprai, H. K., Pilkington, G. J., and Fillmore, H. L. (2004) Neuro-oncol. 6 188-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, A., Lehti, K., Wang, X., Weiss, S. J., Keski-Oja, J., and Pei, D. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13693-13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uekita, T., Itoh, Y., Yana, I., Ohno, H., and Seiki, M. (2001) J. Cell Biol. 155 1345-1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakahara, H., Howard, L., Thompson, E. W., Sato, H., Seiki, M., Yeh, Y., and Chen, W. T. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7959-7964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato, T., del Carmen Ovejero, M., Hou, P., Heegaard, A. M., Kumegawa, M., Foged, N. T., and Delaisse, J. M. (1997) J. Cell Sci. 110 589-596 [DOI] [PubMed] [Google Scholar]
- 17.Galvez, B. G., Matias-Roman, S., Albar, J. P., Sanchez-Madrid, F., and Arroyo, A. G. (2001) J. Biol. Chem. 276 37491-37500 [DOI] [PubMed] [Google Scholar]
- 18.Nyalendo, C., Michaud, M., Beaulieu, E., Roghi, C., Murphy, G., Gingras, D., and Beliveau, R. (2007) J. Biol. Chem. 282 15690-15699 [DOI] [PubMed] [Google Scholar]
- 19.Pertz, O., and Hahn, K. M. (2004) J. Cell Sci. 117 1313-1318 [DOI] [PubMed] [Google Scholar]
- 20.Giepmans, B. N., Adams, S. R., Ellisman, M. H., and Tsien, R. Y. (2006) Science 312 217-224 [DOI] [PubMed] [Google Scholar]
- 21.Wang, Y., Botvinick, E. L., Zhao, Y., Berns, M. W., Usami, S., Tsien, R. Y., and Chien, S. (2005) Nature 434 1040-1045 [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, A. W., and Daugherty, P. S. (2005) Nat. Biotechnol. 23 355-360 [DOI] [PubMed] [Google Scholar]
- 23.Rozanov, D. V., Deryugina, E. I., Monosov, E. Z., Marchenko, N. D., and Strongin, A. Y. (2004) Exp. Cell Res. 293 81-95 [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita, T., Sato, H., Takino, T., Itoh, M., Akizawa, T., and Seiki, M. (1996) Cancer Res. 56 2535-2538 [PubMed] [Google Scholar]
- 25.Carter, R. E., and Sorkin, A. (1998) J. Biol. Chem. 273 35000-35007 [DOI] [PubMed] [Google Scholar]
- 26.Hotary, K. B., Allen, E. D., Brooks, P. C., Datta, N. S., Long, M. W., and Weiss, S. J. (2003) Cell 114 33-45 [DOI] [PubMed] [Google Scholar]
- 27.Tan, J. L., Liu, W., Nelson, C. M., Raghavan, S., and Chen, C. S. (2004) Tissue Eng. 10 865-872 [DOI] [PubMed] [Google Scholar]
- 28.Lehti, K., Allen, E., Birkedal-Hansen, H., Holmbeck, K., Miyake, Y., Chun, T. H., and Weiss, S. J. (2005) Genes Dev. 19 979-991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiley, H. S., and Burke, P. M. (2001) Traffic 2 12-18 [DOI] [PubMed] [Google Scholar]
- 30.Zhai, Y., Hotary, K. B., Nan, B., Bosch, F. X., Munoz, N., Weiss, S. J., and Cho, K. R. (2005) Cancer Res. 65 6543-6550 [DOI] [PubMed] [Google Scholar]
- 31.Hotary, K. B., Yana, I., Sabeh, F., Li, X. Y., Holmbeck, K., Birkedal-Hansen, H., Allen, E. D., Hiraoka, N., and Weiss, S. J. (2002) J. Exp. Med. 195 295-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.deFazio, A., Chiew, Y. E., Donoghue, C., Lee, C. S., and Sutherland, R. L. (1992) J. Biol. Chem. 267 18008-18012 [PubMed] [Google Scholar]
- 33.Koshikawa, N., Giannelli, G., Cirulli, V., Miyazaki, K., and Quaranta, V. (2000) J. Cell Biol. 148 615-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varon, C., Tatin, F., Moreau, V., Van Obberghen-Schilling, E., FernandezSauze, S., Reuzeau, E., Kramer, I., and Genot, E. (2006) Mol. Cell Biol. 26 3582-3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori, H., Tomari, T., Koshikawa, N., Kajita, M., Itoh, Y., Sato, H., Tojo, H., Yana, I., and Seiki, M. (2002) EMBO J. 21 3949-3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, J., Zhang, Z., Lin, J., Lu, J., Liu, B. F., Zeng, S., and Luo, Q. (2007) Biochim. Biophys. Acta 1773 400-407 [DOI] [PubMed] [Google Scholar]
- 37.Nagai, T., Yamada, S., Tominaga, T., Ichikawa, M., and Miyawaki, A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10554-10559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santin, A. D., Cane, S., Bellone, S., Bignotti, E., Palmieri, M., De Las Casas, L. E., Anfossi, S., Roman, J. J., O'Brien, T., and Pecorelli, S. (2003) Cancer 98 1898-1904 [DOI] [PubMed] [Google Scholar]
- 39.Jessani, N., Liu, Y., Humphrey, M., and Cravatt, B. F. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 10335-10340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann, U. B., Westphal, J. R., Van Kraats, A. A., Ruiter, D. J., and Van Muijen, G. N. (2000) Int. J. Cancer 87 12-19 [DOI] [PubMed] [Google Scholar]
- 41.Zaidel-Bar, R., Cohen, M., Addadi, L., and Geiger, B. (2004) Biochem. Soc. Trans. 32 416-420 [DOI] [PubMed] [Google Scholar]
- 42.Cabodi, S., Moro, L., Bergatto, E., Boeri Erba, E., Di Stefano, P., Turco, E., Tarone, G., and Defilippi, P. (2004) Biochem. Soc. Trans. 32 438-442 [DOI] [PubMed] [Google Scholar]
- 43.Miyamoto, S., Teramoto, H., Gutkind, J. S., and Yamada, K. M. (1996) J. Cell Biol. 135 1633-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leu, T. H., and Maa, M. C. (2003) Front. Biosci. 8 s28-38 [DOI] [PubMed] [Google Scholar]
- 45.Arias-Salgado, E. G., Lizano, S., Sarkar, S., Brugge, J. S., Ginsberg, M. H., and Shattil, S. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13298-13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, X., Gan, B., Yoo, Y., and Guan, J. L. (2005) Dev. Cell 9 185-196 [DOI] [PubMed] [Google Scholar]
- 47.Labrecque, L., Nyalendo, C., Langlois, S., Durocher, Y., Roghi, C., Murphy, G., Gingras, D., and Beliveau, R. (2004) J. Biol. Chem. 279 52132-52140 [DOI] [PubMed] [Google Scholar]
- 48.Wells, A., Ware, M. F., Allen, F. D., and Lauffenburger, D. A. (1999) Cell Motil Cytoskeleton 44 227-233 [DOI] [PubMed] [Google Scholar]
- 49.Lippincott-Schwartz, J., and Smith, C. L. (1997) Curr. Opin. Neurobiol. 7 631-639 [DOI] [PubMed] [Google Scholar]
- 50.Lidke, D. S., Lidke, K. A., Rieger, B., Jovin, T. M., and Arndt-Jovin, D. J. (2005) J. Cell Biol. 170 619-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zucker, S., Hymowitz, M., Conner, C. E., DiYanni, E. A., and Cao, J. (2002) Lab. Investig. 82 1673-1684 [DOI] [PubMed] [Google Scholar]
- 52.Klein, S., Kaszkin, M., Barth, H., and Kinzel, V. (1997) Biochem. J. 322 937-946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beningo, K. A., Dembo, M., and Wang, Y. L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 18024-18029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf, K., and Friedl, P. (2006) Br. J. Dermatol. 154 (Suppl. 1) 11-15 [DOI] [PubMed] [Google Scholar]
- 55.Ghajar, C. M., Blevins, K. S., Hughes, C. C., George, S. C., and Putnam, A. J. (2006) Tissue Eng. 12 2875-2888 [DOI] [PubMed] [Google Scholar]
- 56.Cukierman, E., Pankov, R., Stevens, D. R., and Yamada, K. M. (2001) Science 294 1708-1712 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.