Abstract
Overexpression of enhancer of zeste homologue 2 (EZH2) occurs in various malignancies and is associated with a poor prognosis, especially because of increased cancer cell proliferation. In this study we found an inverse correlation between EZH2 and RUNX3 gene expression in five cancer cell lines, i.e. gastric, breast, prostate, colon, and pancreatic cancer cell lines. Chromatin immunoprecipitation assay showed an association between EZH2 bound to the RUNX3 gene promoter, and trimethylated histone H3 at lysine 27, and HDAC1 (histone deacetylase 1) bound to the RUNX3 gene promoter in cancer cells. RNA interference-mediated knockdown of EZH2 resulted in a decrease in H3K27 trimethylation and unbound HDAC1 and an increase in expression of the RUNX3 gene. Restoration of RUNX3 expression was not associated with any change in DNA methylation status in the RUNX3 promoter region. RUNX3 was repressed by histone deacetylation and hypermethylation of a CpG island in the promoter region and restored by trichostatin A or/and 5-aza-2′-deoxycytidine. Immunofluorescence staining confirmed restoration of expression of the RUNX3 protein after knockdown of EZH2 and its restoration resulted in decreased cell proliferation. In vivo, an inverse relationship between expression of the EZH2 and RUNX3 proteins was observed at the individual cell level in gastric cancer patients in the absence of DNA methylation in the RUNX3 promoter region. The results showed that RUNX3 is a target for repression by EZH2 and indicated an underlying mechanism of the functional role of EZH2 overexpression on cancer cell proliferation.
Three members of the Runt-related (RUNX) family of genes, RUNX1, RUNX2, and RUNX3, play pivotal roles in normal development and neoplasia. All RUNX family members share the central Runt domain, which is well conserved and recognizes a specific DNA sequence, but each has relatively divergent N- and C-terminal regions (1). RUNX3 is involved in neurogenesis (2, 3) and thymopoiesis (4, 5) and functions as a tumor suppressor gene in gastric cancer (6, 7). Failure to express RUNX3 because of a combination of hemizygous deletion and DNA hypermethylation of the RUNX3 promoter region has been found in about 60% of primary gastric cancer specimens (7). RUNX3-R122C is a mutation located in the conserved Runt domain that was discovered in a case of gastric cancer and it abolishes the tumor suppressive activity of RUNX3 (7). Subsequent studies have revealed that RUNX3 inactivation is not limited to gastric cancer, and frequent inactivation of RUNX3 due to DNA hypermethylation has been reported in various other cancers, including lung cancer (8), hepatocellular carcinoma (9), breast cancer (10), colon cancer (11), pancreatic cancer (12), bile duct cancer (12), prostate cancer (13), and laryngeal cancer (10). Thus, RUNX3 is primarily inactivated by epigenetic silencing, rather than by mutations or deletions, suggesting that RUNX3 can be reactivated and serve as a good gene for drug targeting.
Enhancer of zeste homologue 2 (EZH2)2 is one of the polycomb group proteins involved in the regulation of proliferation and cell cycle progression (14). More specifically, EZH2 is a histone methyltransferase controlled by the E2F transcription factors that regulate the transition from G2 to the mitotic phase of the cell cycle through nucleosome modification, chromatin remodeling, and interaction with other transcription factors (15). Disruption of EZH2 expression in senescent fibroblasts retards cell proliferation and induces cell cycle arrest at the G2 to mitosis transition (16), whereas overexpression of EZH2 in cultured mouse embryonic fibroblasts shortens the G1 phase of the cell cycle and leads to accumulation of cells in the S phase (17). EZH2 expression has been found to be linked to the progression of prostate and breast cancer (18, 19), and because it is a biomarker of tumor progression, EZH2 has also been suggested to be an oncogene that leads to tumor development. EZH2 competes with histone deacetylase in binding to retinoblastoma protein 2/p130 and subsequently reduces the transcriptional repression of the CyclinA promoter, suggesting a molecular mechanism linking elevated EZH2 expression to malignant transformation (20). However, the downstream signaling and molecular mechanism for the aberrant expression of EZH2 in cancer has been poorly understood.
RUNX3 has been found to up-regulate p21WAF1/Cip1, an important factor in cyclin-dependent kinase inhibition and cell cycle control, and has been found to do so in collaboration with Smads downstream of transforming growth factor-β in gastric cancer (21). RUNX3 is also responsible for transcriptional up-regulation of Bim in transforming growth factor-β-induced apoptosis (22). Thus, RUNX3 plays a critical role in the induction of apoptosis as well as in the regulation of cell growth arrest, suggesting that RUNX3 is a significant tumor suppressor gene in carcinogenesis. In the present study, we investigated the mechanism of the role that EZH2 plays in cancer cell proliferation in several different cancer cell lines and found that EZH2 is a transcriptional repressor of RUNX3 expression and acts synergistically with DNA methylation.
EXPERIMENTAL PROCEDURES
Cell Culture—Five human cancer cell lines, i.e. gastric cancer cell line MKN28, breast cancer cell line MCF-7, prostate cancer cell line LNCap, colon cancer cell line DLD1, and pancreatic cancer cell line MiaPaca2, were maintained at 37 °C in RPMI 1640 or Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum and 1% glutamine in a 5% CO2 atmosphere.
Tissue Specimens—A total of 17 gastric cancer specimens (stage IA, 3; IB, 4; II, 2; IIIA, 3; IIIB, 2; and IV, 3; 8 intestinal-type adenocarcinomas and 9 diffuse-type adenocarcinomas) and corresponding non-neoplastic gastric mucosa were studied. The specimens were obtained from 8 males and 9 females (mean age 62.8 years; range 52–77 years) by surgical resection at the National Cancer Hospital East and were immediately frozen in liquid nitrogen and stored at -80 °C until examined. The histological diagnosis was confirmed by microscopic analysis of a section of each frozen specimen before DNA extraction. None of the patients had any preoperative treatment, such as radiation or chemotherapy. All patients agreed to enrollment in the study and gave their informed consent. The institutional review board of the National Cancer Center approved all protocols after obtaining the patients' consent. All clinicopathological data were according to the TNM classification (UICC) and obtained from the clinical and pathology records.
RNA Interference—Two different 21-nucleotides duplex siRNAs for EZH2 and one negative control siRNA were synthesized by Ambion (EZH2; siRNA ID 107417 and 214022). Twenty-four hours after plating, the cells were transfected with EZH2 siRNA or control siRNA using the DharmaFECT transfection reagent (DHARMACON) according to the manufacturer's instructions. At various time points after transfection, cells were harvested and subjected to several assays, including real-time PCR and Western blotting analysis.
RNA Isolation and Real-time RT-PCR—Total RNA from the five different cell lines was isolated with TRIzol Reagent (Invitrogen) and reverse transcribed to cDNA with ExScript RT Reagent (Takara). Real-time RT-PCR was carried out with specific primers for EZH2 and RUNX3 and Smart Cycler (Cepheid, Sunnyvale, CA). GAPDH expression was used to normalize for variance. Real-time fluorescence monitoring of the PCR products was performed with SYBR Green I fluorescent dye (Takara). The expression levels of specific genes are reported as ratios of expression of GAPDH in the same master reaction. The PCR primer pairs (5′ to 3′) used for each gene were: EZH2, CCCTGACCTCTGTCTTACTTGTGGA and ACGTCAGATGGTGCCAGCAATA; RUNX3, TCTGTAAGGCCCAAAGTGGGTA and ACCTCAGCATGACAATATGTCACAA; GAPDH, GCACCGTCAAGGCTGAGAAC and ATGGTGGTGAAGACGCCAGT.
Chromatin Immunoprecipitation (ChIP) Assay—The ChIP assay was performed as previously described (23). The PCR conditions for the RUNX3 gene promoter were applied with the following two primer pairs (5′ to 3′): ChIP primer 1, TGTCCCGGGATCCTCTTCT and TAGAGACGTTGGTGCGGAAAT and ChIP primer 2, CTCTCTGCTCTCCCCTCAAAAC and GGACCGTGGTTACATGCGTAA. These primer sets were designed to encompass the transcriptional start site of RUNX3 variant 2 in the CpG island. A 5 μg amount of each antibody was used in this assay. The antibodies used were EZH2 antibody and dimethyl H3 (Lys9) antibody, purchased from Abcam Inc., and HDAC1 antibody and trimethyl H3 (Lys27) antibody, purchased from Upstate. Individual ChIP assays were repeated at least twice to confirm the reproducibility of the PCR-based experiment. Preliminary PCR were performed to determine the optimal PCR conditions to assure linear amplification of DNA. PCR products were electrophoresed on a 6% polyacrylamide gel, stained with ethidium bromide, and photographed. To measure the levels of EZH2, HDAC1, and histone methylation (K9 and K27) in each immunoprecipitate, the ratios were calculated by measuring the intensity of the PCR product in immunoprecipitated DNA versus input DNA (total chromatin) amplified by PCR in a linear range. The ratios were calculated by performing a DNA 1000 assay with the Agilent 2100 bioanalyzer and using DNA chips for electrophoresis (Agilent Technologies). The MYT1 (myelin transcription factor 1) gene was used as a positive control to validate each ChIP assay. The MYT1 gene has been found to be a target gene of EZH2 (24).
DNA Extraction—Genomic DNA from the five cancer cell lines and the 17 gastric cancer specimens and corresponding non-neoplastic gastric mucosa was extracted with a DNeasy tissue kit (Qiagen).
Analysis of the Methylation Status of Genomic DNA—Bisulfite treatment of the DNA for the methylation assays was performed as previously described (25). Methylation of RUNX3 was determined by a methylation-specific polymerase chain reaction (MSP). Briefly, 2 μl of bisulfite-treated DNA was used as the PCR template, and primer sets specific for methylated and unmethylated alleles were used. The MSP primers for RUNX3 used in this study have been described previously (7). The PCR products from the methylated and unmethylated reactions were electrophoresed on 6% polyacrylamide gels and visualized by ethidium bromide staining.
Immunofluorescence—Cells grown on coverslips were fixed with 3% paraformaldehyde in phosphate-buffered saline and processed for immunofluorescence. Rabbit antibody against EZH2 (1:125) (Zymed Laboratories) and mouse antibody against RUNX3 (1 μg/ml) (R3–6E9) (26) were used. The donkey secondary antibody was anti-rabbit IgG-Alexa Fluor 488, and the goat secondary antibody was anti-mouse IgG-Alexa Fluor 546 (Molecular Probes). Cells were examined with a Zeiss LSM5 PASCAL microscope and nuclear contour ratios were computed.
Western Blot Analysis—Cells were lysed with whole cell lysis buffer (50 mm HEPES, 150 mm NaCl, 1.5 mm MgCl2, 0.5 mm EDTA, 10% glycerol, 1% Triton X-100, 10 mm sodium fluoride, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride), and then frozen at -80 °C and thawed three times to rupture the cell membranes. Samples of the lysates were incubated for 30 min on ice to lyse the nuclei and then centrifuged at 8900 × g. The protein concentration of each sample was determined by a standard Bradford assay. Equal amounts of protein (20 μg) from each cell line were subjected to Western blot analysis. The probing antibodies were EZH2 antibody (1:1000) (BD Transduction Laboratories), RUNX3 antibody (1 μg/ml) (R3–5G4) (26), Histone H3 antibody (0.5 μg/ml) (Upstate), and β-actin antibody (1:50) (Santa Cruz Biotechnology).
Cell Treatment with Trichostatin A (TSA) and 5-Aza-dC— Preliminary experiments were performed to determine the maximum concentration tolerated of TSA and 5-aza-dC by each cell line, and TSA concentrations in the 330 nm to 5 μm range and 5-aza-dC concentrations in the 800 nm to 1 μm range were optimal for the cancer cell lines. TSA concentrations of 330 nm were used for MCF-7 and MiaPaca2 cells, 1 μm for LNCap and DLD1 cells, and 5 μm for MKN28 cells, and the 5-aza-dC concentrations used were 800 nm for MCF-7 cells and 1 μm for MKN28, LNCap, DLD1, and MiaPaca2 cells. Cells were seeded at low density in a 35-mm tissue culture dish and incubated at 37 °C for a total of 72 h. Cells were incubated for 24 h prior to treatment with the chemicals. Mock treatment with an identical volume of absolute ethanol or water was used as a control. The 5-aza-dC was added after 24 h of incubation and cells were incubated for 48 h after it was added. TSA was added to the medium after 24 h of incubation, and cells were incubated with TSA for 48 h. When 5-aza-dC and TSA were both added, they were added after incubation for 24 h, and cells were incubated for 48 h. The culture medium was exchanged every 24 h for 5-aza-dC, TSA, and the combined treatment. Total cellular RNA was extracted for real-time PCR analysis.
Cell Proliferation Assay—The five cancer cell lines were transfected with EZH2 siRNA or control siRNA 48 h before the cell proliferation assay. The cell proliferation assay was started by seeding the cells at a density of 1.0 × 105 per dish. The assay was subsequently processed by incubating the cells at 37 °C in a tissue culture incubator for 72, 96, 120, and 144 h after transfection, and counting the cells to plot a cell growth curve.
Immunohistochemistry—The same 17 gastric adenocarcinoma specimens as used for MSP analysis were used for immunohistochemical analysis. Immunohistochemistry for EZH2 (1:25) (BD Transduction Laboratories) and RUNX3 (1 μg/ml) (R3–6E9) (26) was performed on formalin-fixed, paraffin-embedded tissue sections by steam heat-induced or microwave-induced epitope retrieval and with the Dako Envision detection system. Specimens were scored negative for overexpression of EZH2 when 0–20% of the cells were positive, and positive for overexpression when >21% cells were positive. Specimens were scored positive for RUNX3 expression and negative. For RUNX3 expression, specimens were also evaluated with the localization of RUNX3 expression, nucleus or cytoplasm. Appropriate positive and negative internal controls were used to validate immunohistochemical staining.
RESULTS
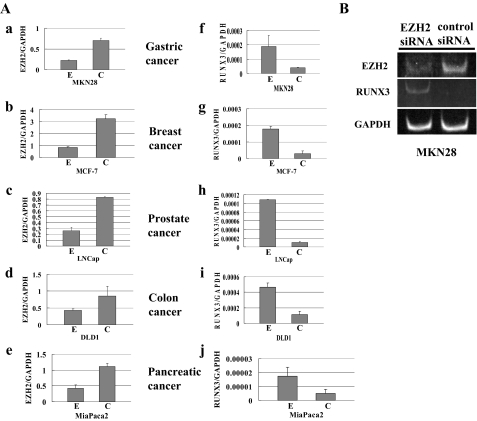
Inhibitory Effect of EZH2 on RUNX3 Gene Expression—To determine whether EZH2 is a negative modulator of RUNX3 gene expression in the five cancer cell lines, i.e. gastric cancer line MKN28, breast cancer line MCF-7, prostate cancer line LNCap, colon cancer line DLD1, and pancreatic cancer line MiaPaca2, EZH2 mRNA was suppressed with EZH2 siRNA in all five cancer cell lines, in which RUNX3 is silenced (7, 11, 12, 27, 28). The results of RT-PCR showed significantly decreased levels of EZH2 mRNA in all five cancer cell lines within 48–96 h after transfection with EZH2 siRNA (the percentage of EZH2 mRNA expression level by EZH2 siRNA transfection to that by control siRNA transfection; MKN28, 31.6%; MCF-7, 25.7%; LNCap, 31.5%; DLD1, 50.0%; MiaPaca2, 47.0%) (Fig. 1A, a–e). A marked decrease in EZH2 protein level in all five cancer cell lines was observed by Western blot 96 h after transfection (data not shown), an increased level of RUNX3 mRNA was in every line. Real-time RT-PCR was performed to quantitatively measure the restored RUNX3 mRNA expression level after knockdown of EZH2 (Fig. 1A, f–j). RUNX3 mRNA had increased from 3.5 to 10.4-fold after EZH2 knockdown in the cancer cell lines (MKN28, 4.7-fold; MCF-7, 6.1-fold; LNCap, 10.4-fold; DLD1, 4.1-fold; MiaPaca2, 3.5-fold) (Fig. 1A, f–j). However, the transcriptional expression level of GAPDH mRNA was unchanged in all five cancer cell lines after EZH2 knockdown, suggesting that GAPDH is not a target of EZH2 (data not shown). By non-real-time RT-PCR, restoration of RUNX3 mRNA expression in the MKN28 cells was visualized on 6% PAGE (Fig. 1B).
FIGURE 1.
Restoration of RUNX3 mRNA levels after knockdown of EZH2 in five cancer cell lines, MKN28, MCF-7, LNCap, DLD1, and MiaPaca2. A, the level of EZH2 mRNA after knockdown by siRNA transfection and the restored level of RUNX3 mRNA expression were quantified by real-time RT-PCR. EZH2 mRNA expression levels are shown in a–e, and RUNX3 mRNA expression levels in f–j. E, EZH2 siRNA; C, control siRNA. B, the restored RUNX3 mRNA levels in MKN28 cells were analyzed by RT-PCR and visualized by electrophoresis on 6% polyacrylamide gel.
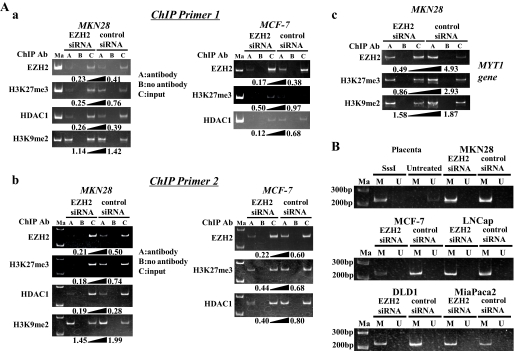
ChIP Assay—To demonstrate a direct interaction between the EZH2 complex and promoter region of the RUNX3 gene, a ChIP assay was performed using the same MKN28 and MCF-7 cells. Two PCR primer sets that spanned the transcriptional start site were used to monitor binding of EZH2 to the promoter that drives expression of RUNX3 mRNA and was up-regulated after loss of EZH2. The MYT1 gene was used as a positive control to validate each ChIP assay (25). The ChIP assay revealed EZH2 binding to the RUNX3 promoter region in the control MKN28 and MCF-7 cells (Fig. 2A, a and b), and the amount of the EZH2 recruited to the promoter region of RUNX3 was inversely correlated with the RUNX3 mRNA expression levels in both cell lines. The level of H3-Lys27 trimethylation in the RUNX3 promoter was significantly reduced in MKN28 and MCF-7 cells transfected with EZH2 siRNA, and the decrease in H3-Lys27 trimethylation was inversely correlated with the increase in expression of the RUNX3 gene. The amount of HDAC1 bound to the RUNX3 promoter was also significantly reduced in MKN28 and MCF-7 cells transfected with EZH2 siRNA, and the decrease in HDAC1 was inversely correlated with an increase in expression of the RUNX3 gene, suggesting that EZH2 forms a transcriptional repressive complex with HDAC1. A decreased level of H3-Lys9 dimethylation was also detected in MKN28 cells transfected with EZH2 siRNA in comparison with MKN28 cells transfected with control siRNA, suggesting that H3-Lys9 status may also be involved in modulation of the RUNX3 gene promoter activity by EZH2. Based on all of the above findings, we concluded that RUNX3 gene silencing in cancer cells is mediated by EZH2 increasing the H3-Lys27 methylation level. Fig. 2A, c, shows that the binding levels of EZH2 and H3K27me3 (H3-Lys27 trimethylation) in the MYT1 promoter region, which is known to be a target of EZH2 (25), clearly decreased after knockdown of EZH2, suggesting the ChIP assay was valid. The means and standard deviations of ChIP experiments for each antibody were shown as diagrams (supplemental Fig. S1).
FIGURE 2.
Restoration of RUNX3 mRNA in gastric and breast cancer cell lines (MKN28 and MCF-7) after EZH2 knockdown accompanied by chromatin remodeling with no change of DNA methylation status in the promoter region of RUNX3 gene. A, a and b, ChIP assay was performed by using the DNA-protein complex isolated from MKN28 and MCF-7 cells transfected with EZH2 siRNA for 96 h and immunoprecipitated with various antibodies, including EZH2 antibody, H3K27me3 antibody (H3-Lys27 trimethylation), HDAC1 antibody, and H3K9me2 antibody (H3-Lys9 dimethylation). The two primer sets for ChIP assay were designated ChIP primer set 1 (a) and ChIP primer set 2 (b). The PCR products of each immunoprecipitated DNA and input DNA were visualized by electrophoresis on a 6% polyacrylamide gel. The number under each gel is the ratio of immunoprecipitated DNA to input DNA quantified (A, antibody; B, no antibody; C, input). c, the MYT1 gene, which is known to be a target of EZH2, was used to validate ChIP assays in the present study. B, MSP analyses of the DNA of the five cancer cell lines transfected with EZH2 siRNA or negative control siRNA, using primer sets that specifically amplify either methylated DNA(M) or unmethylated DNA (U). Control templates from human genomic placenta DNA treated or untreated with SssI methylase are shown. Ma, 200-bp and 300-bp DNA ladder markers.
MSP Analysis—As previously reported, hypermethylation in the RUNX3 promoter region was observed in all five cancer cell lines in the absence of RUNX3 expression (Fig. 2B) (7, 11, 12, 27). We then investigated whether the status of DNA methylation in the RUNX3 promoter region changed after knockdown of EZH2 and resulted in restoration of RUNX3 gene expression. Although RUNX3 was up-regulated after EZH2 siRNA transfection, as shown in Fig. 2B the MSP analysis showed no decrease in DNA methylation of the RUNX3 promoter region. Hypermethylation of the promoter of the RUNX3 gene occurred before and after EZH2 siRNA transfection in all five cancer cell lines examined. DNA hypermethylation in the promoter region is known to be associated with silencing of the RUNX3 gene, however, the above findings suggest that histone modification by EZH2 also played an important role in the regulation of RUNX3 in all five cancer cell lines.
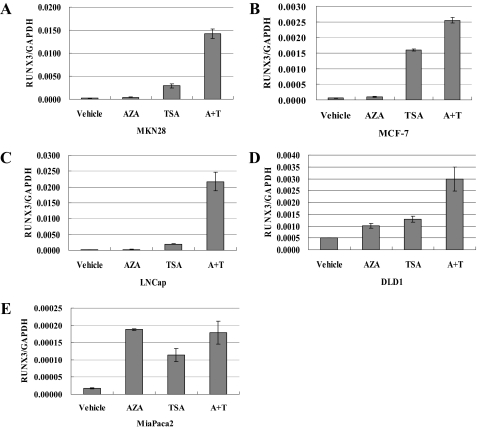
Cell Treatment with TSA and 5-Aza-dC—To determine the relative contribution of DNA methylation and histone deacetylation to RUNX3 silencing, we investigated the effect of a demethylating agent, 5-aza-2′-deoxycytidine (5-aza-dC), and a histone deacetylase inhibitor, TSA, in all five cancer cell lines. Real-time RT-PCR showed that exposure to 5-aza-dC resulted in less restoration of RUNX3 expression than exposure to TSA in four of the cancer cell lines, the exception being the MiaPaca2 line (Fig. 3, A–E), but MSP analysis showed hypermethylation of CpG in all five cancer cell lines (Fig. 2B). Although the histone deacetylase inhibitor TSA alone was sufficient to strongly restore RUNX3 expression (Fig. 3, A–E), the combination of DNA demethylation and inhibition of histone deacetylation restored RUNX3 expression synergistically (Fig. 3, A–E). In other words, the effect of histone deacetylation on RUNX3 transcriptional repression was greater than that of DNA methylation, except in cell line MiaPaca2. Thus, both histone deacetylation and DNA methylation play a synergistic role in silencing RUNX3 expression. Neither 5-aza-dC nor TSA changed the EZH2 expression levels in any of the cancer cell lines (data not shown). As stated above, the ChIP assay showed that down-regulation of EZH2 by siRNA transfection reversed the binding of HDAC1 to the promoter region of RUNX3, which is restoration of RUNX3 expression. These findings could explain why knockdown of EZH2 by siRNA transfection was capable of restoring RUNX3 expression.
FIGURE 3.
Restoration of RUNX3 expression in five cancer cell lines after treatment with 5-aza-dC, TSA, or a combination of 5-aza-dC and TSA. RUNX3 expression in the five lines was evaluated by quantitative real-time RT-PCR. A, MKN28; B, MCF-7; C, LNCap; D, DLD1, and E, MiaPaca2. The concentrations of 5-aza-dC and TSA to which the MCF-7 line was exposed were the same as used by Lau et al. (26). Vehicle, no treatment control; AZA, 5-aza-dC; A+T, combination of 5-aza-dC and TSA.
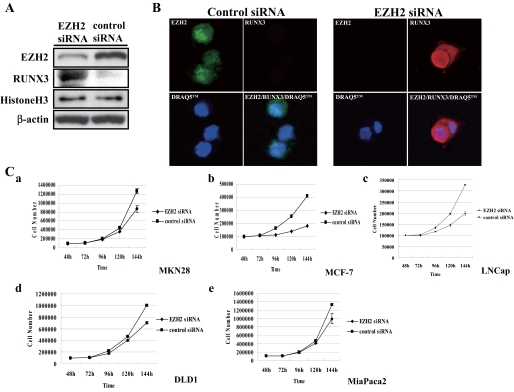
Knockdown of EZH2-induced RUNX3 Protein Expression in Gastric Cancer Cells—To determine whether EZH2 down-regulates the RUNX3 protein, we used immunofluorescence to investigate whether a decrease in the EZH2 protein level would restore RUNX3 protein expression. A Western blot analysis in the MKN28 cells following knockdown of EZH2 by siRNA transfection showed that RUNX3 protein was significantly up-regulated (Fig. 4A). Immunofluorescence staining confirmed that expression of the RUNX3 protein had been restored in both the nucleus and cytoplasm (Fig. 4B), and this finding demonstrated that EZH2 overexpression down-regulates RUNX3 protein in cancer cells.
FIGURE 4.
Knockdown of EZH2-restored RUNX3 expression and decreased cell proliferation by the cancer cell lines. A, RUNX3 re-expression in MKN28 cells was detected by Western blot analysis at 96 h after EZH2 knockdown. B, the nuclear and cytoplasmic distribution of RUNX3 was restored in MKN28 cells transfected with EZH2 siRNA (EZH2, green; RUNX3, red). DRAQ5™ (1,5-bis[[2-(dimethylamino)ethyl]amino]-4,8-dihydroxyanthracene-9,10-dione]) was used to stain the nuclei. C, 48 h after transfection with EZH2 siRNA or control siRNA, cells were re-seeded in new dishes at a density of 1.0 × 105 cells and counted. Points are mean of data from three independent experiments; bars, S.D. Cell proliferation by all five cell lines decreased after transfection with EZH2 siRNA. a, MKN28; b, MCF-7; c, LNCap; d, DLD1; e, MiaPaca2.
EZH2 Knockdown Had an Effect on Cancer Cell Proliferation— At least part of the tumor suppressor activity of RUNX3 is thought to be associated with its ability to induce p21 expression (21). Loss of RUNX3 is thought to play a critical role in the process of tumor cell proliferation, and because EZH2 down-regulates RUNX3 expression, we investigated the effect of EZH2 on cell proliferation by testing the growth inhibitory effect of EZH2 siRNA in all five cancer cell lines, gastric cancer, breast cancer, prostate cancer, colon cancer, and pancreatic cancer lines. At 48 h after transfection with EZH2 siRNA or control siRNA, cells were re-seeded in new plates and observed for cell growth. As shown in Fig. 4C, cell growth started 24 h after re-seeding (72 h after transfection), and there was significantly less cell growth by the EZH2 siRNA-treated cells than by the control cells (Fig. 4C) (t test) (p < 0.05).
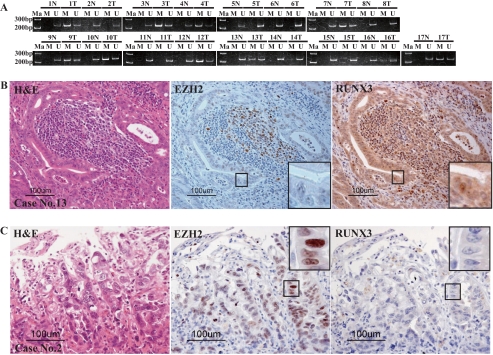
EZH2 Overexpression Also Correlated with Loss of RUNX3 in Gastric Adenocarcinoma Tissue—Because EZH2 overexpression correlated with loss of RUNX3 expression in all five cancer cell lines examined in this study and HDAC1-mediated histone modification by EZH2 acted synergistically with DNA methylation in the promoter region to silence RUNX3 expression, we investigated cancer tissue specimens for such a correlation to determine whether this repressive synergism would silence RUNX3 expression in a clinicopathologically relevant context (Fig. 5, A–C). To determine whether the RUNX3 silencing pathway by EZH2 exists in vivo, immunohistochemical staining was used to examine 17 gastric adenocarcinomas and adjacent non-cancerous gastric mucosa for DNA methylation in the RUNX3 promoter, overexpression of EZH2 protein, and RUNX3 expression. To test for DNA methylation and/or histone modification by EZH overexpression, we investigated individual cases with gastric cancer in vivo. The patients' profiles and clinicopathological data are shown in Table 1. The MSP analysis showed DNA hypermethylation in 64.7% (11/17) of the gastric adenocarcinomas (Fig. 5A). Immunohistochemical staining showed a slight difference in the distribution of RUNX3-positive cells in the fundic gland and pyloric gland portions. In the fundic gland portion, RUNX3-positive cells were observed mainly in a deeper zone of the fundic glands, corresponding to chief cells morphologically. A slight amount of RUNX3 protein was also detected in the surface epithelial cells in the gastric pits. In the pyloric gland portion, scattered RUNX3-positive cells were observed in the lower half of the antral mucosa near the generative zone. In contrast to the surrounding non-cancerous epithelial cells, weak to moderate cytoplasmic and/or nuclear positivity was observed in the gastric cancer cells in some of the specimens (5/17, 29.4%). EZH2 is normally expressed in the neck region of the gastric foveolae, which is the proliferative zone for gastric mucosa (data not shown) and in the germinal center follicular lymphocytes (Fig. 5B, center) (29). The percentage of specimens that showed overexpression of EZH2 (described as “positive” in Table 1) according to the results of immunohistochemical staining was 82.3% (14/17). Interestingly, all (case numbers 13, 15, and 17) of three specimens with DNA methylation in the promoter of RUNX3 and negativity for EZH2 overexpression showed RUNX3 expression (Fig. 5B). On the other hand, four (case numbers 2, 5, 6, and 8) of five specimens without both RUNX3 expression and DNA methylation in the promoter of RUNX3 showed EZH2 overexpression (Fig. 5C). Even in some specimens without DNA hypermethylation of the RUNX3 gene, RUNX3 protein expression was lost when EZH2 was overexpressed. This suggests that the loss of RUNX3 expression is mediated by histone methylation at Lys27 by EZH2 overexpression in vivo. Moreover, no DNA methylation or EZH2 overexpression was observed in one specimen (case number 15) showed RUNX3 expression. These results seem to mean that the effects on RUNX3 expression described above are not limited to cultured cells but occur in vivo as well. However, RUNX3 expression was detected in one specimen (case number 7) despite the presence of both DNA methylation and EZH2 overexpression. This finding seems to suggest the existence of an unknown mechanism of RUNX3 repression and further study will be necessary.
FIGURE 5.
DNA methylation of the RUNX3 promoter, and expression of EZH2 protein and RUNX3 protein detected by immunohistochemical staining in tissue specimens from 17 gastric cancers. A, DNA methylation of the RUNX3 promoter was detected by MSP analysis in 11 of the 17 specimens (64.7%). Primer sets that specifically amplified either methylated DNA (M) or unmethylated DNA (U) were the same as used in the MSP analysis of the five cancer cell lines in Fig. 2B. The upper number is the case number. N, non-cancerous tissue; T, tumor tissue. Ma, 200- and 300-bp DNA ladder markers. B and C, histological appearance of the gastric cancer stained with H&E (hematoxylin and eosin) and immunohistochemically for EZH2 and RUNX3. The representative cases are 13 (B) and 2 (C). Immunohistochemical stainings for EZH2 and RUNX3 proteins were performed using two continuous thin tissue sections to show the inverse correlation of expression of EZH2 and RUNX3 proteins in individual cancer cells. The areas of cancer tissue surrounded by square boxes were magnified to show the expression or repression of EZH2 protein and RUNX3 protein of individual cancer cells. In B (case number 13), EZH2 is normally expressed in the germinal center follicular lymphocytes, whereas RUNX3 is normally expressed in the lymphocytes. In B (case number 13), DNA methylation in the RUNX3 promoter was detected in MSP analysis (A), but RUNX3 expression is observed in gastric cancer cells. On the other hand, in C (case number 2), loss of RUNX3 is observed in gastric cancer cells, but no DNA methylation of the promoter region was detected in MSP analysis (A).
TABLE 1.
Relationship between RUNX3 methylation, overexpression of EZH2 protein, and loss of RUNX3 protein in gastric cancer tissue
| Case No. | Age | Gender | Histologya | pT | pN | pm | pTNM | MSP-RUNX3b | EZH2c | RUNX3d | RUNX3-localizatione |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 61 | M | Sig | T1 | N0 | M0 | IA | Methylated | Positive | Negative | |
| 2 | 52 | F | Tub | T3 | N1 | M0 | IIIA | Unmethylated | Positive | Negative | |
| 3 | 74 | F | Tub | T3 | N1 | M0 | IIIA | Methylated | Positive | Negative | |
| 4 | 55 | F | Por | T3 | N2 | M0 | IIIB | Methylated | Positive | Negative | |
| 5 | 57 | F | Por | T4 | N1 | M0 | IV | Unmethylated | Positive | Negative | |
| 6 | 58 | M | Por | T1 | N1 | M0 | IB | Unmethylated | Positive | Negative | |
| 7 | 75 | M | Tub | T1 | N0 | M0 | IA | Methylated | Positive | Positive | C |
| 8 | 54 | F | Sig | T1 | N1 | M0 | IB | Unmethylated | Positive | Negative | |
| 9 | 65 | M | Sig | T2a | N0 | M0 | IB | Methylated | Positive | Negative | |
| 10 | 77 | M | Tub | T2a | N1 | M0 | II | Methylated | Positive | Negative | |
| 11 | 58 | F | Tub | T1 | N0 | M0 | IA | Unmethylated | Positive | Positive | C |
| 12 | 60 | M | Tub | T2b | N3 | M0 | IV | Methylated | Positive | Negative | |
| 13 | 66 | F | Por | T2b | N1 | M0 | II | Methylated | Negative | Positive | N and C |
| 14 | 68 | F | Por | T2b | N0 | M0 | IB | Methylated | Positive | Negative | |
| 15 | 66 | M | Tub | T3 | N2 | M0 | IIIB | Unmethylated | Negative | Positive | N and C |
| 16 | 65 | F | Tub | T2b | N2 | M0 | IIIA | Methylated | Positive | Negative | |
| 17 | 57 | M | Por | T1 | N0 | M1 | IV | Methylated | Negative | Positive | C |
Histology: sig, signet-ring cell carcinoma; tub, tubular adenocarcinoma; por, poorly differentiated adenocarcinoma
Methylation status of RUNX3 by methylation-specific PCR (MSP)
EZH2 overexpression was evaluated by immunohistochemical staining
RUNX3 expression was evaluated by immunohistochemical staining
RUNX3 protein was localized in nucleus (N) or/and cytoplasm (C)
DISCUSSION
The Polycomb group (PcG) protein EZH2, a histone methyltransferase and component of the PRC2 complex, is the product of a well known oncogene and a marker of poor prognosis in various cancers, including prostate cancer and breast cancer (18, 19). The mechanism underlying EZH2 overexpression associated with poor prognosis and the target genes down-regulated by EZH2, which lead to oncogenic activity, are still unknown. The mechanism of regulation of RUNX3 by EZH2 reported above may shed some light on the mechanism underlying EZH2 overexpression.
Polycomb target genes are often silenced by histone deacetylation and DNA methylation of CpG islands (30), and this is explained by the ability of PcG proteins to bind histone deacetylases and recruit DNA methyltransferases. We used TSA, a cell permeable inhibitor of histone deacetylases, to investigate whether expression of the RUNX3 gene is also controlled by histone deacetylation, and exposure of the five cancer cell lines to TSA for 48 h actually resulted in a 2.5–26.2-fold increase in the RUNX3 transcript level (Fig. 3, A–E). Because TSA did not affect the expression level of EZH2, these findings strongly indicate that the RUNX3 gene is also silenced by histone deacetylation. To test in detail, we performed ChIP experiments with antibodies against HDAC1. After transfection with control siRNA, both regions of the RUNX3 promoter examined were bound to HDAC1 in the MKN28 and MCF-7 cell lines (Fig. 2A), both of whose Runx3 transcript levels were restored only by TSA treatment for 48 h. By contrast, after transfection of these two cancer cell lines with EZH2 siRNA, less HDAC1 was bound to both regions of the RUNX3 promoter examined when RUNX3 expression was restored (Fig. 2A). This suggests that down-regulation of RUNX3 may be mediated by both H3K27 trimethylation by EZH2 and histone deacetylation by HDAC1. A previous study also showed restoration of both RUNX3 transcript and RUNX3 protein levels in breast cancer cell lines including MCF-7, T47D, and MDA-MB-231 by TSA alone (27). Moreover, the results of our study showed that EZH2 and HDAC1 act synergistically to down-regulate RUNX3 expression.
Hypermethylation of RUNX3 had been found to be common in many types of cancer cell lines and to correlate with loss of RUNX3 expression, and in the present study we found that 5-aza-dC, an inhibitor of DNA methyltransferase, promoted expression of the RUNX3 gene in five cancer cell lines by 1.5–10.9-fold (Fig. 3, A–E), indicating that DNA methylation also contributed to the repression of RUNX3. A comparison between DNA methylation and HDAC1 binding (histone deacetylation) to determine which was more effective in repressing RUNX3 showed that the level of RUNX3 transcript restoration by TSA was much greater than by 5-aza-dC in four of the cancer cell lines, the exception being the MiaPaca2 line (Fig. 3, A–E). In our study, EZH2 knockdown by transient siRNA transfection restored the RUNX3 transcript level 3.5–10.4-fold without any change in the DNA methylation status of the RUNX3 promoter region (Fig. 1A). This finding indicates that histone modification by EZH2 plays a key role in down-regulation of RUNX3, in addition to DNA methylation in the promoter.
EZH2 expression has been proposed as a marker of invasion and aggressive tumors (18, 19, 31), and experimental data have indicated a role of EZH2 in cell cycle regulation and proliferation. For example, disruption of EZH2 expression retards cell proliferation and induces cell cycle arrest at the G2-M transition (16), and overexpression of EZH2 in cultured mouse embryonic fibroblasts has been found to shorten the G1 phase of the cell cycle and lead to accumulation of cells in the S phase (17). A significant association between EZH2 and tumor cell proliferation, as estimated in human tumor tissue by Ki-67 expression and mitotic count, was recently demonstrated in clinical specimens including specimens of cutaneous melanoma and cancers of the endometrium, prostate, and breast (32). By contrast, RUNX3 up-regulates p21WAF1/Cip1, an important factor in cyclin-dependent kinase inhibition and cell cycle control (21). It was suggested that down-regulation of RUNX3 by EZH2 overexpression shown in the present study may be one pathway by which EZH2 affects tumor cell proliferation.
In summary, in this study, we showed that increased expression of EZH2 results in H3K27 trimethylation of the RUNX3 gene, indicating that RUNX3 is a novel EZH2 target gene. The identification of RUNX3 as an EZH2 target gene can support an influence of EZH2 overexpression on increasing tumor cell proliferation. Our findings suggest that specific inhibitors of EZH2 may be useful in the treatment of several types of cancers because such inhibitors are expected to reverse the down-regulation of the tumor suppressor RUNX3.
Supplementary Material
Acknowledgments
We thank Mai Okumoto for technical assistance.
This work was supported by Ministry of Education, Culture, Sports, Science and Technology, Japan, Grant 19590417 (to S. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Author's Choice – Final version full access
Footnotes
The abbreviations used are: EZH2, enhancer of zeste homologue 2; siRNA, small interfering RNA; RT, reverse transcriptase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ChIP, chromatin immunoprecipitation; MYT1, myelin transcription factor 1; MSP, methylation-specific polymerase chain reaction; TSA, trichostatin A; 5-aza-dC, 5-aza-2′-deoxycytidine; HDAC1, histone deacetylase 1.
References
- 1.Ito, Y. (2004) Oncogene 23 4198-4208 [DOI] [PubMed] [Google Scholar]
- 2.Inoue, K., Ozaki, S., Shiga, T., Ito, K., Masuda, T., Okado, N., Iseda, T., Kawaguchi, S., Ogawa, M., Bae, S. C., Yamashita, N., Itohara, S., Kudo, N., and Ito, Y. (2002) Nat. Neurosci. 5 946-954 [DOI] [PubMed] [Google Scholar]
- 3.Levanon, D., Bettoun, D., Harris-Cerruti, C., Woolf, E., Negreanu, V., Eilam, R., Bernstein, Y., Goldenberg, D., Xiao, C., Fliegauf, M., Kremer, E., Otto, F., Brenner, O., Lev-Tov, A., and Groner, Y. (2002) EMBO J. 21 3454-3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniuchi, I., Osato, M., Egawa, T., Sunshine, M. J., Bae, S. C., Komori, T., Ito, Y., and Littman, D. R. (2002) Cell 111 621-633 [DOI] [PubMed] [Google Scholar]
- 5.Woolf, E., Xiao, C., Fainaru, O., Lotem, J., Rosen, D., Negreanu, V., Bernstein, Y., Goldenberg, D., Brenner, O., Berke, G., Levanon, D., and Groner, Y. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7731-7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo, W. H., Weng, L. Q., Ito, K., Chen, L. F., Nakanishi, H., Tatematsu, M., and Ito, Y. (2002) Oncogene 21 8351-8355 [DOI] [PubMed] [Google Scholar]
- 7.Li, Q. L., Ito, K., Sakakura, C., Fukamachi, H., Inoue, K., Chi, X. Z., Lee, K. Y., Nomura, S., Lee, C. W., Han, S. B., Kim, H. M., Kim, W. J., Yamamoto, H., Yamashita, N., Yano, T., Ikeda, T., Itohara, S., Inazawa, J., Abe, T., Hagiwara, A., Yamagishi, H., Ooe, A., Kaneda, A., Sugimura, T., Ushijima, T., Bae, S. C., and Ito, Y. (2002) Cell 109 113-124 [DOI] [PubMed] [Google Scholar]
- 8.Li, Q. L., Kim, H. R., Kim, W. J., Choi, J. K., Lee, Y. H., Kim, H. M., Li, L. S., Kim, H., Chang, J., Ito, Y., Youl, Lee, K., and Bae, S. C. (2004) Biochem. Biophys. Res. Commun. 314 223-228 [DOI] [PubMed] [Google Scholar]
- 9.Xiao, W. H., and Liu, W. W. (2004) World J. Gastroenterol. 10 376-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, T. Y., Lee, H. J., Hwang, K. S., Lee, M., Kim, J. W., Bang, Y. J., and Kang, G. H. (2004) Lab. Investig. 84 479-484 [DOI] [PubMed] [Google Scholar]
- 11.Ku, J. L., Kang, S. B., Shin, Y. K., Kang, H. C., Hong, S. H., Kim, I. J., Shin, J. H., Han, I. O., and Park, J. G. (2004) Oncogene 23 6736-6742 [DOI] [PubMed] [Google Scholar]
- 12.Wada, M., Yazumi, S., Takaishi, S., Hasegawa, K., Sawada, M., Tanaka, H., Ida, H., Sakakura, C., Ito, K., Ito, Y., and Chiba, T. (2004) Oncogene 23 2401-2407 [DOI] [PubMed] [Google Scholar]
- 13.Kang, G. H., Lee, S., Lee, H. J., and Hwang, K. S. (2004) J. Pathol. 202 233-240 [DOI] [PubMed] [Google Scholar]
- 14.Laible, G., Wolf, A., Dorn, R., Reuter, G., Nislow, C., Lebersorger, A., Popkin, D., Pillus, L., and Jenuwein, T. (1997) EMBO J. 16 3219-3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon, J. A., and Tamkun, J. W. (2002) Curr. Opin. Genet. Dev. 12 210-218 [DOI] [PubMed] [Google Scholar]
- 16.Tang, X., Milyavsky, M., Shats, I., Erez, N., Goldfinger, N., and Rotter, V. (2004) Oncogene 23 5759-5769 [DOI] [PubMed] [Google Scholar]
- 17.Bracken, A. P., Pasini, D., Capra, M., Prosperini, E., Colli, E., and Helin, K. (2003) EMBO J. 22 5323-5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varambally, S., Dhanasekaran, S. M., Zhou, M., Barrette, T. R., Kumar-Sinha, C., Sanda, M. G., Ghosh, D., Pienta, K. J., Sewalt, R. G., Otte, A. P., Rubin, M. A., and Chinnaiyan, A. M. (2002) Nature 419 624-629 [DOI] [PubMed] [Google Scholar]
- 19.Kleer, C. G., Cao, Q., Varambally, S., Shen, R., Ota, I., Tomlins, S. A., Ghosh, D., Sewalt, R. G., Otte, A. P., Hayes, D. F., Sabel, M. S., Livant, D., Weiss, S. J., Rubin, M. A., and Chinnaiyan, A. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11606-11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tonini, T., Bagella, L., D'Andrilli, G., Claudio, P. P., and Giordano, A. (2004) Oncogene 23 4930-4937 [DOI] [PubMed] [Google Scholar]
- 21.Chi, X. Z., Yang, J. O., Lee, K. Y., Ito, K., Sakakura, C., Li, Q. L., Kim, H. R., Cha, E. J., Lee, Y. H., Kaneda, A., Ushijima, T., Kim, W. J., Ito, Y., and Bae, S. C. (2005) Mol. Cell. Biol. 25 8097-8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yano, T., Ito, K., Fukamachi, H., Chi, X. Z., Wee, H. J., Inoue, K., Ida, H., Bouillet, P., Strasser, A., Bae, S. C., and Ito, Y. (2006) Mol. Cell. Biol. 26 4474-4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii, S., Luo, R. Z., Yuan, J., Kadota, M., Oshimura, M., Dent, S. R., Kondo, Y., Issa, J. P., Bast, R. C., Jr., and Yu, Y. (2003) Hum. Mol. Genet. 12 1791-1800 [DOI] [PubMed] [Google Scholar]
- 24.Herman, J. G., Graff, J. R., Myohanen, S., Nelkin, B. D., and Baylin, S. B. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirmizis, A., Bartley, S. M., Kuzmichev, A., Margueron, R., Reinberg, D., Green, R., and Farnham, P. J. (2004) Genes Dev. 18 1592-1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito, K., Liu, Q., Salto-Tellez, M., Yano, T., Tada, K., Ida, H., Huang, C., Shah, N., Inoue, M., Rajnakova, A., Hiong, K. C., Peh, B. K., Han, H. C., Ito, T., Teh, M., Yeoh, K. G., and Ito, Y. (2005) Cancer Res. 65 7743-7750 [DOI] [PubMed] [Google Scholar]
- 27.Lau, Q. C., Raja, E., Salto-Tellez, M., Liu, Q., Ito, K., Inoue, M., Putti, T. C., Loh, M., Ko, T. K., Huang, C., Bhalla, K. N., Zhu, T., Ito, Y., and Sukumar, S. (2006) Cancer Res. 66 6512-6520 [DOI] [PubMed] [Google Scholar]
- 28.Fowler, M., Borazanci, E., McGhee, L., Pylant, S. W., Williams, B. J., Glass, J., Davis, J. N., and Meyers, S. (2006) J. Cell. Biochem. 97 1-17 [DOI] [PubMed] [Google Scholar]
- 29.Raaphorst, F. M., van Kemenade, F. J., Blokzijl, T., Fieret, E., Hamer, K. M., Satijn, D. P., Otte, A. P., and Meijer, C. J. (2000) Am. J. Pathol. 157 709-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viré, E., Brenner, C., Deplus, R., Blanchon, L., Fraga, M., Didelot, C., Morey, L., Van Eynde, A., Bernard, D., Vanderwinden, J. M., Bollen, M., Esteller, M., Di Croce, L., de Launoit, Y., and Fuks, F. (2006) Nature 439 871-874 [DOI] [PubMed] [Google Scholar]
- 31.Raaphorst, F. M., Meijer, C. J., Fieret, E., Blokzijl, T., Mommers, E., Buerger, H., Packeisen, J., Sewalt, R. A., Otte, A. P., and van Diest, P. J. (2003) Neoplasia (Bratisl.) 5 481-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachmann, I. M., Halvorsen, O. J., Collett, K., Stefansson, I. M., Straume, O., Haukaas, S. A., Salvesen, H. B., Otte, A. P., and Akslen, L. A. (2006) J. Clin. Oncol. 24 268-273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.