Abstract
The 140-nucleotide trp leader RNA, which is formed by transcription termination under conditions of high intracellular tryptophan, was used to study RNA turnover in Bacillus subtilis. We showed in vivo that the amount of endonuclease cleavage at ∼nucleotide 100 is decreased under conditions where RNase J1 concentration is reduced. In addition, under these conditions the level of 3′-terminal RNA fragments, which contain the strong transcription terminator structure, increases dramatically. These results implicated RNase J1 in the initiation of trp leader RNA decay as well as in the subsequent steps leading to complete turnover of the terminator fragment. To confirm a direct role for RNase J1, experiments were performed in vitro with various forms of trp leader RNA and 3′-terminal RNA fragments. Specific endonuclease cleavages, which were restricted to single-stranded regions not bound by protein, were observed. Degradation of the 3′-terminal fragment by the 5′ to 3′-exonuclease activity of RNase J1 was also demonstrated, although the presence of strong secondary structure impeded RNase J1 processivity to some extent. These results are consistent with a model for mRNA decay in Bacillus subtilis whereby the downstream products of RNase J1 endonucleolytic cleavage become substrates for the 5′ to 3′-exoribonuclease activity of the enzyme.
The number of documented ribonuclease activities in Bacillus subtilis has grown rapidly in the last few years, with at least 15 enzymes known (1). Of those that have been identified recently, perhaps the most relevant ribonuclease for RNA turnover is RNase J1, encoded by the rnjA (formerly ykqC) gene. RNase J1 was originally identified as an endoribonuclease that shares characteristics with Escherichia coli RNase E (2), suggesting that RNase J1 may be the primary nuclease activity responsible for regulating mRNA decay in B. subtilis as RNase E is thought to be in E. coli (3, 4). Indeed, Putzer and co-workers (2) reported that global mRNA half-life was increased slightly in an RNase J1 mutant that was grown under conditions where RNase J1 was depleted. More recently, the surprising finding was reported that RNase J1 also has 5′ to 3′-exoribonuclease activity (5), an activity not previously known to exist in bacteria. The presence of a 5′ to 3′-exoribonuclease activity in B. subtilis provides a rationale for the often observed 5′-end dependence of mRNA decay in B. subtilis (6). RNase J1 has also been shown to be important for processing of two stable RNAs, scRNA (7) and 16S rRNA (8). Another endoribonuclease, RNase J2, encoded by the rnjB (formerly ymfA) gene, shares characteristics with RNase J1 (2); however, whereas RNase J1 is essential, RNase J2 is not.
RNase J1 activity is sensitive to the phosphorylation state of the 5′-end, with severalfold greater activity observed with monophosphorylated or hydroxylated 5′-ends than with triphosphorylated 5′-ends (2, 5, 9). E. coli RNase E also shows a strong preference for monophosphorylated versus triphosphorylated 5′-ends, but, unlike RNase J1, it is poorly active on a hydroxylated 5′-end (10, 11). It has been suggested that the 5′-end-dependent action of RNase E on E. coli RNAs is preceded by a pyrophosphatase activity that converts the 5′-triphosphate end of the transcription product to a 5′-monophosphate end (12). The activity in E. coli responsible for this conversion has been identified recently as the pyrophosphohydrolase RppH (formerly NudH) (13). No such enzyme has yet been identified in B. subtilis.
To study RNA processing and turnover in B. subtilis, we have been using the 140-nucleotide (nt)2 trp leader RNA as a model. The B. subtilis trp operon is regulated at the level of transcription termination/antitermination (14, 15), which is controlled by binding of the trp RNA-binding attenuation protein (TRAP, encoded by the mtrB gene) to trp leader RNA. In the absence of TRAP binding, when intracellular tryptophan concentration is low, an antiterminator structure forms (Fig. 1A) that precludes formation of the terminator structure and transcription proceeds into the trp structural genes, which encode tryptophan biosynthetic enzymes. When the intracellular supply of tryptophan is sufficient, the 11-mer TRAP protein complex is activated by binding of tryptophan, allowing the complex to bind to 11 trinucleotide repeats (GAG or UAG) located between nt 36–91 of the trp leader sequence (Fig. 1B). TRAP binding results in formation of a stem-loop structure (nt 108–133) that induces transcription termination, generating the 140-nt trp leader RNA. The small size of this RNA makes it a useful tool for studying in detail the activity of ribonucleases in vivo and in vitro.
FIGURE 1.
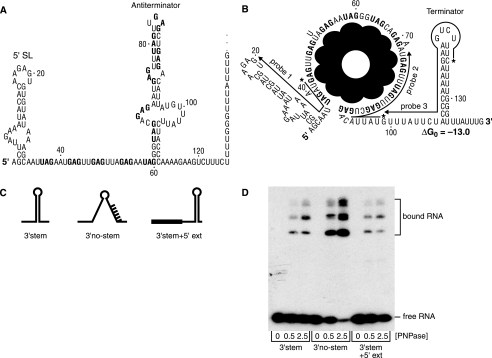
Sequence and secondary structure of trp leader RNA without (A) and with (B) bound TRAP. Numbering is from the natural start site of trp transcription. GAG and UAG triplet repeats at which TRAP binds are in bold. 5′-SL denotes the 5′-stem-loop structure. In B, lines indicate complementarity of probes 1, 2, and 3 to trp leader RNA sequences, with the 5′-labeled end of the probe indicated by an asterisk. The ΔG0 (kcal mol–1) for the trp leader RNA 3′-end structure, calculated according to the default conditions of the mfold version 3.2 site, is shown. C, schematic diagrams of RNA oligonucleotides used to probe RNase J1 activity on the 3′-terminal fragment. D, binding of purified PNPase to RNA oligonucleotides. Concentration (nm) of PNPase is indicated below each lane.
We have recently shown that initiation of trp leader RNA decay is by an endonuclease cleavage at ∼nt 100 of trp leader RNA (16). Turnover of the 5′-cleavage fragment, which is essential for release and ongoing activity of TRAP, is mediated by polynucleotide phosphorylase (PNPase), a phosphorolytic 3′ to 5′-exoribonuclease (17). Here we show that the endonuclease cleavage at nt 100 is RNase J1-dependent in vivo and that cleavage at this site, as well as 5′ to 3′-exonucleolytic degradation of the resulting 3′-terminal fragment, is catalyzed directly by RNase J1 in vitro.
EXPERIMENTAL PROCEDURES
Bacterial Strains—The “wild-type” B. subtilis strain for in vivo experiments was BG581, which is trpC2 thr-5 and carries plasmid pGD5, a high copy number plasmid that contains the trp leader region and the mtrB gene (16). For experiments with RNase J1 and J2 mutants, BG581 was transformed with pMAP65, which carries the lacI gene (18), to give BG598. BG598 was transformed with chromosomal DNA isolated from the RNase J1 conditional mutant and the RNase J2 deletion mutant described previously (8) to give BG600 (RNase J1 mutant) and BG601 (RNase J2 mutant).
Purification of RNase J1 and H76A Mutant Protein—Cloning and overexpression of the rnjA gene, as well as the H76A mutant derivative, were as described (8).
Preparation of RNA Substrates—trp leader RNA was synthesized using T7 RNA polymerase transcription (Ambion T7 MAXIscript kit) of a PCR fragment that contained the trp leader sequence with a T7 promoter included in the upstream primer. The transcription product was purified from a 6% denaturing polyacrylamide gel as described (19), using a diffusion buffer that contained 1 m ammonium acetate, 2 mm magnesium acetate, 1 mm EDTA, and 0.2% SDS. Quantitation of unlabeled trp leader RNA was done spectrophotometrically. For 5′-triphosphate end labeling of trp leader RNA, transcription was done in the presence of [γ-32P]GTP. For 5′-monophosphate end labeling of trp leader RNA, cold trp leader RNA was treated with calf intestine alkaline phosphatase (New England Biolabs) and labeled with polynucleotide kinase using [γ-32P]ATP. For 3′-end labeling, cytidine 3′,5′-bis(phosphate),[5′-32P] (referred to as “pCp”) (New England Nuclear) was ligated to the 3′-end of trp leader RNA using T4 RNA ligase (New England Biolabs) for 6 h at 18 °C. Labeled RNA was purified from a 6% denaturing polyacrylamide gel as above. RNA oligonucleotides, bearing a 5′-hydroxyl end, were obtained from Integrated DNA Technologies and were labeled at the 5′-end (monophosphate) with polynucleotide kinase and at the 3′-end by T4 RNA ligase, as above for full-length trp leader RNA, and were purified from 9% denaturing polyacrylamide gels. For the RNA oligonucleotide with the labeled triphosphate end, RNA was synthesized by T7 RNA polymerase transcription in the presence of [γ-32P]GTP and purified from a 9% polyacrylamide gel.
Assay of RNase J1 Activity—Reactions were done using 10 nm RNA substrate, of which 10% was labeled, and 1.4 μm RNase J1. The buffer contained 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.1 mm dithiothreitol, 5 mm MgCl2. For analysis of full-length trp leader RNA, 2.5 mm tryptophan was present, with or without 37.5 nm TRAP. For experiments where TRAP was present, RNA was incubated with TRAP for 15 min at 22 °C before addition of RNase J1. Initially, incubation with RNase J1 was done at two temperatures, 22 and 37 °C. However, no difference was observed in the results, so subsequent experiments were performed at 22 °C. The reactions were stopped by addition of EDTA to 10 mm and an equal volume of gel loading buffer (Ambion) and were put immediately on dry ice. Samples were loaded on either an 8 or a 22.5% denaturing polyacrylamide gel. For thin layer chromatography, reactions were stopped by addition of 10 mm EDTA and aliquots were applied to a polyethyleneimine-cellulose sheet (Merck) and developed in 1 m LiCl. Quantitation of decay products was done with a Storm 860 PhosphorImager (GE Healthcare) or a Typhoon TRIO variable mode imager (GE Healthcare).
PNPase Binding Assay—Binding of PNPase to the 3′-end of RNA oligonucleotides was performed as described (16).
Northern Blot Analysis—RNA isolation from B. subtilis strains grown in 2× YT (1% yeast extract, 2% tryptone, 1% NaCl) and Northern blot analysis were performed as described (17). To control for RNA loading in Northern blot analyses, membranes were stripped and probed for 5S rRNA as described (20). High resolution Northern blot analysis was performed using a 7% denaturing polyacrylamide gel and electroblotting at 20 volts overnight, followed by an additional 60 min at 30 volts. 5′-end-labeled oligonucleotides were used as probes, except for the ΔermC riboprobe (20) used for the Northern blot in Fig. 7.
FIGURE 7.
Northern blot analysis of accumulation of 3′-terminal fragments under conditions of reduced RNase J1 for rpsO mRNA (A) and ΔermC mRNA (B). Migration of fragments that accumulate are indicated by brackets on the right of the gel. A diagram of the predicted secondary structure at the 3′-end of these mRNAs is shown, along with the calculated ΔG0 (kcal mol–1). WT, wild type.
RESULTS
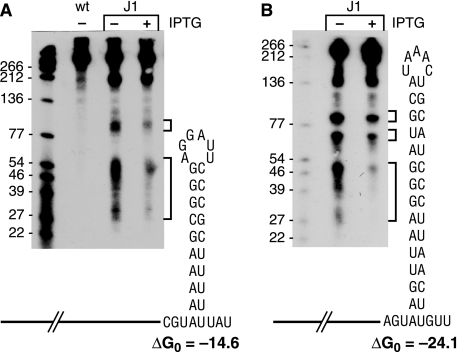
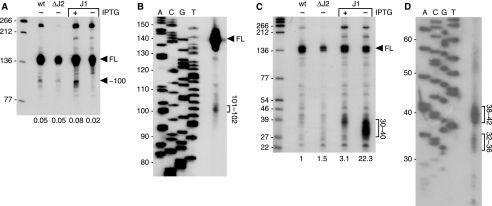
We had shown previously that endonuclease cleavage around nt 100 of trp leader RNA was responsible for initiation of decay (16). To study whether RNase J1 was involved in trp leader RNA turnover, strains were constructed that carried the high copy number plasmid pGD5 (16) containing the trp leader region and a copy of the mtrB gene and that conditionally expressed RNase J1 under control of the IPTG-inducible pspac promoter or had a deletion of the gene encoding RNase J2. Expression of the chromosomal copy of the trp leader is low, and the presence of pGD5 results in a 30-fold increase in trp leader RNA levels, allowing easier detection of processing products. To ensure low level expression of RNase J1 in the absence of IPTG, the strains also contained pMAP65, a plasmid that carries the lacI gene, providing extra copies of the lac repressor (18). Northern blot analysis of trp leader RNA was performed using a probe that was complementary to sequences upstream of the endonuclease cleavage site (probe 1 in Fig. 1B). The results (Fig. 2A) showed that there was a 3-fold (average of five experiments) decrease in the ∼100-nt fragment when the RNase J1 conditional strain was grown in the absence of IPTG compared with the presence of IPTG. Additional faint bands were detected above the 100-nt band. These are likely minor alternative endonuclease cleavage products or the result of exonuclease decay of full-length RNA. Deletion of the RNase J2-encoding gene had no effect on trp leader RNA processing (Fig. 2A) (For unknown reasons, we consistently observed a lower quantity of trp leader RNA in the RNase J2 deletion strain. However, the amount of ∼100-nt fragment relative to full-length trp leader RNA was virtually the same in the RNase J2 deletion strain and the wild-type strain.) A high resolution Northern blot analysis using probe 2 showed that the major endonuclease cleavages occurred at about nt 100–102 (Fig. 2B). Northern blot analysis was also done using a probe that was complementary to the stem-loop terminator sequence (probe 3 in Fig. 1B). In this case, the results showed a large increase (∼20-fold) in the amount of small RNA fragments ranging in size from ∼30–40 nt (Fig. 2C; the stem-loop structure itself is 26 nt). Probing of a high resolution Northern blot with probe 3 showed two clusters of fragments from about 32–36 and 38–42 nt (Fig. 2D). The increased level of 3′-terminal fragments observed in Fig. 2C is consistent with these fragments being generated by endonuclease cleavages at ∼nt 100 and resisting further decay in conditions of reduced RNase J1.
FIGURE 2.
Northern blot analysis of trp leader RNA in RNase J1 and J2 mutants. A, Northern blot probed with probe 1. Migration of full-length (FL) trp leader RNA and a major RNase J1 cleavage product of ∼100 nt are indicated on the right. Below the gel are the ratios of the intensity of the ∼100-nt RNA relative to FL RNA. RNAs that are larger than trp leader RNA (140 nt) are read-through transcripts, as demonstrated previously (16). Leftmost lanes in parts A and C contain 5′-end-labeled TaqI fragments of plasmid pSE420 DNA (29), and the sizes of these fragments are indicated on the left of the gel. B, high resolution Northern blot analysis of RNA isolated from the wild-type strain, probed with probe 2. Migration of prominent RNA fragments and their sizes (nt) are indicated at the right. Sequencing ladder on the left was generated on M13mp18 single-stranded DNA. C, Northern blot probed with probe 3. The cluster of 3′-terminal fragments is indicated on the right as running between 30–40 nt. Below the gel are the amounts, relative to the amount in wild type, of these fragments. D, high resolution Northern blot analysis of RNA isolated from the wild-type strain, probed with probe 3. Migration of prominent RNA fragments and their sizes (nt) are indicated at the right. WT, wild type.
These results suggested that the endonuclease activity of RNase J1 played a role in the initiation of trp leader RNA decay and that the exonuclease and/or endonuclease activity of RNase J1 was important in the subsequent degradation of the 3′-terminal fragment. We therefore sought to characterize RNase J1 activity on trp leader RNA in vitro to determine whether the observed in vivo effects could be attributed directly to RNase J1.
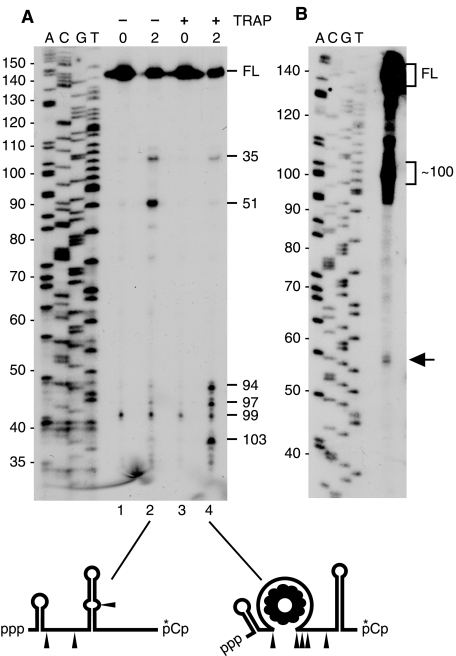
trp leader RNA transcripts were prepared to contain a labeled triphosphate 5′-end, which was incubated with RNase J1 in the presence or absence of purified TRAP protein. (Note that to ensure efficient transcription by T7 RNA polymerase the in vitro transcripts contained three extra Gs at the 5′-end. Thus, 5′-end-containing fragments observed in vitro are 3 nucleotides longer than the corresponding fragments in vivo. Numbering of the fragment sizes in Fig. 3 is according to the numbering of the natural trp leader RNA as shown in Fig. 1.) As a control, labeled RNAs were also incubated with the RNase J1 H76A mutant protein, which has severely reduced nuclease activity (8). The results in Fig. 3, A and B, showed that trp leader RNA was indeed cleaved endonucleolytically by RNase J1 and that this cleavage occurred differently depending on the presence or absence of TRAP. In the absence of TRAP, RNase J1 cleaved broadly in the single-stranded region between the 5′-stem-loop structure and the antiterminator structure (nt 33–59; see Fig. 1A). The sizes of the ladder of fragments seen in the high resolution (“sequencing”) gel in Fig. 3B, bottom lane 2, corresponded well with multiple endonuclease cleavages in this single-stranded region. In the presence of TRAP, however, cleavage was observed at ∼nt 100 as well as in a smaller number of sites in the region between the 5′-stem-loop and the beginning of the TRAP binding site (Fig. 3A and 3B, lane 4). Virtually no cleavage activity was observed with the H76A mutant protein. The rate of cleavage appeared to be slower in the presence of TRAP, and this is likely due to binding of TRAP to sites that cover 40% of the RNA sequence, thus reducing the available number of cleavage sites. Also, note that in addition to the products of RNase J1 activity that are visible on the gel in Fig. 3A, other small endonuclease fragments, as well as a low level of single nucleotide (as determined by thin layer chromatography), are produced (data not shown).
FIGURE 3.
Analysis of RNase J1 endonuclease activity on trp leader RNA bearing a triphosphate 5′-end (A) or a monophosphate 5′-end (C). Times of incubation (min) are indicated on top of each lane. Lanes M, substrate incubated with H76A mutant RNase J1 protein. The general locations of cleavage sites are indicated by arrowheads on the trp leader RNA schematics below each gel. ppp, triphosphate; p, monophosphate; *, labeled phosphate. B, sequencing gel to determine precise sizes of endonuclease cleavage fragments generated from trp leader RNA bearing a 5′-triphosphate end. Range of sizes of RNA fragments is indicated at the right. D, quantitation of accumulation of endonuclease fragments generated for 5′-triphosphorylated substrate (filled squares) and 5′-monophosphorylated substrate (open triangles) in the presence or absence of TRAP. Data are the average of three experiments ± S.D. FL, full length.
When trp leader RNA was synthesized to contain a 5′-monophosphate end (Fig. 3C), the relative amount of endonuclease cleavage was reduced by about half (Fig. 3D). Also, in the absence of TRAP, cleavage at a distal site (at about 54 nt) was preferred, unlike the broad range of cleavages in this region seen with the triphosphorylated substrate (Fig. 3A and see “Discussion”).
trp leader RNA transcripts (i.e. bearing a 5′-triphosphate) were also labeled at the 3′-end by addition of [32P]pCp (see “Experimental Procedures”) in order to observe the generation of 3′-end-containing fragments. In this case, the 5′-exonuclease activity of RNase J1 made it difficult to assess a rate or exact site of initial cleavage, because downstream products of endonuclease cleavage were subsequently degraded from their 5′-ends. Nevertheless, the approximate location of endonuclease cleavages could be determined by short incubation with RNase J1 and resolution of the products on a sequencing gel (Fig. 4A, lanes 2 and 4). (Note that the numbering in Fig. 4A takes into account the extra nucleotide added by addition of pCp and is according to the numbering of the natural trp leader RNA shown in Fig. 1.) The strongest cleavages in the absence of TRAP were at nt 35, which is a few nucleotides after the 5′-stem-loop, and at nt 51, which is several nucleotides upstream of the antiterminator stem. A cleavage at nt 99 was also observed, which is in a bulged region on the downstream side of the antiterminator stem. Additional cleavages may have occurred closer to the 3′-end, but these were not detected on this gel. In the presence of TRAP, there was a cleavage at nt 35 and a weak cleavage at nt 51, but the stronger cleavages were in the single-stranded region immediately downstream of the TRAP binding site. The results with the 3′-labeled substrate were consistent with the cleavages observed with the 5′-labeled substrate (Fig. 3). The ladder seen between nt 35 and 54 with the 5′-labeled RNA in the absence of TRAP (Fig. 3B) is replaced in the case of the 3′-labeled RNA with more discrete species at the boundaries of this zone (nt 35 to 51). This suggests that the species generated by endonucleolytic cleavage are rapidly nibbled down to nt 51 by the 5′ to 3′-exonuclease activity of RNase J1.
FIGURE 4.
A, Sequencing gel of endonuclease products generated from 3′-end-labeled trp leader RNA incubated with RNase J1 for 0 or 2 min, with and without TRAP. At right are the sites of endonuclease cleavage in accordance with the numbering in Fig. 1. The general location of these cleavage sites is indicated by arrowheads in the schematics below the gel. ppp, triphosphate 5′-end. The small fragment representing cleavage at nt 99 is seen here even at time zero. This is the product of rapid RNase J1 cleavage, as it is not seen when the 3′-end-labeled RNA is incubated in the absence of RNase J1 (data not shown). B, high resolution Northern blot analysis of in vivo RNA (15 μg) for detection of the trp leader RNA fragment that is predicted to result from RNase J1 endonuclease cleavage upstream and downstream of the TRAP binding site. Arrow at right indicates the detected band that is the predicted size (57–58 nt) for this RNA fragment. FL, full length.
If the in vitro results obtained with TRAP-bound RNA reflected events occurring in vivo, we predicted that, in a strain grown in the presence of tryptophan, RNase J1 would cleave in the single-stranded regions of trp leader RNA on either side of the TRAP binding site (nt 36–91), generating an RNA fragment about the size of this region (56 nt). We probed for such a fragment using probe 2 (Fig. 1B). As can be seen in the high resolution Northern blot in Fig. 4B, a doublet of ∼56–57 nt was detected by this probe. This fragment was not detected by the upstream probe 1 (data not shown) and so is likely to consist of the TRAP binding region. Thus, the observed RNase J1 cleavages in vitro appear to take place in vivo, suggesting that this enzyme is directly responsible for decay-initiating RNA turnover.
The observed effect of reduced RNase J1 levels on 3′-terminal fragment turnover (Fig. 2C) prompted an in vitro investigation of RNase J1 activity on this RNA. An RNA oligonucleotide, designated “3′-stem,” was used that contained the sequence of the 3′-end of trp leader RNA (nt 99–140; Fig. 1C). As controls, two other RNAs were used, “3′-no-stem,” which was similar to 3′-stem RNA except that six of the complementary nucleotides in the downstream side of the stem were changed such that the strong secondary structure could not form, and “3′-stem+5′-ext,” which had the same sequence as 3′-stem RNA plus a 24-nt extension added at the 5′-end (Fig. 1C). (For technical reasons, 3′-stem+5′-ext RNA had only two nucleotides downstream of the stem structure rather than the six nucleotides in 3′-stem RNA. We assumed this difference was irrelevant under in vitro conditions where no 3′ to 5′-exonuclease was present.) As a test of whether the predicted structure was forming at the 3′-end of these RNA oligonucleotides, PNPase binding experiments were done. We have shown previously that PNPase binds well to substrates with 3′-single-stranded tails but binds poorly to substrates with 3′-terminal secondary structure (16). From the binding assay shown in Fig. 1D, it was clear that the level of PNPase binding to 3′-stem RNA and 3′-stem+5′-ext RNA was much lower than the level of binding to 3′-no-stem RNA. Quantitation of the results with 2.5 nm PNPase showed that 80% of 3′-no-stem RNA was bound as opposed to only 15% of 3′-stem and 3′-stem+5′-ext RNA. These percentages are consistent with our previous data on binding of PNPase to trp leader RNA without TRAP present (single-stranded RNA at 3′-end) and with TRAP present (secondary structure at 3′-end) (16). Thus, although it is likely that the RNA oligonucleotide substrates assume alternative conformations to some extent, the PNPase binding data suggest that the RNA oligonucleotides are mostly in the predicted conformations shown in Fig. 1C.
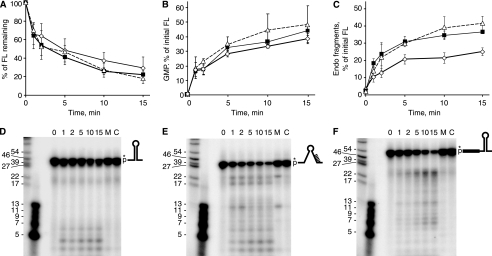
These RNAs were labeled at their 5′-ends by addition of 32P-monophosphate and were incubated in the presence of RNase J1. Aliquots were removed at various times after addition of RNase J1 and were run on (a) an 8% denaturing polyacrylamide gel, to measure loss of label at the 5′-end; (b) a polyethyleneimine-cellulose sheet (thin layer chromatography), to measure release of labeled GMP by the 5′ to 3′-exonuclease activity; and (c) a 22.5% denaturing polyacrylamide gel, to measure accumulation of small fragments arising from endonucleolytic cleavage. Experiments were performed in quadruplicate. In Fig. 5, A–C, are shown graphs of the data from these assays on the three RNA substrates. Primary data for the assays of endonuclease activity on 22.5% denaturing polyacrylamide gels are shown in Fig. 5, D–F.
FIGURE 5.
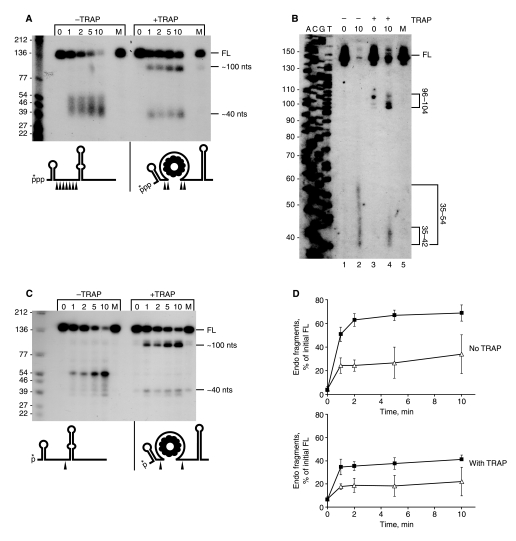
Analysis of RNase J1 activity on RNA oligonucleotides representing 3′-terminal RNA fragments. A–C, data from an 8% denaturing polyacrylamide gel (A), from thin layer chromatography (B), and from a 22.5% denaturing polyacrylamide gel (C) were for 3′-stem RNA (open diamonds), 3′-no-stem RNA (filled squares), and 3′-stem+5′-ext RNA (open triangles). D–F, primary data from 22.5% denaturing polyacrylamide gels for the three substrates, as indicated by the schematic diagrams. Control lanes C, RNA substrate incubated without RNase J1. Lanes M, RNA substrate incubated with mutant RNase J1. Very small size markers (second lane from left) were 5′-end-labeled DNA oligonucleotides of the indicated sizes. Data are the average of four experiments ± S.D. FL, full length.
Disappearance of full-length RNA occurred at similar rates for the three substrates (Fig. 5A), and these were inversely proportional to accumulation of labeled GMP (Fig. 5B). Thus, all three substrates were equally susceptible to the 5′ to 3′-exonuclease activity of RNase J1. However, the accumulation of small 5′-end-containing fragments, representing endonuclease activity, differed significantly among the three substrates. Quantitatively, there was a lower level of endonuclease fragments from the 3′-stem RNA than from the 3′-no-stem and the 3′-stem+5′-ext RNAs (Fig. 5C). Qualitatively, the 5′-end-containing fragments were of different sizes for the three substrates: for 3′-stem RNA (Fig. 5D), small fragments about 3–7 nt accumulated, with little accumulation of larger fragments; for 3′-no-stem RNA (Fig. 5E), fragments in a range from 3 to 20 nt were observed; for 3′-stem+5′-ext RNA, the pattern was shifted to mostly larger fragments (Fig. 5F). These observations could be explained in light of the structures of the three substrates. 3′-stem RNA has a 9-nt single-stranded region that precedes the stem structure. The data suggested that RNase J1 makes endonucleolytic cleavages in this single-stranded sequence but does not attack the stem structure. The small single-stranded target size and/or the proximity of the stem structure to the 5′-end in this substrate RNA reduces the rate of RNase J1 endonucleolytic cleavage relative to the other substrates (Fig. 5C). In the case of 3′-nostem RNA, RNase J1 can cleave at a wider range of phosphodiester bonds, as there is no inhibitory secondary structure present. For 3′-stem+5′-ext RNA, the single-stranded region preceding the stem structure is 33 nt long, and cleavage distal to the 5′-end is preferred.
To analyze the effect of secondary structure on 5′ to 3′-processivity, the RNA oligonucleotide substrates were labeled at their 3′-ends by addition of [32P]pCp and were incubated in the presence of RNase J1. Note that the 3′-end-labeled substrates in these experiments had a 5′-hydroxyl end rather than the 5′-monophosphate end in previous experiments. Based on the published results describing 5′ to 3′-exonuclease activity of RNase J1 (5), we expected that the activity of RNase J1 on 5′-hydroxylated RNA would be very similar to its activity on 5′-monophosphorylated RNA.
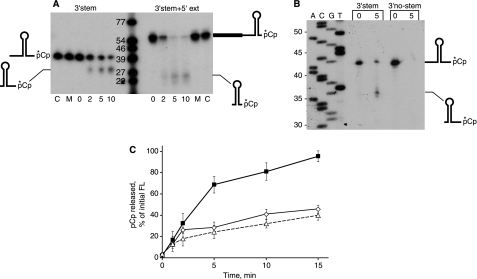
The 3′-stem substrate showed an accumulation with time of a product that was several nucleotides shorter than the initial substrate (Fig. 6A). This product constituted 21.5% (average of four experiments) of the full-length RNA at time zero. From a sequencing gel, the size of the accumulated fragment was found to be 35–37 nt, with most of the product at 36 nt (Fig. 6B). Taking into account the added C residue at the 3′-end, this corresponded to a fragment whose 5′-end was 3 nt upstream of the base of the stem structure, thus suggesting that RNase J1 exonucleolytic processivity is hindered when it reaches within a few nucleotides of strong secondary structure. The product of the block to RNase J1 processivity was not observed with the 3′-no-stem substrate (Fig. 6B). We considered the possibility that RNase J1 processivity was negatively affected by the short single-stranded “ramp” (only 9 nt) of 3′-stem RNA and that perhaps providing a longer single-stranded region would allow more progress through the stem. To test this, the 3′-stem+5′-ext substrate was used. As shown in Fig. 6A, there was a hindrance to processivity with the 3′-stem+5′-ext RNA as well, with the upstream end of the substrate occurring the same distance from the stem-loop structure as in 3′-stem RNA. This was confirmed by separation of the products of digestion on a sequencing gel (not shown) that demonstrated that the sizes of the 3′-stem+5′-ext products were 4–5 nt shorter than the 3′-stem products, which was the expected size because the 3′-stem+5′-ext RNA has 4 less nt at the 3′-end, as mentioned above. A quantitative assessment of the release of labeled pCp is shown in Fig. 6C. Complete degradation of 3′-no-stem RNA approached 100%, whereas degradation of the RNAs with strong secondary structure leveled out at ∼40%. Thus, RNase J1 is able to digest through structured RNA in the 5′ to 3′-direction, although it is sensitive to the presence of a relatively strong structure (–13 kcal mol–1).
FIGURE 6.
A, degradation of 3′-end-labeled RNA oligonucleotides in the presence of RNase J1 for the indicated times (in min). Schematics of RNA substrates and products are on either side of the gel. Lanes C, RNA substrate incubated without RNase J1. Lanes M, RNA substrate incubated with mutant RNase J1. B, sizing of products on an 8% denaturing polyacrylamide gel. Schematics of RNA substrate and processed product are at the right of the gel. C, quantitation of pCp released from 3′-stem RNA (open diamonds), 3′-no-stem RNA (filled squares), and 3′-stem+5′-ext RNA (open triangles) after incubation with RNase J1 up to 15 min. Data are the average of three experiments ± S.D. FL, full length.
To determine whether the accumulation of 3′-terminal fragments in conditions of RNase J1 depletion (Fig. 2C) was unique to trp leader RNA, we probed for 3′-terminal fragments from two other mRNAs that we have studied previously: rpsO, a 388-nt monocistronic RNA encoding ribosomal protein S15, and ΔermC, a 254-nt RNA encoded by a deletion derivative of ermC (21). Fig. 7 shows the Northern blot analysis of steady-state rpsO mRNA (A) and ΔermC mRNA (B) in the presence and absence of IPTG-induced RNase J1 expression. 3′-Terminal fragments between 25–80 nt in length also accumulated for these two mRNAs in conditions of decreased RNase J1 levels.
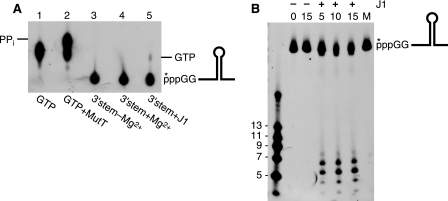
It has been observed previously that the 5′ to 3′-exonuclease activity of RNase J1 is sensitive to the phosphorylation state of the 5′-end (5). To determine whether degradation of the 3′-terminal fragment in the manner described above was dependent on prior endonucleolytic cleavage by RNase J1 to generate a 5′-monophosphate end, we examined the RNase J1 degradation pattern on 3′-stem RNA containing a 5′-triphosphate group. (Note that the 3′-stem RNA bearing the 5′-triphosphate includes two additional G residues at the 5′-end to allow efficient transcription by T7 RNA polymerase.) Lane 5 of the thin layer chromatogram in Fig. 8A shows that RNase J1 had little 5′-exonuclease activity on the 3′-stem substrate containing a triphosphate 5′-end; only ∼5% of labeled GTP was released. This was ∼8-fold less than the activity observed for 5′-monophosphorylated 3′-stem RNA (Fig. 5B). The chromatogram was used to determine whether RNase J1 might also have pyrophosphatase activity, similar to that of E. coli RppH (13). As a control, [γ-32P]GTP was treated with purified E. coli MutT protein (22) to generate labeled pyrophosphate (Fig. 8A, lane 2). It was clear from the results in Fig. 8A that treatment with RNase J1 did not result in pyrophosphate release. On the other hand, RNase J1 did cleave the 5′-triphosphorylated 3′-stem RNA endonucleolytically (Fig. 8B), showing cleavage products that were the same size as those observed with the monophosphorylated 3′-stem RNA (Fig. 5D). In addition, the bands in Fig. 8B were sharper, likely because there is little 5′-exonuclease activity on the small fragments. In the case of the monophosphorylated 3′-stem RNA (Fig. 5D), endonucleolytic cleavage could be followed by 5′ to 3′-exonuclease degradation. The results with the 5′-triphosphorylated 3′-stem RNA suggest that a primary transcript bearing a 5′-triphosphate would be resistant to 5′ to 3′-exonuclease activity but subject to downstream endonuclease cleavage.
FIGURE 8.
Analysis of RNase J1 activity on 3′-stem RNA carrying a 5′-triphosphate end. A, thin layer chromatography using labeled GTP and labeled GTP treated with MutT pyrophosphatase as controls. Migration of pyrophosphate is indicated at the left and migration of GTP is indicated at the right. B, detection of endonuclease fragments from 3′-stem RNA bearing a 5′-triphosphate end after incubation with RNase J1 for the indicated times (min). Lane M, RNA substrate incubated with mutant RNase J1. ppp, triphosphate 5′-end.
DISCUSSION
In this study we attempted to correlate the observed in vivo effects of RNase J1 depletion on trp leader RNA processing with the in vitro activity of RNase J1. The in vivo results obtained with RNA isolated under conditions of depleted RNase J1 and using probe 1 showed a 3-fold decrease in the ∼100-nt RNA (Fig. 2A). This -fold decrease is similar to the effect of RNase J1 depletion on scRNA processing that we observed previously (7) and likely reflects the persistence of low levels of RNase J1 even in the absence of IPTG induction. The approximate size of the upstream product of RNase J1 cleavage is consistent with a recognition site in the AU-rich region around nt 100 (Fig. 1B). Cleavage at similar AU-rich, single-stranded sequences has been shown to take place in other RNAs, including thrS and thrZ mRNAs (2) and scRNA (7). In these characteristics, RNase J1 resembles E. coli RNase E, which cleaves mostly in single-stranded, AU-rich regions (23). In fact, RNase E and J1 cleave at the same site in the thrS leader (24). The multiple cleavages in the single-stranded region between the 5′-stem-loop and the antiterminator structure that were observed in vitro (Fig. 3B) also occur in a highly AU-rich sequence (74% AU).
In addition to cleavage at around nt 100, we observed cleavage in vitro of TRAP-bound trp leader RNA at nt 35–39 (Fig. 3A). The result of this cleavage together with cleavage at nt 100 gave the predicted 57–58-nt fragments in vivo (Fig. 4B). However, earlier experiments suggest that cleavage at around nt 35 is a low frequency event in vivo: in a PNPase deletion strain, the remaining 3′ to 5′-exonucleases were unable to digest RNA that is TRAP-bound (17). In this situation, we observed massive accumulation of a 95-nt RNA, which is consistent with RNase J1 cleavage at around nt 100, followed by trimming in the 3′ to 5′-direction back to the downstream edge of the TRAP binding site. If RNase J1 cleavage at the upstream edge of the TRAP binding site would occur efficiently in vivo, we would expect an ∼60-nt RNA fragment to accumulate, rather than the observed 95-nt RNA. It is likely that, under conditions where TRAP is bound in vivo, the short single-stranded region between the 5′-stem-loop and the TRAP binding site is recognized poorly by RNase J1.
The pattern of endonucleolytic cleavage on 5′-monophosphorylated and 5′-triphosphorylated trp leader RNA, in the absence of TRAP, was similar (Fig. 3, A and C). Multiple cleavages were observed in the single-stranded region between the 5′-stem-loop and the antiterminator structure. However, there appeared to be some difference in that there was a preference for distal cleavage in the substrate with the 5′-monophosphate end. That such a difference was observed is surprising. The recently solved structure of Thermus thermophilus RNase J (9) explains the 5′-end dependence of the 5′ to 3′-exonuclease activity: a nucleotide binding pocket large enough for a 5′-monophosphate, but not a triphosphate, is located only one nucleotide distance from the catalytic site. However, the structure does not easily account for a 5′-end effect on endonuclease activity. One possible interpretation of our observation is that RNase J1 may have a second 5′-end binding pocket, more distant from the catalytic site (similar to that proposed for RNase E; (11, 25) that allows for RNA looping and the positioning of the endonucleolytic cleavage site RNA in the catalytic site. Clearly, more experiments will be needed to address this issue.
3′-Terminal fragments of trp leader RNA accumulated to high levels under conditions of RNase J1 depletion (Fig. 2C). It would appear at first that a decrease in the level of the upstream 100-nt fragment and an increase in the level of downstream terminator stem fragments, both in response to reduced RNase J1 levels, is contradictory: if reduced RNase J1 in the cell resulted in less endonuclease cleavage at the site located at around nt 100, one would expect to observe less, not more, of the downstream product of this cleavage. However, our in vitro data show that RNase J1 could be involved not only in the endonuclease cleavage to generate the downstream product but also in its turnover. The most likely explanation for the accumulation of 3′-terminal fragments under RNase J1 depletion conditions is the high degree of stability of these fragments, which contain the strong transcription terminator structure. We observed that reduction of RNase J1 levels results in accumulation of 3′-terminal fragments for two other RNAs, rpsO mRNA and ΔermC mRNA (Fig. 7). If these results are representative for mRNAs generally, then the 5′ to 3′-exonuclease activity of RNase J1 might be the solution to the long-standing problem of how the cell deals with structurally stable 3′-terminal fragments, which are resistant to degradation by 3′ to 5′-exonucleases (26, 27). Although RNase R has been shown to be less sensitive to secondary structure, it requires a single-stranded tail of five or more nucleotides for efficient processing (28). This could be provided by addition of a poly(A) tail, but the extent of mRNA polyadenylation in a cell may not be sufficient, or may not occur fast enough, to ensure rapid turnover of 3′-terminal fragments. On the other hand, a combination of endonuclease cleavage (by RNase J1 itself or other endonucleases) upstream of the 3′-terminator structure, followed by RNase J1 5′ to 3′-exonuclease degradation of the 3′-RNA product, would accomplish this turnover quickly without reliance on an extra step of poly(A) addition. The existence of a 5′ to 3′-exonuclease activity acting on 3′-terminal fragments would help explain the low abundance of such RNAs in the cell,3 even though the difficulty that 3′ to 5′-exonucleases have in getting through such structured RNAs would cause one to expect these RNAs to be present in large amounts.
In this respect, our results with the 3′-stem RNA oligonucleotide are pertinent. We find that, although RNase J1 processivity is impeded to some degree by the trp leader RNA terminator structure (Fig. 6, A–B), it can degrade through this structure to a large extent, as evidenced by the release of nearly 40% of labeled pCp from the 3′-end (Fig. 6C). It is quite possible that the in vivo activity of RNase J1 on 3′-terminal fragments is even more efficient, being enhanced by some other factor as has been suggested previously (8) or by an associated RNA helicase activity. In an earlier study (16), we assessed the resistance of the trp leader 3′-terminal RNA structure to PNPase, a relatively strong 3′ to 5′-exonuclease. We found that 60% of the substrate was protected against PNPase digestion when the 3′-terminator stem-loop structure was present, even when a 17-nt poly(A) tail was added to the 3′-end. In the current study, we found that only 25% of the 3′-end-labeled 3′-stem RNA that had been attacked by RNase J1 remained in the terminator structure. Although these were in vitro results obtained under conditions that are likely suboptimal for both enzymes, they nevertheless suggest the possibility that RNase J1 is perhaps a more important player in 3′-terminal fragment turnover than the many 3′ to 5′-exonucleases found in B. subtilis.
The 5′ to 3′-exonuclease activity of RNase J1 was extremely low when presented with a 5′-triphosphorylated substrate (Fig. 8A). Thus, decay of mRNA by processive 5′-exonuclease activity must depend on the presence of a 5′-monophosphate end, which could be generated either by pyrophosphatase activity at the native 5′-triphosphate end or by endonuclease cleavage at a downstream site. Because the endonuclease activity of RNase J1 on trp leader RNA was quite efficient in the presence of a 5′-triphosphate (Fig. 8B), these data are consistent with a model whereby the presence of two ribonuclease activities in RNase J1 allows it to act in mRNA decay by a combination of endonuclease cleavage some distance from the 5′-end, followed by 5′-exonuclease degradation on the downstream fragment (5, 9). According to this model, neither dephosphorylation of the 5′-end nor polyadenylation at the 3′-end would be required for efficient RNA turnover.
Acknowledgments
We thank Paul Babitzke for purified TRAP protein, Joel Belasco for purified E. coli MutT protein, and Olivier Pellegrini for purification of wild-type and mutant RNase J1. The rpsO Northern blot and the ΔermC Northern blot in Fig. 7 were performed by Shiyi Yao and Joshua B. Blaustein, respectively.
This work was supported, in whole or in part, by National Institutes of Health U. S. Public Health Service Grant GM48804 (to D. H. B.). It was also supported by funds from the CNRS (UPR 9073), Université Paris VII-Denis-Diderot, and the Agence Nationale de la Recherche (to C. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: nt, nucleotide; TRAP, trp RNA-binding attenuation protein; pCp, cytidine 3′,5′-bis(phosphate),[5′-32P]; IPTG, isopropyl-1-thio-β-d-galactopyranoside.
S. Yao and D. H. Bechhofer, unpublished results.
References
- 1.Condon, C. (2007) Curr. Opin. Microbiol. 10 271–278 [DOI] [PubMed] [Google Scholar]
- 2.Even, S., Pellegrini, O., Zig, L., Labas, V., Vinh, J., Brechemmier-Baey, D., and Putzer, H. (2005) Nucleic Acids Res. 33 2141–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coburn, G. A., and Mackie, G. A. (1999) Prog. Nucleic Acids Res. Mol. Biol. 62 55–5108 [DOI] [PubMed] [Google Scholar]
- 4.Kushner, S. R. (2002) J. Bacteriol. 184 4658–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathy, N., Benard, L., Pellegrini, O., Daou, R., Wen, T., and Condon, C. (2007) Cell 129 681–692 [DOI] [PubMed] [Google Scholar]
- 6.Condon, C. (2003) Microbiol. Mol. Biol. Rev. 67 157–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao, S., Blaustein, J. B., and Bechhofer, D. H. (2007) Nucleic Acids Res. 35 4464–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britton, R. A., Wen, T., Schaefer, L., Pellegrini, O., Uicker, W. C., Mathy, N., Tobin, C., Daou, R., Szyk, J., and Condon, C. (2007) Mol. Microbiol. 63 127–138 [DOI] [PubMed] [Google Scholar]
- 9.de la Sierra-Gallay, I. L., Zig, L., Jamalli, A., and Putzer, H. (2008) Nat. Struct. Mol. Biol. 15 206–212 [DOI] [PubMed] [Google Scholar]
- 10.Mackie, G. A. (1998) Nature 395 720–723 [DOI] [PubMed] [Google Scholar]
- 11.Jiang, X., and Belasco, J. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9211–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celesnik, H., Deana, A., and Belasco, J. G. (2007) Mol. Cell 27 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deana, A., Celesnik, H., and Belasco, J. G. (2008) Nature 451 355–358 [DOI] [PubMed] [Google Scholar]
- 14.Babitzke, P., and Gollnick, P. (2001) J. Bacteriol. 183 5795–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkin, T. M., and Yanofsky, C. (2002) BioEssays 24 700–707 [DOI] [PubMed] [Google Scholar]
- 16.Deikus, G., and Bechhofer, D. H. (2007) J. Biol. Chem. 282 20238–20244 [DOI] [PubMed] [Google Scholar]
- 17.Deikus, G., Babitzke, P., and Bechhofer, D. H. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2747–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petit, M. A., Dervyn, E., Rose, M., Entian, K. D., McGovern, S., Ehrlich, S. D., and Bruand, C. (1998) Mol. Microbiol. 29 261–273 [DOI] [PubMed] [Google Scholar]
- 19.Alberta, J. A., Rundell, K., and Stiles, C. D. (1994) J. Biol. Chem. 269 4532–4538 [PubMed] [Google Scholar]
- 20.Sharp, J. S., and Bechhofer, D. H. (2003) J. Bacteriol. 185 5372–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drider, D., DiChiara, J. M., Wei, J., Sharp, J. S., and Bechhofer, D. H. (2002) Mol. Microbiol. 43 1319–1329 [DOI] [PubMed] [Google Scholar]
- 22.Bhatnagar, S. K., Bullions, L. C., and Bessman, M. J. (1991) J. Biol. Chem. 266 9050–9054 [PubMed] [Google Scholar]
- 23.Cohen, S. N., and McDowall, K. J. (1997) Mol. Microbiol. 23 1099–1106 [DOI] [PubMed] [Google Scholar]
- 24.Condon, C., Putzer, H., Luo, D., and Grunberg-Manago, M. (1997) J. Mol. Biol. 268 235–242 [DOI] [PubMed] [Google Scholar]
- 25.Callaghan, A. J., Marcaida, M. J., Stead, J. A., McDowall, K. J., Scott, W. G., and Luisi, B. F. (2005) Nature 437 1187–1191 [DOI] [PubMed] [Google Scholar]
- 26.McLaren, R. S., Newbury, S. F., Dance, G. S., Causton, H. C., and Higgins, C. F. (1991) J. Mol. Biol. 221 81–95 [PubMed] [Google Scholar]
- 27.Spickler, C., and Mackie, G. A. (2000) J. Bacteriol. 182 2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng, Z.-F., and Deutscher, M. P. (2005) Mol. Cell 17 313–318 [DOI] [PubMed] [Google Scholar]
- 29.Brosius, J. (1992) Methods Enzymol. 216 469–483 [DOI] [PubMed] [Google Scholar]