Abstract
Human checkpoint kinase 1 (Chk1) is an essential kinase required for cell cycle checkpoints and for coordination of DNA synthesis. To gain insight into the mechanisms by which Chk1 carries out these functions, we used mass spectrometry to identify previously uncharacterized interacting partners of Chk1. We describe a novel interaction between Chk1 and proliferating cell nuclear antigen (PCNA), an essential component of the replication machinery. Binding between Chk1 and PCNA was reduced in the presence of hydroxyurea, suggesting that the interaction is regulated by replication stress. A highly conserved PCNA-interacting protein (PIP) box motif was identified in Chk1. The intact PIP box is required for efficient DNA damage-induced phosphorylation and release of activated Chk1 from chromatin. We find that the PIP box of Chk1 is crucial for Chk1-mediated S-M and G2-M checkpoint responses. In addition, we show that mutations in the PIP box of Chk1 lead to decreased rates of replication fork progression and increased aberrant replication. These findings suggest an additional mechanism by which essential components of the DNA replication machinery interact with the replication checkpoint apparatus.
The Chk1 kinase is required for the intra-S, S-M and G2-M checkpoint responses in vertebrate cells (1–8). In addition, there is mounting evidence that Chk1 has a critical role in regulating the timing and progression of replication in unperturbed cell cycles (9–11). Chk15 is essential during mouse embryogenesis and for proliferation of somatic cells (4, 7). A cell line that lacks Chk1 was generated in chicken DT40 lymphoma cells; although viable, these cells proliferate slowly and display increased spontaneous cell death (12). DT40 cells that lack Chk1 (DT40Chk1-/-) are exquisitely sensitive to replication stress because they fail to maintain functional replication fork structures and do not appropriately suppress origin firing (9, 11, 13). Thus, in both mammalian and chicken cells, Chk1 regulates origin firing during normal S-phase, protects against replication-associated DNA breaks, and prevents the firing of late origins when early origins are stalled (10, 11, 14, 15). The mechanisms by which Chk1 inhibits cell cycle progression are fairly well understood; however, despite the increasing evidence implicating Chk1 directly in replication control (for review, see Ref. 16), it is unclear exactly how Chk1 regulates and is regulated by different S-phase processes.
Activation of Chk1 requires direct phosphorylation by ATR and ATM on consensus Ser-Gln motifs (2, 17–19). In mammals Chk1 phosphorylation occurs primarily on two serine residues, Ser-317 and Ser-345, in the carboxyl terminus. Autophosphorylation, conformational changes, and changes in subcellular location of Chk1 are also thought to contribute to its full activation and are necessary for function (20–24).
In addition to the kinases that phosphorylate Chk1 on Ser-Gln sites, several other checkpoint proteins are required for Chk1 function in vivo. These include TopBP1, Rad17, the Rad9·Rad1·Hus1 (9-1-1) complex, and Claspin (25–30). Claspin functions as an adapter protein that is required for the efficient phosphorylation of Chk1 by ATR (31–34). The replication factor C-related checkpoint protein Rad17 is required to load the 9-1-1 complex onto chromatin, and it interacts with Claspin to promote the phosphorylation of Chk1 (30, 35, 36). Rad9 binds to and recruits TopBP1 to sites of DNA damage to allow efficient phosphorylation of ATR substrates, including Chk1 (37). TopBP1 is a general regulator of ATR function required to stimulate phosphorylation of several substrates (28). Like TopBP1, the Timeless-Tipin complex is a replication-associated factor that is required for Chk1 function (38–41). The interactions that these factors mediate, between core components of the replication machinery and Chk1, enhance the mechanism by which aberrations in ongoing replication promote activation of Chk1.
To gain insight into the mechanisms of regulation and function of Chk1, we sought to identify novel interacting proteins by mass spectrometry-based methods. This analysis revealed a previously uncharacterized interaction between Chk1 and PCNA. We discovered a conserved PCNA-interacting protein (PIP) box in the carboxyl terminus of Chk1 that is required for efficient DNA damage-induced phosphorylation and chromatin release of Chk1. We demonstrate that the PIP box is critical for both the Chk1-mediated S-M and G2-M checkpoints and for appropriate regulation of DNA replication.
EXPERIMENTAL PROCEDURES
Cell Lines and Treatments—HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% bovine calf serum, penicillin, and streptomycin at 37 °C and 5% CO2. Cells were treated with 2 mm hydroxyurea (Sigma-Aldrich) where indicated. DT40 B lymphoma cells (12) were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 1% chicken serum, 10 μm β-mercaptoethanol, penicillin, and streptomycin, at 37 °C. Cells were irradiated with 10 gray using a cesium source or treated with 20 μm aphidicolin or 3.3 nm nocodazole (Sigma) where indicated. DT40Chk1-/- cells were as described in Zachos et al. (12). To generate DT40Chk1TRAA, DT40Chk1-/- were transfected with avian Chk1TRAA expressed in pcDNA3.1zeo (Invitrogen) and selected in 500 μg/ml zeocin (Invitrogen). Drug-resistant clones were screened for Chk1 expression by Western blotting.
Plasmids and Constructs—GST fusion Chk1 proteins were made by annealing the following oligonucleotides and cloning into pGEX using standard cloning procedures: TRFF (wild type), 5′-AATTCGGCAGCGGTTGGTCAAAAGAATGACACGATTCTTTACCAAATTGTAAGATATC-3′ and 5′-CATGGATATCTTACAATTTGGTAAAGAATCGTGTCATTCTTTTGACCAACCCGCTGCCG-3′; ARFF (Thr-378 to Ala), 5′-AATTCGGCAGCGGTTGGTCAAAAGAATGGCACGATTCTTTACCAAATTGTAAGATATC-3′ and 5′-CATGGATATCTTACAATTTGGTAAAGAATCGTGCCATTCTTTTGACCAACCGCTGCCG-3′; TRAA (Phe-380 and Phe-381 both to Ala), 5′-AATTCGGCAGCGGTTGGTCAAAAGAATGACACGAGCCGCTACCAAATTGTAAGATATC-3′ and 5′-CATGGATATCTTACAATTTGGTAGCGGCTCGTGTCATTCTTTTGACCAACCGCTGCCG-3′; TREE (Phe-380 and Phe-381 both to Glu), 5′-AATTCGGCAGCGGTTGGTCAAAAGAATGACACGAGAAGAAACCAAATTGTAAGATATC-3′ and 5′-CATGGATATCTTACAATTTGGTTTCTTCTCGTGTCATTCTTTTGACCAACCGCTGCCG-3′. For FLAG-Chk1 constructs the altered 3′ end of Chk1 was cloned into the coding region in pcDNA3.1 using an internal BamHI site. All mutations were verified by sequencing.
Oligonucleotides and Transfections—Plasmids were transfected into HeLa cells using Effectene (Qiagen) and small interfering RNAs transfected using Oligofectamine (Invitrogen). Claspin small interfering RNA duplexes (CCUUGCUUAGAGCUGAGUCdTdT) were from Dharmacon Research. For small interfering RNA, HeLa cells were transfected twice 24 h apart, and cells were harvested 48 h after initial transfection. The final concentration of the small interfering RNA duplex in media was 75 nm.
Antibodies—Antibodies to HA (12CA5) and FLAG (M2) were from Sigma-Aldrich. Monoclonal Chk1 (G-4) and monoclonal PCNA (PC10) antibodies were from Santa Cruz. Polyclonal Orc2 antibody was from BD Pharmingen. Polyclonal phospho-Ser-345 and phospho-Ser-317 Chk1 antibodies were from Cell Signaling. Polyclonal antibodies to Claspin (ab3720) were from Abcam. A polyclonal rabbit antiserum specific for avian Chk1 was generated using a synthetic peptide corresponding to the carboxyl-terminal 47-amino acids of chicken Chk1 and was used for immunoprecipitation of avian Chk1 (12).
Western Blotting and Immunoprecipitations—Cell extracts were prepared in radioimmune precipitation assay buffer (50 mm Tris pH 7.5, 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, 2 mm EDTA, 50 mm NaF, 0.1% β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin/pepstatin and 0.1 μm okadaic acid). For PCNA and Chk1 immunoprecipitates, radioimmune precipitation buffer lysates were incubated with protein A/G beads (Amersham Biosciences), 1 μg of monoclonal PCNA or Chk1 antibody, and 1 μg/ml ethidium bromide for 2 h at 4 °C. For FLAG immunoprecipitates, cells were extracted with FLAG lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 10 mm NaF, 1 μg/ml leupeptin/pepstatin, 1 mm phenylmethylsulfonyl fluoride, and 0.1 μm okadaic acid). Immunoprecipitations were performed for 2–4 h at 4 °C using FLAG M2 beads (Sigma). To prepare samples for mass spectrometry, FLAG-bead complexes were washed in FLAG lysis buffer, and proteins were eluted with 100 μg/ml 3×FLAG peptide (Sigma) in FLAG lysis buffer for 4 h at 4 °C. Eluted proteins were precipitated with trichloroacetic acid and subjected to Multidimensional Protein Identification Technology (MudPIT) analysis.
MudPIT—MudPIT analysis was performed as described previously (42) with the following modifications. Samples were resuspended in 8 m urea, 100 mm Tris, pH 8.5, reduced, alkylated, diluted to 2 m urea, 1 mm CaCl2, 100 mm Tris, pH 8.5, and digested with trypsin. Tryptic digests were supplemented with formic acid to 5% and loaded onto a desalting column (250 μm diameter, 125 A pore size, 5 μm Aqua C18 resin (Phenomenex)) fitted with a 2-μm filtered union. This was placed in-line with 9 cm of 3 μm Aqua C18 resin of 125A pore size (Phenomenex) and 4 cm of 5 μm Partisphere SCX resin of 120A pore size (Whatman). MS analysis was performed on a LTQ mass spectrometer (Thermo-Fisher) using a 6-step MudPIT method with increasing salt pulses at 0, 10, 20, 40, 60, and 100% buffer C (250 mm ammonium acetate, 5% high performance liquid chromatography-grade acetonitrile, 0.02% heptafluorobutyric acid). Each full MS scan was followed by 5 MS/MS scans. The MS/MS spectra were searched with SEQUEST (43) against a combined RefSeq data base of human, mouse, and rat proteins with a 3-atomic mass unit mass tolerance window and no enzyme specificity. The SEQUEST outputs were filtered with DTASelect 1.8 (44) with the following parameters: Xcorr ≥ 1.8 (+1), 2.5 (+2), or 3.5 (+3), ΔCN ≥ 0.08; only half or fully tryptic peptides were considered. Proteins with at least two peptides that passed the DTASelect filter were considered real hits. The false positive rate of identified proteins was below 5%.
In Vitro Binding Assays—Expression of fusion proteins was induced in log-phase BL21 by the addition of 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 2–3 h at 37 °C. Cells were pelleted and resuspended in lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 5 mm EDTA, 10% glycerol, 0.1% Nonidet P-40, 0.1% β-mercaptoethanol). GST proteins were purified from cleared lysates on GSH beads (Amersham Biosciences). GSH beads were washed and incubated with HeLa or DT40 cell supernatants at 4 °C for 2–4 h. Bound proteins were released by boiling in SDS-loading buffer and analyzed by SDS-PAGE and Western blotting.
Kinase Assays—FLAG-Chk1 expressed in HeLa cells or Gallus gallus (Gg) Chk1 in DT40 cells was immunoprecipitated using specific antibodies. Kinase assays were performed by incubating immunoprecipitated proteins in kinase buffer (50 mm imidazole, pH7.4, 10 mm MgCl2) and 5 μCi of ATP at 30 °C for 15 min. Reactions were terminated by the addition of SDS loading buffer. Samples were run on SDS-PAGE gels, and kinase activity was monitored using a Storm 840 PhosphorImager.
Chromatin Fractionation—Chromatin fractionation was performed essentially as described in Smits et al. (21). Cytoplasmic proteins (S1) and soluble nuclear proteins (S2) were pooled into one fraction (S1 + S2, soluble proteins). Isolated chromatin (P2) was resuspended in 150 μl of Laemmli sample buffer (P2) and briefly sonicated. Protein concentrations were determined by the Bradford method. 1× cell equivalents of S1 + S2 and 5× cell equivalents of P2 were loaded on SDS-PAGE gels.
Fluorescence-activated Cell Sorter Analysis—Cells were fixed in 70% ethanol, phosphate-buffered saline at 4 °C. Cells were labeled with antibody to Ser(P)10-H3 and fluorescein isothiocyanate-conjugated secondary antibody, counterstained with propidium iodide, and analyzed on a BD Biosciences FACScan flow cytometer as described previously (45).
Replication Labeling and DNA Fiber Spreads—Replication tracks were labeled by adding 25 μm bromodeoxyuridine (BrdUrd) (Sigma) to culture media for 10, 20, or 40 min. For dual labeling, cells were pulsed with 25 μm BrdUrd for 10 min, washed twice with phosphate-buffered saline and twice with media and then labeled with 250 μm iododeoxyuridine (IdUrd) (Sigma) for 20 min. DNA spreads were prepared as described in Maya-Mendoza et al. (11). BrdUrd-containing tracks were detected with sheep anti-BrdUrd antibody (Biodesign; M20107S) and Cy3-conjugated donkey anti-sheep secondary (Jackson ImmunoResearch). In dual labeling experiments, BrdUrd-labeled tracks were detected with rat anti-BrdUrd (Accurate; BU 1/75) and Alexa-fluor488-conjugated chicken anti-rat secondary (Invitrogen) followed by IdUrd detection using mouse anti-BrdUrd/IdUrd (BD Biosciences) and Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch). Fibers were examined using a Nikon Eclipse E800 microscope using a 100× lens. Labeled track lengths were measured using ImageJ and converted to base pairs using the conversion 1 μm = 2.59 kilobases (46). Measurements were made in randomly selected fields of discrete, untangled fibers.
RESULTS
Chk1 and PCNA Interact in Vivo—To identify novel interacting partners of Chk1, HeLa cells were transfected with a plasmid encoding FLAG-tagged Chk1, and associated proteins were immunopurified using FLAG M2 affinity gel. The protein mix was digested with trypsin, and the resulting peptides were separated and identified using mass spectrometry-based MudPIT (47, 48). This resulted in the identification of known Chk1 interactors (e.g. Claspin, 14-3-3) as well as several novel Chk1 interacting proteins (supplemental Table 1). PCNA, a homotrimeric ring protein that functions as a processivity factor for replicative DNA polymerases (49), was identified as a novel interacting partner of Chk1 and was chosen for further study.
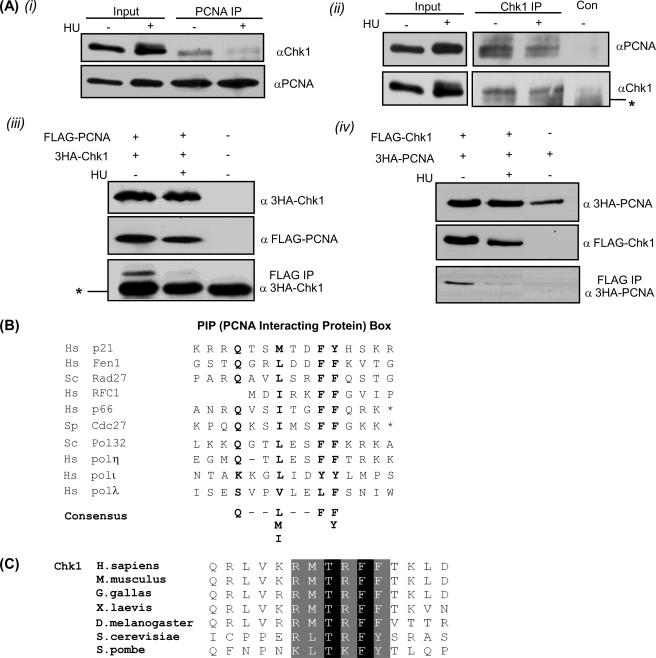
To confirm the interaction between Chk1 and PCNA in vivo, we performed co-immune precipitation experiments. The immunoprecipitation experiments were done in the presence of ethidium bromide to disrupt DNA-mediated interactions (50). Immunoprecipitates of endogenous PCNA contained Chk1 (Fig. 1A, top left), and immune precipitates of endogenous Chk1 contained PCNA (Fig. 1A, top right). PCNA was not present in pre-immune control precipitates. As a control for antibody specificity and because tagged-Chk1 was needed for analysis of site specific mutants, the Chk1-PCNA interaction was reconfirmed using tagged versions of Chk1 and PCNA. As shown in Fig. 1A, immune complexes containing FLAG-tagged PCNA also contained 3HA-Chk1 (Fig. 1A, bottom left panel). The interaction between these proteins was decreased after hydroxyurea (HU) treatment. Similarly, 3HA-PCNA was detected in an immune precipitate of FLAG-Chk1, and again the interaction was decreased when cells were cultured in the presence of HU (Fig. 1A, bottom right panel).
FIGURE 1.
Chk1 and PCNA interact in vivo and interaction is reduced after HU. A, top panels, interaction of endogenous proteins in HeLa cells. i, Chk1 was detected in PCNA immune -precipitates (IP) from HeLa cells. ii, similarly, Chk1 monoclonal antibodies, but not control antibodies (Con), immunoprecipitate PCNA from HeLa cells. The asterisk indicates cross-reaction with IgG heavy chain. Bottom panels, HeLa cells were transfected with PCNA and Chk1 as indicated above each panel. iii, 3HA-Chk1 was detected in FLAG-immune precipitates from cells that express FLAG-PCNA. The asterisk indicates IgG heavy chain. iv, in a reciprocal experiment, 3HA-PCNA was detected in FLAG immune precipitates from cells that express FLAG-Chk1. In all cases association was reduced in samples that had been cultured in 2 mm HU for 16 h before harvesting. B, an alignment of PIP box sequences from known PCNA binding proteins highlights a potential PIP box in the carboxyl terminus of Chk1. C, alignment of Chk1 carboxyl termini from diverse species indicates high conservation of the PIP box. Black indicates identical residues, and gray indicates similar residues.
Characterization of the PIP Box of Chk1—Many proteins that interact with PCNA contain a PCNA binding motif (QXXΨXXF(F/Y), where Ψ is M/L/I) known as a PIP box (51). Analysis of the amino acid sequence of Chk1 revealed a consensus PIP box in the carboxyl-terminal region encompassing residues 374–381 (Fig. 1B). This putative PIP box contains a sequence, TRFF, which is highly conserved between species (Fig. 1C); this sequence is one of the few regions in the carboxyl terminus of Chk1 that is highly conserved. To investigate whether the TRFF motif of Chk1 is a bona fide PIP box, we created GST fusion proteins containing the predicted minimal PCNA binding region (GST-Chk1PIP). The design of these constructs was based on the PCNA binding region of p21, an extensively characterized PCNA-binding protein (52, 53). The importance of the two phenylalanine residues that are crucial for PCNA binding in other PCNA-interacting proteins (53–56) was tested by generating constructs in which these residues were changed either to alanine (creating TRAA) or to glutamate (creating TREE). In addition, threonine 378 was mutated to alanine (ARFF), generating a mutation that was not expected to disrupt PCNA binding. This mutation was of interest because a similar substitution in Xenopus Chk1 had been shown to result in hyperactivation of Chk1 (57).
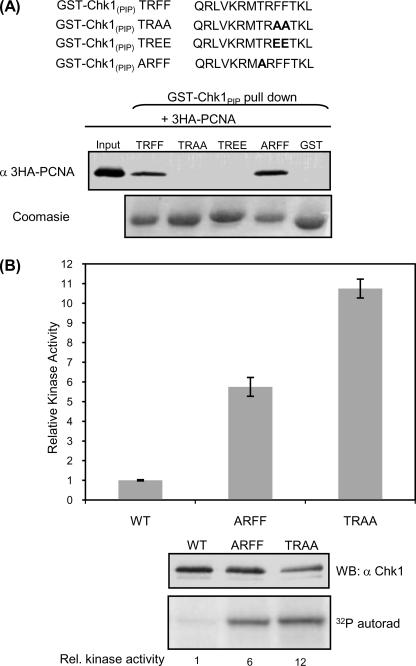
Purified GST-Chk1PIP was examined for the ability to bind 3HA-PCNA expressed in HeLa cells. As predicted, the wild type TRFF-containing sequence binds to PCNA (Fig. 2A). Mutation of threonine 378 to alanine did not affect binding; however, mutation of the conserved phenylalanine residues (Phe-380 and Phe-381) either to alanine or to glutamic acid disrupted binding to PCNA (Fig. 2A).
FIGURE 2.
Mutations within the PIP box activate Chk1 kinase activity. A, wild type or mutant versions of the PIP box (amino acids 371–384) from Chk1 were fused to GST and were probed for the ability to interact with 3HA-PCNA expressed in HeLa cells. GST-Chk1PIP wild type (TRFF) and ARFF sequences bind to PCNA, whereas mutation of conserved hydrophobic residues either to neutral (TRAA) or to acidic residues (TREE) abolishes PCNA binding. B, the kinase activity of FLAG-Chk1ARFF and FLAG-Chk1TRAA is increased compared with FLAG-Chk1WT. Kinase assays were performed on the indicated FLAG-Chk1 proteins immunoprecipitated from HeLa cells. Kinase activity was normalized to protein expression levels. The graph represents the average Chk1 kinase activity measured in four independent experiments. Error bars represent S.E. A representative Western blot (WB) and autoradiograph of indicated FLAG-Chk1 proteins is shown below the graph.
To investigate the role of the PIP box in Chk1 function, we introduced the TRAA mutation into FLAG-tagged Chk1. As shown in Fig. 2B, both this mutation and mutation of the nearby threonine (in ARFF) led to increased Chk1 kinase activity. FLAG-Chk1ARFF has an ∼6-fold increase in autophosphorylation compared with wild type protein. Likewise, mutation of the conserved phenylalanine residues to create FLAG-Chk1TRAA increased Chk1 kinase activity ∼10-fold. Similar increases in activity were seen when the ability to phosphorylate an extrinsic substrate, GST-Cdc25254–316, was monitored (data not shown). Mutation and deletions in the carboxyl-terminal region of Chk1 have previously been shown to increase intrinsic kinase activity and are thought to result in reduced interaction between the amino terminus and the catalytic domain (22, 57, 58). The increased intrinsic activity of FLAG-Chk1TRAA and FLAG-Chk1ARFF is likely to be explained by the same intramolecular mechanism (22).
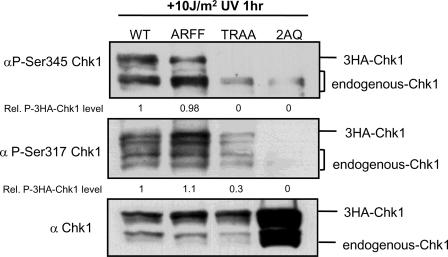
The PIP Box Is Essential for Efficient DNA Damage-induced Phosphorylation of Chk1—Given that the TRFF motif of Chk1 is a PIP box and is highly conserved between species, we sought to determine how mutations in this domain affect Chk1 function and regulation. To determine whether the PIP box is required for DNA-damage-induced phosphorylation, we examined Ser-Gln phosphorylation of Chk1 after UV and HU. FLAG-Chk1WT and FLAG-Chk1ARFF were efficiently phosphorylated on both Ser-317 and Ser-345 after UV (Fig. 3). In contrast, phosphorylation of FLAG-Chk1TRAA was markedly decreased after UV or HU treatment (Fig. 3 and data not shown). Background staining was determined by comparison to FLAG-Chk12AQ in which both Ser-317 and Ser-345 are mutated to alanine (Fig. 3; FLAG-Chk12AQ).
FIGURE 3.
The PIP box of Chk1 is required for efficient damage-induced Chk1 phosphorylation. HeLa cells expressing 3HA-Chk1WT, 3HA-Chk1ARFF, 3HA-Chk1TRAA, and 3HA-Chk12AQ (S317A, S345A) were irradiated with 10 J/m2 UV and harvested 1 h later. The extent of Chk1 phosphorylation was monitored using anti-Ser(P)-345 and anti-Ser(P)-317. Migration of exogenous 3HA-Chk1 and endogenous Chk1 was as indicated. Levels of phosphorylated 3HA-Chk1 were quantified using ImageJ and are shown below the relevant lanes. Excess 3HA-Chk12AQ was used to monitor the extent to which these antibodies react with unphosphorylated Chk1.
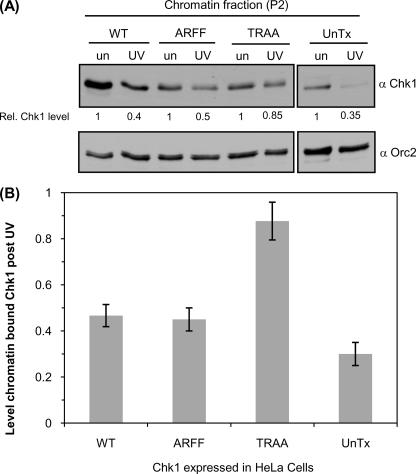
The PIP Box of Chk1 Is Required for DNA Damage-induced Chromatin Dissociation—A fraction of Chk1 binds to chromatin in unperturbed cells, and phosphorylation of Chk1 on Ser-317 and Ser-345 triggers dissociation from chromatin upon DNA damage (18, 21). To investigate whether the defective Ser-Gln phosphorylation of FLAG-Chk1TRAA affects chromatin dissociation after DNA damage, we fractionated proteins from untreated or UV-irradiated cells. As expected, 65–70% of chromatin-bound endogenous Chk1 and 50–60% of chromatin-bound FLAG-Chk1WT was consistently found to dissociate after UV treatment (Fig. 4, A and B). Mutation of threonine 378 to alanine did not disrupt DNA damage-induced chromatin release, and like the wild type protein, FLAG-Chk1ARFF was efficiently released upon UV irradiation. By contrast, disruption of the PIP box prevented FLAG-Chk1TRAA from efficiently dissociating from chromatin after DNA damage with only ∼15% being released after irradiation (Fig. 4A and quantified in Fig. 4B). Together these data indicate that mutations which specifically disrupt the PIP box of Chk1 cause defective DNA damage-induced phosphorylation of Chk1 and inefficient dissociation from chromatin.
FIGURE 4.
The PIP box of Chk1 is required for efficient dissociation of Chk1 from chromatin. A, HeLa cells expressing FLAG-Chk1WT, FLAG-Chk1ARFF, FLAG-Chk1TRAA, or untransfected cells (UnTx) were irradiated with 50 J/m2 UV (UV) or left untreated (un). Two hours after UV treatment subcellular fractions were prepared. The level of Chk1 retained on chromatin (P2) was quantified relative to untreated samples using ImageJ and is shown below the relevant lanes. A longer exposure of the same blot was used to analyze retention of endogenous Chk1 in untransfected cells (right hand panels). Orc2 was used to verify loading of the chromatin fraction. B, quantification showing a defect in release of FLAG-Chk1TRAA from chromatin. The level of Chk1 retained on chromatin after 50J/m2 UV was quantified relative to untreated samples using ImageJ. The graph represents a fraction of chromatin-bound Chk1 measured in three independent experiments. Error bars represent S.E.
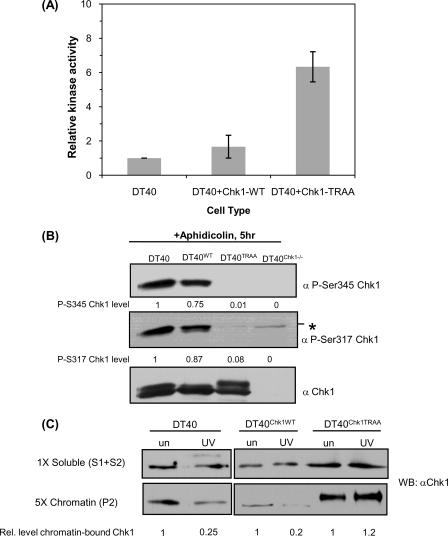
Characterization of GgChk1 PIP Box—The increased kinase activity of FLAG-Chk1TRAA (Fig. 2B) might be expected to result in enhanced checkpoint function (22, 57, 58). On the other hand, the reduced Ser-Gln phosphorylation and inefficient chromatin release of FLAG-Chk1TRAA might result in reduced DNA damage-induced checkpoint responses. These possibilities were difficult to address in mammalian cells in which Chk1 is essential (4, 7); therefore, the phenotypic consequences of Chk1 PIP box mutations were examined in DT40 chicken cells in which both copies of the gene encoding Chk1 had been removed by homologous recombination (12). Accordingly, DT40Chk1-/- cells were reconstituted with either wild type Chk1 (DT40Chk1WT) or Chk1TRAA (DT40Chk1TRAA). Untagged GgChk1 protein was used to complement Chk1-deficient cells as in Zachos et al. (12). Initially, we determined the kinase activity of GgChk1 proteins expressed in DT40 cells. This analysis showed that, similar to the human protein, GgChk1TRAA has ∼7-fold increased activity relative to the wild type kinase (Fig. 5A).
FIGURE 5.
PIP-disrupted chicken Chk1 is hyperactive but is not efficiently phosphorylated after DNA damage and is not efficiently released from chromatin after DNA damage. A, the kinase activity of GgChk1TRAA is increased compared with endogenous GgChk1 (DT40) and wild type Chk1 (DT40Chk1WT). The graph represents average Chk1 kinase activity measured in three independent experiments. Error bars represent S.E. B, DT40 cells and DT40Chk1-/- reconstituted with GgChk1WT (DT40Chk1WT) or GgChk1TRAA (DT40Chk1TRAA) were cultured in 20 μm aphidicolin for 5 h. The extent of Chk1 phosphorylation was monitored using anti-Ser(P)-345 and anti-Ser(P)-317. Levels of phospho-Chk1 were quantified using ImageJ and are shown below the relevant lanes. A background/nonspecific band (*) was detected using the P-S317 but not P-S345 antibody. C, DT40, DT40WT, DT40TRAA, and DT40Chk1-/- cells were irradiated with 50 J/m2 UV (UV) or left untreated (un). Two hours later subcellular fractions were prepared. The levels of Chk1 retained on chromatin were quantified relative to untreated samples using ImageJ and are shown below the relevant lanes.
DNA damage-induced phosphorylation of GgChk1 and GgChk1TRAA was examined in cells treated with aphidicolin or UV light. Wild type GgChk1 protein was efficiently phosphorylated on both Ser-317 and Ser-345 after aphidicolin treatment (Fig. 5B) and UV irradiation (data not shown). In contrast and consistent with data from human cells, GgChk1TRAA showed significantly reduced DNA damage-induced phosphorylation on both sites (Fig. 5B).
The damage-induced chromatin release of endogenous GgChk1, GgChk1WT, and GgChk1TRAA was investigated by fractionation of DT40, DT40Chk1WT, and DT40Chk1TRAA cells. Cells were either untreated or irradiated with 50 Jm-2 of UV light. Two hours after irradiation cells were harvested and separated into soluble and chromatin-bound fractions following the same protocol as in Fig. 3 and (21). Western blotting of the relevant fractions showed that ∼75% of endogenous GgChk1 and 80% wild type GgChk1 was released from chromatin after DNA damage (Fig. 5C). In contrast, GgChk1TRAA was not efficiently released from chromatin after DNA damage (Fig. 5C). The proportion of GgChk1TRAA retained on chromatin was greatly increased (∼5-fold) both in treated and untreated samples. Interestingly, the majority of chromatin-bound GgChk1TRAA migrated more slowly than wild type protein; as shown in supplemental Fig. 1, this was due to autophosphorylation. Despite being hyperphosphorylated, DNA damage-induced chromatin release of GgChk1TRAA was significantly and consistently impaired (Fig. 5C). These data indicate that mutations within the PIP box of both chicken and human Chk1 lead to reduced phosphoinositide 3-kinase-related kinase-dependent phosphorylation and defective DNA damage-induced chromatin release.
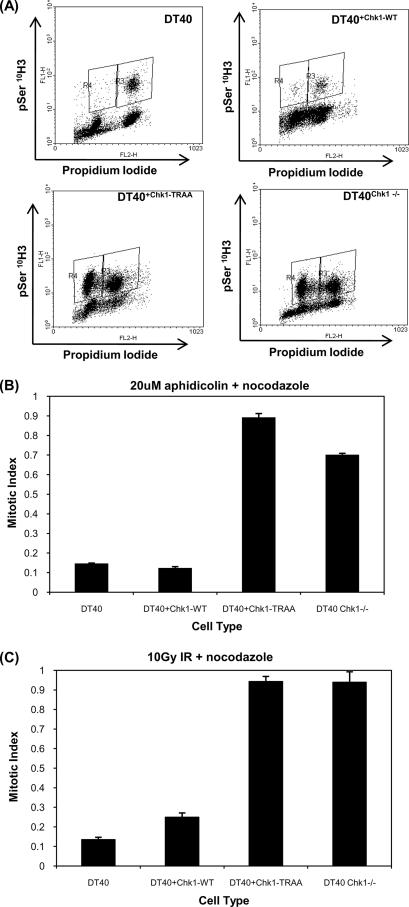
The PIP Box of Chk1 Is Critical for DNA Damage-induced Checkpoint Function—The phenotypic consequences of disrupting the PIP box were examined by analyzing aphidicolin and irradiation-induced checkpoint responses in complemented DT40Chk1-/- cells. When cultured in the presence of nocodazole, all cell types (DT40, DT40Chk1-/-, DT40Chk1WT, and DT40Chk1TRAA) progressed through the cell cycle and accumulated with phosphorylated histone H3 and a 4N DNA content (Ref. 13 and data not shown). When aphidicolin was added to DT40 or DT40Chk1WT cells, replication was inhibited, and only a small number of cells (∼1.1%) were trapped in mitosis; these cells had a 4N DNA content (Fig. 6A and quantified in Fig. 6B). In contrast, when DT40Chk1TRAA and DT40Chk1-/- cells were cultured in aphidicolin, the number of phospho-H3-positive cells increased (32.8 and 29.7%, respectively), indicating that entry into mitosis was not blocked (Fig. 6A). Most strikingly, there was a significant accumulation of phospho-H3-positive cells that had a 2N DNA content (Fig. 6A, region R4), indicating that cells entered mitosis with unreplicated DNA. These data suggest that DT40Chk1TRAA cells, sim ilar to DT40Chk1-/- cells, lack the checkpoint that prevents the onset of mitosis when DNA replication is blocked.
FIGURE 6.
Chk1-TRAA expressing DT40 cells have defective S-M and G2-M checkpoint responses. A, fluorescence due to phosphorylation of histone H3 (pSer 10H3) and PI staining (DNA content) of DT40 cells cultured in 3. 3 nm nocodazole and 20 μm aphidicolin for 16 h. Region R4 includes cells that have high Ser(P)-10 H3 signals and a 2N content of DNA. B, mitotic indices of DT40 cells incubated in the presence of nocodazole and aphidicolin for 16 h. Mitotic index was calculated from the total number of Ser(P)-10 H3-positive cells in aphidicolin plus nocodazole cultures relative to nocodazole-only control cultures. The graph represents average mitotic index measured in three independent experiments. Error bars represent S.E. C, mitotic indices of irradiated (10 gray γ-IdUrd) DT40 cells incubated in the presence of nocodazole. Mitotic index was calculated from the total number of Ser(P)-10 H3-positive cells in irradiated cultures relative to nocodazole only control cultures. The graph represents the average mitotic index measured in three independent experiments. Error bars represent S.E.
To examine the proficiency of the damage-induced G2-M checkpoint, DT40 cells were incubated in nocodazole with or without prior irradiation, and the effect on cell cycle progression was determined by flow cytometry. Six hours after irradiation with 10 gray, wild type DT40 cells accumulated predominantly with a 4N DNA content. Phospho-H3 staining revealed that few mitotic cells were present (quantified in Fig. 6C). DT40 cells lacking Chk1 failed to arrest in G2, and the number of phospho-H3-positive cells increased regardless prior irradiation (Fig. 6C, DT40Chk1-/-). Loss of the G2-M checkpoint was attributable to loss of Chk1, since DT40Chk1 -/- cells reconstituted with wild type Chk1 (DT40Chk1WT) arrested (Ref. 13 and quantified in Fig. 6C). Similar to Chk1 null cells, cells expressing Chk1TRAA (DT40Chk1TRAA) exhibited no detectable irradiated-induced arrest and continued to enter mitosis, indicating G2-M checkpoint failure in these cells (Fig. 6C). Together these data show that, despite the hyperactivity of Chk1TRAA, inefficient DNA damage-induced phosphorylation and defective chromatin dissociation prevent appropriate checkpoint responses.
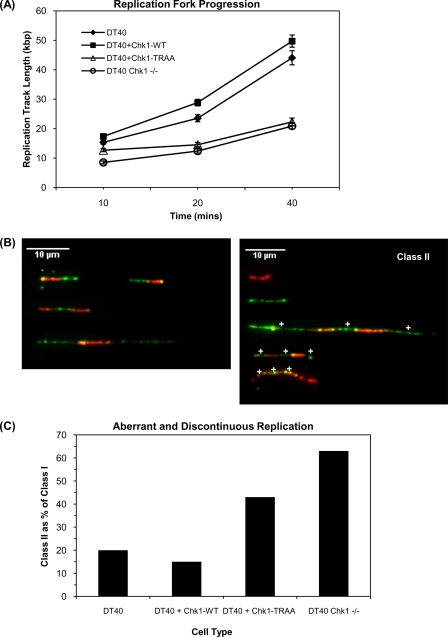
The PIP Box of Chk1 Is Important for Chk1-dependent Replication Functions—Recent studies have shown that Chk1 is required for normal progression of replication forks and regulates origin activation (9, 11). To determine whether the PIP box of Chk1 is also required for Chk1-dependent replication functions, we examined individual replication fibers from DT40, DT40Chk1WT, DT40Chk1TRAA, and DT40Chk1-/- using DNA combing (46). To measure fork rates, active replicons were labeled with BrdUrd for 10, 20, or 40 min, and the mean length of at least 100 labeled tracks was calculated for each time period. Analysis of labeled track lengths revealed that replication forks in Chk1 null DT40 cells progress at ∼50% that of the rate of wild type cells (Fig. 7A). Decreased replication fork progression was also observed in DT40Chk1TRAA cells, with forks progressing ∼65% more slowly than in wild type cells (Fig. 7A).
FIGURE 7.
The PIP box of Chk1 is required for appropriate progression of replication forks. A, the indicated DT40 cells were labeled with 25 μm BrdUrd for 10, 20, or 40 min before preparing DNA fibers. The mean length of at least 100 replication tracks is plotted for each time point. Error bars indicate S.E. B, examples of dual labeled replication tracks. Class I includes all ongoing replication (red-green patterns). Class II represents discontinuous and aberrant replication tracks (red only, green only, or interspersed patterns). White crosses show active origins. C, DT40 cells were labeled with 25 μm BrdUrd for 10 min followed by 250 μm IdUrd for 20 min before preparing DNA fibers. Replication tracks were assigned as either class I (ongoing) or class II (discontinuous and aberrant) as in B. At least 100 replication tracks were counted, and class II replication structures were quantified as a percentage of ongoing tracks.
To establish which parameters of DNA replication were affected by mutation of the PIP box, we used dual labeling. Cells were pulse-labeled with BrdUrd followed by IdUrd, and sites of incorporation on isolated fibers were detected with antibodies that preferentially recognized the different halogenated nucleotides. Replication tracks were classified into two categories; continuous BrdUrd-IdUrd (red-green) patterns were classified as ongoing replication (class I), and discontinuous patterns and closely spaced active origins were categorized in class II (Fig. 7B). Discontinuous patterns represent origins that stalled during the first pulse and remained inactive in the second (green only), newly initiated origins (red only) and closely spaced active origins that present as an interspersed pattern (59). Inappropriate origin activation is associated with an increase in interspersed and discontinuous labeling, most often accompanied by a decrease in ongoing replication (11). To quantify abnormal and discontinuous replication events, class II replication structures were quantified as a percentage of ongoing class I replication. This revealed a striking increase in class II replication structures in DT40Chk1-/- cells compared with parental DT40 cells (Fig. 7C). Aberrant replication events were also increased in DT40Chk1TRAA cells compared with parental, but this increase was not as pronounced as in Chk1 null cells, suggesting that PIP mutant Chk1 retains some Chk1-dependent replication functions. These data show that the PIP box of Chk1 is required for proper progression of replication forks and for preventing aberrantly increased initiation of DNA replication and replication fork stalling.
DISCUSSION
Chk1 is an important checkpoint kinase that is required for the S-M and G2-M checkpoint responses (4, 7). It is also required for appropriate progression of replication forks and for maintenance of stable forks (9, 10, 13). Overall rates of replication, origin stalling, and new origin firing are controlled through formation and activation of the replication complex (for review, see Ref. 60); however, the mechanism by which Chk1 monitors and controls events at individual replication forks is currently unclear (61–63). To gain further insight into the function of Chk1, we sought to identify novel Chk1-interacting partners. We discovered a previously uncharacterized interaction between Chk1 and PCNA. PCNA forms a sliding clamp that encircles DNA and enhances the processivity of DNA polymerase by tethering it to DNA (64, 65). Significantly, PCNA binds multiple proteins involved in DNA replication, Okazaki fragment processing, DNA repair, cell cycle control, and chromatin remodeling (49, 66). In this context PCNA functions as a recruiting or coordinating center (48). Based on this, we hypothesized that PCNA acts as a platform that facilitates and coordinates damage-induced phosphorylation of Chk1 in vivo. Indeed, mutation of the PIP box in Chk1 led to inefficient DNA damage-induced phosphorylation.
Many of the factors that interact with PCNA associate with the same small region of the protein, in particular, PIP box-containing proteins are usually found to associate with the interdomain connecting loop (see the supplementary material in data in Moldovan et al. (49)). The binding of so many different partners to the same region of PCNA is thought to result in a form of steric competition that enables PCNA to function as a mediator of multiple processes. A consequence of the reduced Chk1-PCNA interaction seen in HU-treated cells might, therefore, be an increase in the binding of another PCNA partner (49). Whether Chk1 plays a direct role in regulating PCNA functions remains to be determined.
Several studies have demonstrated that other replisome-associated complexes, such as Cdc7-Dbf4 and Timeless/Tipin, are involved in coordinating replication functions with Chk1 activation (39, 40, 67, 68). These interactions together with the Chk1-PCNA interaction described here highlight the importance of direct communication between the replication machinery and Chk1 (29, 61, 68). An interaction between PCNA and Claspin, an adaptor protein required specifically for Chk1 activation, has previously been reported (69). No PIP box has been identified in Claspin, and it is not known if the interaction is direct. In the context of the full-length protein, mutations in the PIP box of Chk1 reduced damaged-induced phosphorylation and prevented the release of Chk1 from chromatin; however, they did not abolish the interaction between full-length Chk1 and PCNA (supplemental Fig. 2). The remaining Chk1-PCNA interaction may be mediated by a non-canonical PCNA binding motif either within Chk1 or within Claspin. Reduced Chk1-PCNA association is seen in cells treated with HU (Fig. 1). Likewise, the association of Claspin with PCNA is reduced in HU-treated cells (69). Together these data infer that the PCNA-Chk1/Claspin association is reduced in response to replication stress.
Several studies have demonstrated that mutations within, or deletion from the carboxyl terminus of Chk1 lead to increased intrinsic kinase activity and hyperactive checkpoint function (22, 57, 58, 70). These data support a model in which the carboxyl terminus of Chk1 interacts with the kinase domain in an autoinhibitory manner (22). We observed that mutations within the PIP box of Chk1 also led to increased intrinsic kinase activity. Because this mutation is predicted to affect interaction with the amino-terminal autoinhibitory domain, hyperactivation is likely due to this effect rather than PIP box disruption (22). Given that the PIP box mutant, Chk1TRAA, is hyperactive in kinase assays, we expected that it would also lead to hyperactive checkpoint responses in vivo (22, 57, 58). However, DT40 cells expressing Chk1TRAA enter mitosis in the presence of a replication block despite not having completed replication. In addition, they fail to induce the G2-M checkpoint after γ-irradiation. Reduced DNA damage-induced phosphorylation and inefficient chromatin release most likely prevent Chk1TRAA from functioning in vivo. Consistent with this idea, previous studies have shown that chromatin-immobilized Chk1 is not functional (21). The observation that a PIP box mutant, but not other carboxyl-terminal hyperactive Chk1 mutants (e.g. ARFF), result in reduced DNA damage-induced Chk1 phosphorylation strongly suggests that the Chk1 PIP is required for efficient activation of Chk1. Furthermore, these data highlight the different levels of regulation required for proper Chk1 function. It is not sufficient that Chk1TRAA has increased kinase activity; efficient phosphorylation by ATM/ATR is additionally required for both the S-M and G2-M checkpoints.
Chk1TRAA is impaired in several aspects of Chk1 function. In addition to checkpoint defects, elevated levels of fork stalling and increased incidence of aberrant replication events are seen in Chk1TRAA-expressing cells. Thus, although it is clear that the PIP box of Chk1 is critical for Chk1-mediated DNA damage responses and Chk1-dependent replication functions, the mechanism by which PCNA influences these responses remains to be established.
Supplementary Material
Acknowledgments
We thank Elizabeth Black and Max Walker for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA077254 (to C. H. M.) and P41 RR011823 and R01 DK074798 (to J. R. Y.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1 and S2.
Footnotes
The abbreviations used are: Chk1, checkpoint kinase 1; GST, glutathione S-transferase; MS, mass spectroscopy; PCNA, proliferating cell nuclear antigen; HU, hydroxyurea; PIP box, PCNA protein-interacting box; HA, hemagglutination antigen; MudPIT, multidimensional protein identification technology; WT, wild type; BrdUrd, bromodeoxyuridine; IdUrd, iododeoxyuridine; Gg, G. gallus; ATM, ataxia telangiectasia mutated; ATR, ATM- and Rad3-related.
References
- 1.Bartek, J., and Lukas, J. (2007) Curr. Opin. Cell Biol. 19 238-245 [DOI] [PubMed] [Google Scholar]
- 2.Capasso, H., Palermo, C., Wan, S., Rao, H., John, U. P., O'Connell, M. J., and Walworth, N. C. (2002) J. Cell Sci. 115 4555-4564 [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z., Xiao, Z., Chen, J., Ng, S. C., Sowin, T., Sham, H., Rosenberg, S., Fesik, S., and Zhang, H. (2003) Mol. Cancer Ther. 2 543-548 [PubMed] [Google Scholar]
- 4.Liu, Q., Guntuku, S., Cui, X. S., Matsuoka, S., Cortez, D., Tamai, K., Luo, G., Carattini-Rivera, S., DeMayo, F., Bradley, A., Donehower, L. A., and Elledge, S. J. (2000) Genes Dev. 14 1448-1459 [PMC free article] [PubMed] [Google Scholar]
- 5.Sorensen, C. S., Syljuasen, R. G., Falck, J., Schroeder, T., Ronnstrand, L., Khanna, K. K., Zhou, B. B., Bartek, J., and Lukas, J. (2003) Cancer Cell 3 247-258 [DOI] [PubMed] [Google Scholar]
- 6.Xiao, Z., Chen, Z., Gunasekera, A. H., Sowin, T. J., Rosenberg, S. H., Fesik, S., and Zhang, H. (2003) J. Biol. Chem. 278 21767-21773 [DOI] [PubMed] [Google Scholar]
- 7.Takai, H., Tominaga, K., Motoyama, N., Minamishima, Y. A., Nagahama, H., Tsukiyama, T., Ikeda, K., Nakayama, K., and Nakanishi, M. (2000) Genes Dev. 14 1439-1447 [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao, H., Watkins, J. L., and Piwnica-Worms, H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14795-14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petermann, E., Maya-Mendoza, A., Zachos, G., Gillespie, D. A., Jackson, D. A., and Caldecott, K. W. (2006) Mol. Cell. Biol. 26 3319-3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syljuasen, R. G., Sorensen, C. S., Hansen, L. T., Fugger, K., Lundin, C., Johansson, F., Helleday, T., Sehested, M., Lukas, J., and Bartek, J. (2005) Mol. Cell. Biol. 25 3553-3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maya-Mendoza, A., Petermann, E., Gillespie, D. A., Caldecott, K. W., and Jackson, D. A. (2007) EMBO J. 26 2719-2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zachos, G., Rainey, M. D., and Gillespie, D. A. (2003) EMBO J. 22 713-723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zachos, G., Rainey, M. D., and Gillespie, D. A. (2005) Mol. Cell. Biol. 25 563-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feijoo, C., Hall-Jackson, C., Wu, R., Jenkins, D., Leitch, J., Gilbert, D. M., and Smythe, C. (2001) J. Cell Biol. 154 913-923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heffernan, T. P., Simpson, D. A., Frank, A. R., Heinloth, A. N., Paules, R. S., Cordeiro-Stone, M., and Kaufmann, W. K. (2002) Mol. Cell. Biol. 22 8552-8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conti, C., Seiler, J. A., and Pommier, Y. (2007) Cell Cycle 6 2760-2767 [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Girona, A., Tanaka, K., Chen, X. B., Baber, B. A., McGowan, C. H., and Russell, P. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11289-11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niida, H., Katsuno, Y., Banerjee, B., Hande, M. P., and Nakanishi, M. (2007) Mol. Cell. Biol. 27 2572-2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, K., Pereira, E., Maxfield, M., Russell, B., Goudelock, D. M., and Sanchez, Y. (2003) J. Biol. Chem. 278 25207-25217 [DOI] [PubMed] [Google Scholar]
- 20.Ng, C. P., Lee, H. C., Ho, C. W., Arooz, T., Siu, W. Y., Lau, A., and Poon, R. Y. (2004) J. Biol. Chem. 279 8808-8819 [DOI] [PubMed] [Google Scholar]
- 21.Smits, V. A., Reaper, P. M., and Jackson, S. P. (2006) Curr. Biol. 16 150-159 [DOI] [PubMed] [Google Scholar]
- 22.Katsuragi, Y., and Sagata, N. (2004) Mol. Biol. Cell 15 1680-1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loffler, H., Bochtler, T., Fritz, B., Tews, B., Ho, A. D., Lukas, J., Bartek, J., and Kramer, A. (2007) Cell Cycle 6 2541-2548 [DOI] [PubMed] [Google Scholar]
- 24.Clarke, C. A., and Clarke, P. R. (2005) Biochem. J. 388 705-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss, R. S., Matsuoka, S., Elledge, S. J., and Leder, P. (2002) Curr. Biol. 12 73-77 [DOI] [PubMed] [Google Scholar]
- 26.Chini, C. C., and Chen, J. (2003) J. Biol. Chem. 278 30057-30062 [DOI] [PubMed] [Google Scholar]
- 27.Sorensen, C. S., Syljuasen, R. G., Lukas, J., and Bartek, J. (2004) Cell Cycle 3 941-945 [PubMed] [Google Scholar]
- 28.Liu, S., Bekker-Jensen, S., Mailand, N., Lukas, C., Bartek, J., and Lukas, J. (2006) Mol. Cell. Biol. 26 6056-6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, J. E., McAvoy, S. A., Smith, D. I., and Chen, J. (2005) Mol. Cell. Biol. 25 10907-10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, X., Zou, L., Lu, T., Bao, S., Hurov, K. E., Hittelman, W. N., Elledge, S. J., and Li, L. (2006) Mol. Cell 23 331-341 [DOI] [PubMed] [Google Scholar]
- 31.Jeong, S. Y., Kumagai, A., Lee, J., and Dunphy, W. G. (2003) J. Biol. Chem. 278 46782-46788 [DOI] [PubMed] [Google Scholar]
- 32.Kumagai, A., and Dunphy, W. G. (2000) Mol. Cell 6 839-849 [DOI] [PubMed] [Google Scholar]
- 33.Lee, J., Gold, D. A., Shevchenko, A., and Dunphy, W. G. (2005) Mol. Biol. Cell 16 5269-5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chini, C. C., and Chen, J. (2004) DNA Repair (Amst) 3 1033-1037 [DOI] [PubMed] [Google Scholar]
- 35.Roos-Mattjus, P., Vroman, B. T., Burtelow, M. A., Rauen, M., Eapen, A. K., and Karnitz, L. M. (2002) J. Biol. Chem. 277 43809-43812 [DOI] [PubMed] [Google Scholar]
- 36.Bermudez, V. P., Lindsey-Boltz, L. A., Cesare, A. J., Maniwa, Y., Griffith, J. D., Hurwitz, J., and Sancar, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1633-1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delacroix, S., Wagner, J. M., Kobayashi, M., Yamamoto, K., and Karnitz, L. M. (2007) Genes Dev. 21 1472-1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gotter, A. L., Suppa, C., and Emanuel, B. S. (2007) J. Mol. Biol. 366 36-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou, D. M., and Elledge, S. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 18143-18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unsal-Kacmaz, K., Chastain, P. D., Qu, P. P., Minoo, P., Cordeiro-Stone, M., Sancar, A., and Kaufmann, W. K. (2007) Mol. Cell. Biol. 27 3131-3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshizawa-Sugata, N., and Masai, H. (2007) J. Biol. Chem. 282 2729-2740 [DOI] [PubMed] [Google Scholar]
- 42.Washburn, M. P., Wolters, D., and Yates, J. R., III (2001) Nat. Biotechnol. 19 242-247 [DOI] [PubMed] [Google Scholar]
- 43.Eng, J. K., McCormack, A. L., and Yates, J. R., III (1994) J. Am. Soc. Mass Spectrom. 5 976-989 [DOI] [PubMed] [Google Scholar]
- 44.Tabb, D. L., McDonald, W. H., and Yates, J. R., III (2002) J. Proteome Res. 1 21-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, B., Kim, S. T., Lim, D. S., and Kastan, M. B. (2002) Mol. Cell. Biol. 22 1049-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson, D. A., and Pombo, A. (1998) J. Cell Biol. 140 1285-1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koch, H. B., Zhang, R., Verdoodt, B., Bailey, A., Zhang, C. D., Yates, J. R., III, Menssen, A., and Hermeking, H. (2007) Cell Cycle 6 205-217 [DOI] [PubMed] [Google Scholar]
- 48.Link, A. J., Eng, J., Schieltz, D. M., Carmack, E., Mize, G. J., Morris, D. R., Garvik, B. M., and Yates, J. R., III (1999) Nature Biotechnol. 17 676-682 [DOI] [PubMed] [Google Scholar]
- 49.Moldovan, G. L., Pfander, B., and Jentsch, S. (2007) Cell 129 665-679 [DOI] [PubMed] [Google Scholar]
- 50.Lai, J. S., and Herr, W. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 6958-6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warbrick, E. (1998) BioEssays 20 195-199 [DOI] [PubMed] [Google Scholar]
- 52.Waga, S., Hannon, G. J., Beach, D., and Stillman, B. (1994) Nature 369 574-578 [DOI] [PubMed] [Google Scholar]
- 53.Warbrick, E., Lane, D. P., Glover, D. M., and Cox, L. S. (1995) Curr. Biol. 5 275-282 [DOI] [PubMed] [Google Scholar]
- 54.Nakanishi, M., Robetorye, R. S., Pereira-Smith, O. M., and Smith, J. R. (1995) J. Biol. Chem. 270 17060-17063 [DOI] [PubMed] [Google Scholar]
- 55.Haracska, L., Johnson, R. E., Unk, I., Phillips, B., Hurwitz, J., Prakash, L., and Prakash, S. (2001) Mol. Cell. Biol. 21 7199-7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gulbis, J. M., Kelman, Z., Hurwitz, J., O'Donnell, M., and Kuriyan, J. (1996) Cell 87 297-306 [DOI] [PubMed] [Google Scholar]
- 57.Wang, S. X., and Dunphy, W. G. (2000) FEBS Lett. 487 277-281 [DOI] [PubMed] [Google Scholar]
- 58.Oe, T., Nakajo, N., Katsuragi, Y., Okazaki, K., and Sagata, N. (2001) Dev. Biol. 229 250-261 [DOI] [PubMed] [Google Scholar]
- 59.Merrick, C. J., Jackson, D., and Diffley, J. F. (2004) J. Biol. Chem. 279 20067-20075 [DOI] [PubMed] [Google Scholar]
- 60.DePamphilis, M. L., Blow, J. J., Ghosh, S., Saha, T., Noguchi, K., and Vassilev, A. (2006) Curr. Opin. Cell Biol. 18 231-239 [DOI] [PubMed] [Google Scholar]
- 61.Liu, P., Barkley, L. R., Day, T., Bi, X., Slater, D. M., Alexandrow, M. G., Nasheuer, H. P., and Vaziri, C. (2006) J. Biol. Chem. 281 30631-30644 [DOI] [PubMed] [Google Scholar]
- 62.Paulsen, R. D., and Cimprich, K. A. (2007) DNA Repair (Amst) 6 953-966 [DOI] [PubMed] [Google Scholar]
- 63.Petermann, E., and Caldecott, K. W. (2006) Cell Cycle 5 2203-2209 [DOI] [PubMed] [Google Scholar]
- 64.Burgers, P. M. (1988) Nucleic Acids Res. 16 6297-6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krishna, T. S., Kong, X. P., Gary, S., Burgers, P. M., and Kuriyan, J. (1994) Cell 79 1233-1243 [DOI] [PubMed] [Google Scholar]
- 66.Warbrick, E. (2000) BioEssays 22 997-1006 [DOI] [PubMed] [Google Scholar]
- 67.Errico, A., Costanzo, V., and Hunt, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14929-14934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heffernan, T. P., Unsal-Kacmaz, K., Heinloth, A. N., Simpson, D. A., Paules, R. S., Sancar, A., Cordeiro-Stone, M., and Kaufmann, W. K. (2007) J. Biol. Chem. 282 9458-9468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brondello, J. M., Ducommun, B., Fernandez, A., and Lamb, N. J. (2007) Biochem. Biophys. Res. Commun. 354 1028-1033 [DOI] [PubMed] [Google Scholar]
- 70.Chen, P., Luo, C., Deng, Y., Ryan, K., Register, J., Margosiak, S., Tempczyk-Russell, A., Nguyen, B., Myers, P., Lundgren, K., Kan, C. C., and O'Connor, P. M. (2000) Cell 100 681-692 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.