Abstract
Human islet amyloid polypeptide (hIAPP) misfolding is thought to play an important role in the pathogenesis of type II diabetes mellitus. It has recently been shown that membranes can catalyze the misfolding of hIAPP via an α-helical intermediate of unknown structure. To better understand the mechanism of membrane-mediated misfolding, we used site-directed spin labeling and EPR spectroscopy to generate a three-dimensional structural model of this membrane-bound form. We find that hIAPP forms a single α-helix encompassing residues 9–22. The helix is flanked by N- and C-terminal regions that do not take up a clearly detectable secondary structure and are less ordered. Residues 21 and 22 are located in a transitional region between the α-helical structure and C terminus and exhibit significant mobility. The α-helical structure presented here has important implications for membrane-mediated aggregation. Anchoring hIAPP to the membrane not only increases the local concentration but also reduces the encounter between peptides to essentially a two-dimensional process. It is significant to note that the α-helical membrane-bound form leaves much of an important amyloidogenic region of hIAPP (residues 20–29) exposed for misfolding. Misfolding of this and other regions is likely further aided by the low dielectric environment near the membrane that is known to promote secondary structure formation. Based upon these considerations, a structural model for membrane-mediated aggregation is discussed.
Protein misfolding and amyloid fibril formation are common characteristics of a number of debilitating human diseases, such as Alzheimer disease, Parkinson disease, and type II diabetes mellitus (1, 2). In type II diabetes mellitus, the primary amyloidogenic agent is human islet amyloid polypeptide (hIAPP),2 a 37-residue peptide that is synthesized in pancreatic islet β-cells and co-secreted with insulin. A number of findings support the notion that hIAPP misfolding plays an important role in disease. Approximately 95% of all patients with type II diabetes mellitus have large extracellular deposits composed of fibrillar hIAPP (2, 3). In vitro studies have shown that hIAPP is toxic when exogenously added to cultured human islet β-cells (4–6) and that overexpression of hIAPP in COS cells results in accumulation of peptide aggregates and cell death (7, 8). Mouse and rat IAPP do not misfold and are not toxic to cultured cells, suggesting that misfolding is a prerequisite for IAPP toxicity. Furthermore, mice and rats do not naturally develop type II diabetes mellitus, but transgenic mice and rats that express hIAPP form amyloid deposits and exhibit signs of diabetes, especially when expression occurs in a background of obesity (9–13).
Like other amyloidogenic peptides and proteins, hIAPP misfolds via a nucleation-dependent aggregation pathway in which small oligomeric assemblies precede the formation of mature amyloid fibrils (14). Biological membranes play two important roles in this process (for review, see Ref. 15). First, membranes are thought to be targets of toxicity because oligomers of hIAPP (as well as other amyloid proteins) disrupt membrane integrity and thereby cause toxicity (4, 16–19). Second, membranes can act as catalysts of misfolding (20–28). It has recently been shown that hIAPP interacts with negatively charged membranes and that this interaction can dramatically accelerate misfolding, which reduces the time of aggregation and fibril formation from days to just a few minutes under optimal conditions (24, 29). The degree of acceleration strongly depends on ionic strength and the density of negatively charged lipids, suggesting that electrostatic interactions are important for membrane-mediated aggregation.
Circular dichroism (CD) and thioflavin T fluorescence data have demonstrated that this membrane-mediated misfolding occurs via an α-helical intermediate (29, 30). It has been estimated that ∼15–20 amino acids participate in α-helix formation. What remains unknown, however, are the structure of this α-helical form, the number of helices that are formed, the specific residues that participate in helix formation, and the membrane topography.
Here, we employed a combination of site-directed spin labeling, EPR spectroscopy, and molecular modeling to generate a three-dimensional model of α-helical, membrane-bound hIAPP. We found that membrane-bound hIAPP has a central helical region from residue 9 to residue 22, whereas the N- and C-terminal regions are much less ordered. This structure leaves much of the highly amyloidogenic stretch from residue 20 to residue 29 exposed for misfolding and β-sheet formation.
EXPERIMENTAL PROCEDURES
Preparation of Spin-labeled hIAPP Derivatives—Spin-labeled hIAPP derivatives were generated through specific cysteine labeling. As in our previous study, the native cysteine residues at positions 2 and 7 were substituted with alanine (31). Using CD and thioflavin T fluorescence, we verified that this Cys-less hIAPP had aggregation properties similar to that of wild type. Single-cysteine mutants were generated using peptide synthesis and obtained from Biomer Technology, LLC (Concord, CA). Lyophilized peptides were dissolved in DMSO and Milli-Q water and were reacted with 3× molar excess of the MTSL spin label ((1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate) for 1 h at room temperature. Excess spin label was removed using a Toyopearl cation exchange chromatography column (Tosoh Corp.) from which hIAPP was eluted with 6 m guanidine HCl (Pierce). Subsequently, C-18 Macro SpinColumns (Harvard Apparatus) were used for desalting, and, finally, hIAPP was eluted with hexafluoroisopropanol. Samples were then aliquoted and lyophilized to achieve the desired final concentrations.
Preparation of Large Unilamellar Vesicles—In the present study, we used large unilamellar vesicles containing 80% 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-l-serine) (POPS) and 20% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (mol/mol) (Avanti Polar Lipids). The individual lipids, dissolved in chloroform, were mixed to obtain a total final concentration of 100 mm after rehydration. Next, the lipid mixture was dried under a stream of nitrogen and desiccated overnight. The dried lipid film was rehydrated in 10 mm phosphate buffer, pH 7, to obtain a concentration of 100 mm lipid. After repeated freeze-thawing (five times), the vesicles were extruded using 0.1 μm polycarbonate membranes (Avanti Polar Lipids) to obtain 100-nm diameter lipid vesicles.
Structural Analysis of Spin-labeled IAPP Derivatives—Lyophilized spin-labeled peptides were incubated with large unilamellar vesicles at a 1:1000 molar ratio in 10 mm phosphate buffer, pH 7. A Jasco circular dichroism J-810 spectrometer (Easton, MD) was used to confirm the formation of α-helical structure for the spin-labeled hIAPP derivatives in the presence of membranes. EPR spectra were recorded on a Bruker (Billerica, MA) EMX spectrometer fitted with an HS resonator at 12-milliwatt incident microwave power. Central line widths were obtained by measuring the peak-to-peak distance for the maxima and minima of the central line. The O2 and chelated nickel (nickel(II)-ethylenediamine-N,N′-diacetic acid) (NiEDDA) accessibilities (ΠO2 and ΠNiEDDA) were determined by employing the commonly used power saturation method (32). The concentration of NiEDDA was 40 mm, and the concentration of O2 was that of air in equilibrium with buffer.
Depth Calibration and Analysis of Φ Parameter—The membrane immersion depth of R1 at different sites was calibrated using the previously established relationship that d[Å] = aΦ + b, where Φ = ln(ΠO2/Π(NiEDDA). The a and b parameters were obtained using calibration with 1-palmitoyl-2-stereoyl-(n-DOXYL)-sn-glycero-3-phosphocholine (Avanti Polar Lipids) harboring spin labels at the 5, 7, and 10 positions on the acyl chains, as described previously (32). Using the spin-labeled phosphatidylcholines specified above, we found that a = 6.5 and b = 3.5.
RESULTS
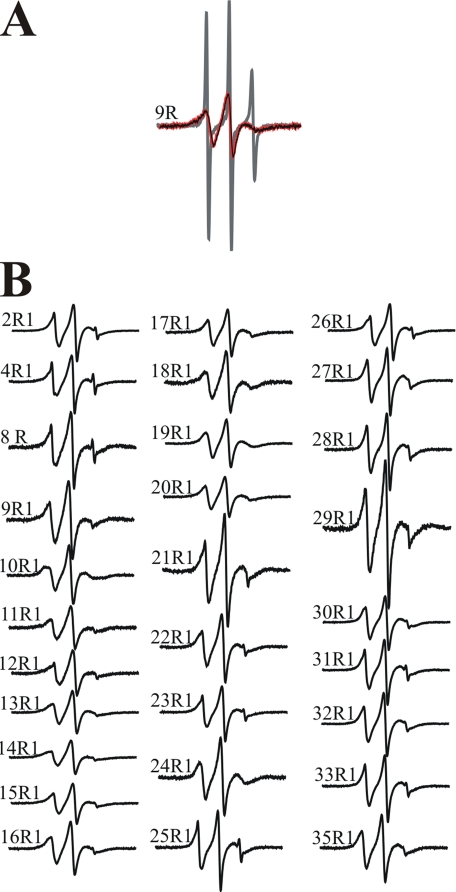
Mobility Analysis Reveals Local Structural Ordering upon Membrane Interaction—To obtain detailed structural information on the α-helical, membrane-bound form of hIAPP, we generated 29 R1-labeled derivatives of hIAPP and recorded the EPR spectra of each spin-labeled derivative in solution and when bound to phospholipid membranes. As shown for the representative example of 9R1 (hIAPP labeled at position 9), the EPR spectra of all freshly dissolved derivatives are characterized by sharp and narrowly spaced lines that are indicative of high mobility (Fig. 1A, gray). These spectra are in agreement with previous data (31) and reflect the highly dynamic structure of the fast tumbling, monomeric hIAPP.
FIGURE 1.
EPR spectra of spin-labeled hIAPP derivatives. A, spectra of hIAPP containing the R1 side chain at residue 9 were obtained in the absence (gray) and in the presence (black) of large unilamellar vesicles. No spectral changes could be detected after 4 h of incubation with the vesicles (red). B, EPR spectra of the indicated hIAPP derivatives in vesicle-bound form. All spectra were collected at a scan width of 100 Gauss and normalized to the same number of spins.
To record the EPR spectra of α-helical, membrane-bound hIAPP, it was important to choose conditions under which no aggregation or β-sheet formation would occur during the time course of the EPR measurements (several minutes). It had previously been shown that the α-helical form can be trapped kinetically and kept stable for several hours at low ionic strength in the presence of highly negatively charged vesicles (29). Under such conditions (80%POPS/20% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), we generated the membrane-bound, α-helical form of all spin-labeled derivatives. As shown for the example of 9R1 in Fig. 1A (black), membrane interaction causes a significant reduction in mobility, as indicated by the broadened lines and the concomitant reduction in signal amplitude. This spectral change reflects reduced tumbling and increased structural ordering upon membrane interaction. No spectral change could be observed over a period of 4 h (Fig. 1A, red), in agreement with CD and thioflavin data indicating that the α-helical structure remains stable for at least 24 h before misfolding and aggregation occurs (data not shown).
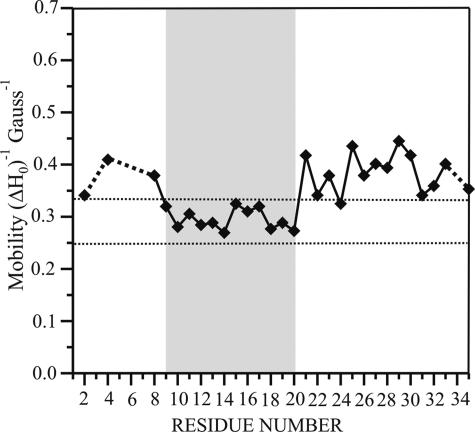
The EPR spectra of the α-helical, membrane-bound forms of all 29 spin-labeled hIAPP derivatives are shown in Fig. 1B. Although membrane interaction causes a reduction in mobility at all sites, the degree of this immobilization varies from site to site. The most pronounced ordering can be observed for residues 9–20. Spectra for these sites have characteristic line shapes and inverse central line width values (between 0.26 and 0.32 Gauss–1) typically observed on surfaces of ordered structures (Fig. 2). In contrast, the soluble peptides have inverse central line width values between 0.62 and 0.68 Gauss–1. Despite the clear decrease in mobility, none of these spectra exhibit any strongly immobilized components that would typically be observed at buried sites. Thus, residues 9–20 do not exhibit strong tertiary or quaternary packing interactions. Although some reduction in mobility can also be observed for sites in the N- and C-terminal regions, those sites still contain components of high mobility, indicating a less ordered state.
FIGURE 2.
Inverse central line width values from the EPR spectra of membrane-bound hIAPP derivatives. The inverse central line width values obtained from the spectra of membrane-bound hIAPP derivatives are given as a function of residue number. The higher values for the N- and C-terminal sites indicate higher mobility upon membrane binding. The central region of the peptide (residues 9–20) exhibits mobility values within the range typically seen for helix surface residues lacking significant tertiary or quaternary packing interactions (43–47) (indicated by dashed vertical lines).
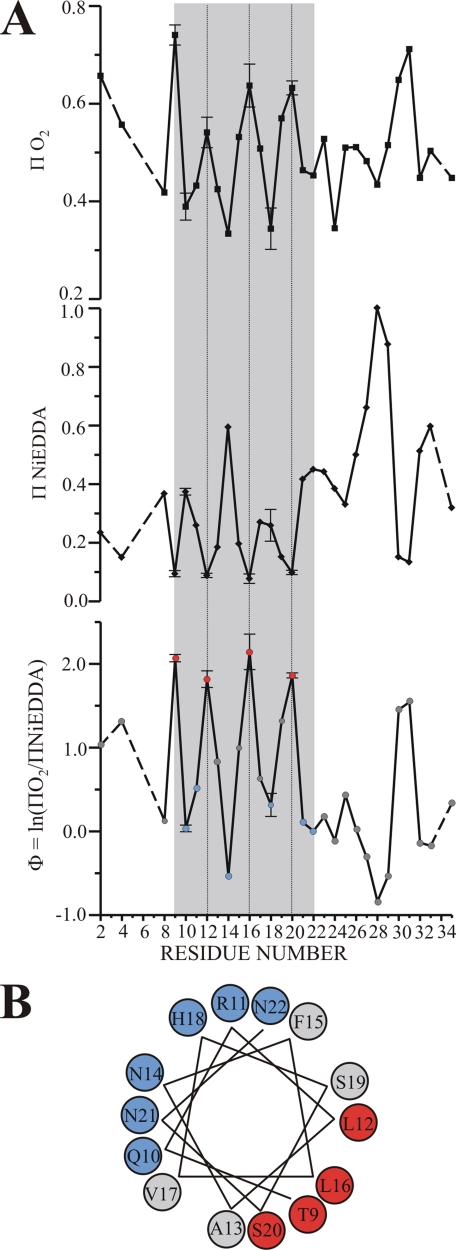
Accessibility Analysis Reveals the Formation of an Amphipathic Helix That Is Located Parallel to the Membrane Surface—Although the mobility data indicate the ordering of a central region, they do not provide information as to secondary structure or membrane topography. To obtain such information, we determined the accessibility of all spin-labeled sites to the paramagnetic colliders O2 and NiEDDA. These measurements are based on the fact that the hydrophobic O2 preferentially partitions into the membrane, whereas the more hydrophilic NiEDDA preferentially partitions into the solvent (33). As a consequence, membrane-exposed sites become more O2-accessible whereas solvent-exposed sites exhibit preferential NiEDDA accessibility.
The accessibilities to O2 and NiEDDA (ΠO2 and ΠNiEDDA, respectively) for all membrane-bound IAPP derivatives are shown in the top panels of Fig. 3A and are summarized by the contrast parameter Φ (Φ = ln(ΠO2/Π (NiEDDA)). Highlighted in gray is a central region from residue 9 to residue 22. The unique feature of this region is the presence of an extensive stretch of highly periodic oscillations that are perfectly out of phase for the O2 and NiEDDA accessibilities. The period of the oscillation indicates an α-helical structure, as maxima and minima are typically spaced three or four residues apart for such structures. The out of phase periodicity of the respective accessibilities reveals an asymmetric solvation in which one face is exposed to the membrane (high ΠO2), while the other face is exposed to the solvent (high Π NiEDDA). The notion of an asymmetrically solvated α-helix is further supported by the Φ plot that also reveals the α-helical oscillations for residues 9–22. When residues 9–22 are plotted on an α-helical wheel, it becomes apparent that the most deeply membrane-embedded residues (Fig. 3B, local Φ maxima, red) fall into the hydrophobic face of an amphipathic α-helix, whereas the solvent-exposed residues fall into the hydrophilic face (local Φ minima, blue). The formation of an α-helical structure is also in good agreement with the mobility analysis that identified residues 9–20 as the most ordered sites. Only residues 21 and 22 exhibit elevated mobility, suggesting that the C-terminal end of the α-helix is likely to be frayed and more dynamic.
FIGURE 3.
Accessibility analysis reveals the formation of an asymmetrically solvated amphipathic helix. A, the two top panels depict the O2 and NiEDDA accessibilities (ΠO2 and ΠNiEDDA) as function of labeling position in membrane-bound hIAPP. The gray shaded area highlights a region of α-helical periodicity in which ΠO2 and ΠNiEDDA are out of phase. The lower panel summarizes the accessibility values using the contrast parameter Φ. Again, helical periodicity is observed in the gray shaded area. Within the α-helical region, high Φ values indicate lipid-exposed sites (red) and low Φ values indicate solvent-exposed sites (blue). Sites with in-between Φ values are in gray. To illustrate the error associated with the present data the S.E. is shown. The error bars were obtained from accessibility measurements that were repeated at least three times for the lipid and solvent-exposed sites in the α-helical region. B, the hIAPP residues were plotted on an α-helical wheel. The lipidexposed residues lie on one side (red), whereas the solvent-exposed sites (blue) are on the opposite side.
It has been well established that Φ values are directly proportional to the membrane insertion depth and that it is possible to calibrate the immersion depth through the use of spin-labeled lipids (32). Using this approach, we have found that the most lipid-exposed sites (positions 9, 12, 16, and 20) are at a relatively constant immersion depth of ∼16 Å. Similar immersion depths are characteristic of a peripherally bound peptide rather than of a transmembrane helix. In the latter case, the immersion depth would be maximal in the center of the helix and would decrease toward the N- and C-terminal regions. Furthermore, the short stretch of ∼14 amino acids from residue 9 to residue 22 would be too short to span the bilayer. Thus, the data are not consistent with the formation of a transmembrane helix but rather with an α-helix that is parallel to the membrane surface.
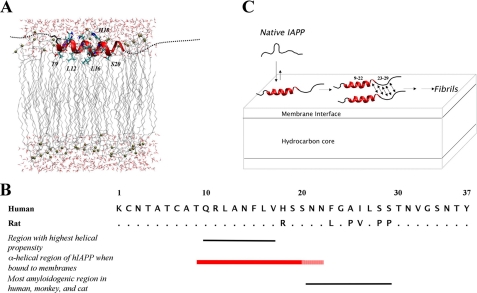
When building structural models, it is important to consider that the measured immersion depths represent those of the N-O moiety of the R1 side chain. According to crystallographic studies, this group is located ∼7–10 Å away from the center of the α-helix (34). We can therefore estimate that the center of the helix is located at an immersion depth of 6–9 Å below the level of the phosphate groups. The structure of residues 9–22 is schematically indicated in Fig. 4A; the N- and C-terminal regions are not shown because the EPR data did not detect any specifically ordered structure in these regions. It is noteworthy that Arg-11 and His-18 (which is likely to be protonated when in the interfacial region) are located in positions that maximize electrostatic interactions with the negatively charged head-groups/phosphates. Moreover, Phe-15 is located close to the level of the phosphates, a position thought to be favored by aromatic side chains of membrane proteins (35). Collectively, these interactions might contribute to the observed immersion depth of the helix.
FIGURE 4.
Structural and mechanistic model of membrane-bound hIAPP. A, schematic model of membrane-bound hIAPP consistent with EPR data on the distribution of secondary structure and placement within the membrane. The thick red ribbon indicates the central helical core (residues 9–20) of the peptide (see “Results” and “Discussion” for details). Residues 21 and 22, which are also found within the helical region but show elevated mobility indicating possible fraying of the C-terminal end of the helix, are shown in a thinner red ribbon. EPR data did not indicate any specific ordered structure for the N- and C-terminal residues (1–8 and 23–37) and are thus shown as dashed lines. The position of the helical core region within the lipid bilayer is depicted, based on EPR depth measurements. Phosphates are depicted as gold spheres. B, primary structure and sequence characteristics of IAPP. All IAPP sequences contain a conserved amidated C terminus and a free N terminus. Rat IAPP differs from the human peptide by six residues, with the most significant variations occurring between residues 23 and 29. The region with highest helical propensity, as predicted by PredictProtein (48), is between residues 10 and 17. The α-helical region of hIAPP when bound to membranes, as determined by this study, spans residues 9–22. This is consistent with the helical regions observed for calcitonin and the calcitonin gene-related peptide in membrane/membrane mimetic environments (49, 50). Calcitonin and calcitonin gene-related peptide belong to the same peptide superfamily as IAPP. The solid red bar indicates the helical core region between residues 9 and 20, and the dashed bar indicates residues 21 and 22. The most amyloidogenic region of IAPP in humans, monkeys, and cats lies between residues 20 and 29. This region also exhibits the largest sequence variation between human and rat IAPP. C, the aggregation of hIAPP in the presence of lipid membranes. Interaction of predominantly unstructured, monomeric, soluble hIAPP with negatively charged lipid membranes results in the formation of an α-helix between residues 9 and 22 whereas the remainder of the peptide stays unstructured. Aggregation of the membrane-bound form of hIAPP is expected due to the formation of secondary structure, initiated by residues within the highly amyloidogenic region, that remain unstructured in the membrane-bound state (residues 23–29).
DISCUSSION
The present study has employed EPR spectroscopy of 29 different spin-labeled hIAPP derivatives and structural modeling to obtain detailed structural information on membrane-bound hIAPP (Fig. 4A). Mobility and accessibility analyses have revealed the formation of an α-helical structure from residue 9 to residue 22, whereas the N- and C-terminal regions are less ordered and do not exhibit a clearly detectable secondary structure. The central amphipathic α-helix runs parallel to the membrane and, according to mobility analysis, shows little tertiary and quaternary packing interactions suggesting the formation of a single, predominantly monomeric helix.
Previous predictions of membrane-bound hIAPP using MPEx revealed that an α-helix composed of residues 10–27 has the highest hydrophobicity and amphipathicity in hIAPP (29). The α-helical region defined in the present study is entirely contained in this previously identified stretch, but, importantly, it lacks the C-terminal amino acids (residues 23–27). A likely explanation for the somewhat shorter helical structure identified in the present study is the local difference in helical propensity. According to PredictProtein, residues 10–17 exhibit the strongest propensity for α-helix formation whereas residues 18–37 have only weak helical propensities. In fact, a recent NMR study of rat IAPP revealed that the helical residues identified in the present study can take up partial helical structure in aqueous solution (36).
The present structural information has important implications for the mechanism of aggregation. Residues 20–29 of hIAPP have long been considered a highly amyloidogenic region, and a peptide containing these residues has been shown to readily form amyloid fibrils (37–39). This region also exhibits the largest sequence variations between hIAPP and rat IAPP, which does not readily form amyloid fibrils due to prolines at positions 25, 28, and 29. Our finding that residues 23–37 do not take up a detectable secondary structure and that residues 21 and 22 exhibit elevated mobility reveals that much of this highly amyloidogenic region (as well as the residual C-terminal region) is poorly folded upon membrane interaction. This result is of particular significance because membrane interaction typically facilitates the formation of secondary structure, including that of β-sheet structure (40). Thus, the formation of α-helical, membrane-bound hIAPP exposes a highly amyloidogenic stretch of hIAPP to an environment that is expected to promote the formation of misfolded β-sheet structures. Moreover, membrane interaction also increases the local concentration of hIAPP and reduces the dimensionality of the encounter between multiple hIAPP molecules, which effectively becomes a two-dimensional process. The combination of these factors is likely to be responsible for the pronounced acceleration of membrane-mediated hIAPP misfolding.
Recently, it was concluded that α-helical, membrane-bound hIAPP can form helical oligomers at high protein-to-lipid ratios, and it was suggested that these α-helical oligomers could then transition into β-sheet-containing misfolded forms (30). When combined with the present data, a likely mechanism for aggregation and misfolding would be the initiation of β-sheet formation in the C terminus of such helical bundles or other transient complexes (Fig. 4C). This notion is further supported by the finding that rat IAPP, which also forms an α-helical structure on membranes, contains three prolines in the critical region (residues 20–29) and does not undergo membrane-mediated misfolding. Similarly, truncated hIAPP containing residues 1–24 readily takes up an α-helical structure but does not readily undergo membrane-mediated aggregation, as judged by thioflavin T fluorescence.3 Subsequent to the formation of the initial, presumably α-helix- and β-sheet-containing, oligomers, the α-helical structure must then be lost, because previous CD analysis shows that gain of β-sheet structure coincides with the loss of α-helical structure. Furthermore, it is now well established that the β-sheet-containing core region of hIAPP fibrils extends well beyond residues 20–29 and includes most, if not all, of the residues that participate in α-helix formation (29, 41).
Our previous study revealed that the most rapid aggregation occurs at a POPS percentage of 25%. These data are consistent with the model in Fig. 4A, wherein the α-helical form represents a transient intermediate. In the concentration range between 0 and 25% POPS, an increase in the amount of POPS increases the amount of α-helix formation and thereby promotes misfolding via the mechanism shown in Fig. 4C. At higher concentrations of POPS, such as those used in the present study, the α-helical form becomes increasingly more stable. This stabilization should inhibit the transition from mixed α/β oligomers to predominantly β-sheet oligomers. In cells, the density of POPS (or other negatively charged lipids) varies significantly but is typically less than 25%. Consequently, we expect that the α-helical form described here will be rather short-lived and, presumably, will readily transition into the misfolded state. Such a mechanism should be particularly important for cytosolic hIAPP (7, 9, 42), and prevention of the misfolding pathway outlined in Fig. 4C could therefore ameliorate the toxic effects of hIAPP.
Acknowledgments
We thank Diana Gegala for assistance during the preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AG027936 (to R. L.). This work was also supported by the Beckman Foundation (to R. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: hIAPP, human islet amyloid polypeptide; POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-l-serine); NiEDDA, nickel(II)-ethylenediamine-N,N′-diacetic acid.
M. Apostolidou and R. Langen, unpublished results.
References
- 1.Soto, C. (2003) Nat. Rev. Neurosci. 4 49–60 [DOI] [PubMed] [Google Scholar]
- 2.Hull, R. L., Westermark, G. T., Westermark, P., and Kahn, S. E. (2004) J. Clin. Endocrinol. Metab. 89 3629–3643 [DOI] [PubMed] [Google Scholar]
- 3.Kahn, S. E., Andrikopoulos, S., and Verchere, C. B. (1999) Diabetes 48 241–253 [DOI] [PubMed] [Google Scholar]
- 4.Janson, J., Ashley, R. H., Harrison, D., McIntyre, S., and Butler, P. C. (1999) Diabetes 48 491–498 [DOI] [PubMed] [Google Scholar]
- 5.Lorenzo, A., Razzaboni, B., Weir, G. C., and Yankner, B. A. (1994) Nature 368 756–760 [DOI] [PubMed] [Google Scholar]
- 6.Ritzel, R. A., and Butler, P. C. (2003) Diabetes 52 1701–1708 [DOI] [PubMed] [Google Scholar]
- 7.O'Brien, T. D., Butler, P. C., Kreutter, D. K., Kane, L. A., and Eberhardt, N. L. (1995) Am. J. Pathol. 147 609–616 [PMC free article] [PubMed] [Google Scholar]
- 8.Hiddinga, H. J., and Eberhardt, N. L. (1999) Am. J. Pathol. 154 1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janson, J., Soeller, W. C., Roche, P. C., Nelson, R. T., Torchia, A. J., Kreutter, D. K., and Butler, P. C. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 7283–7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler, A. E., Jang, J., Gurlo, T., Carty, M. D., Soeller, W. C., and Butler, P. C. (2004) Diabetes 53 1509–1516 [DOI] [PubMed] [Google Scholar]
- 11.Verchere, C. B., D'Alessio, D. A., Palmiter, R. D., Weir, G. C., BonnerWeir, S., Baskin, D. G., and Kahn, S. E. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 3492–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soeller, W. C., Janson, J., Hart, S. E., Parker, J. C., Carty, M. D., Stevenson, R. W., Kreutter, D. K., and Butler, P. C. (1998) Diabetes 47 743–750 [DOI] [PubMed] [Google Scholar]
- 13.Hoppener, J. W., Oosterwijk, C., Nieuwenhuis, M. G., Posthuma, G., Thijssen, J. H., Vroom, T. M., Ahren, B., and Lips, C. J. (1999) Diabetologia 42 427–434 [DOI] [PubMed] [Google Scholar]
- 14.Caughey, B., and Lansbury, P. T. (2003) Annu. Rev. Neurosci. 26 267–298 [DOI] [PubMed] [Google Scholar]
- 15.Jayasinghe, S. A., and Langen, R. (2007) Biochim. Biophys. Acta 1768 2002–2009 [DOI] [PubMed] [Google Scholar]
- 16.Anguiano, M., Nowak, R. J., and Lansbury, P. T., Jr. (2002) Biochemistry 41 11338–11343 [DOI] [PubMed] [Google Scholar]
- 17.Arispe, N., Pollard, H. B., and Rojas, E. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 10573–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volles, M. J., Lee, S. J., Rochet, J. C., Shtilerman, M. D., Ding, T. T., Kessler, J. C., and Lansbury, P. T., Jr. (2001) Biochemistry 40 7812–7819 [DOI] [PubMed] [Google Scholar]
- 19.Lashuel, H. A., and Lansbury, P. T. (2006) Q. Rev. Biophys. 1–35 [DOI] [PubMed]
- 20.Kurganov, B., Doh, M., and Arispe, N. (2004) Peptides 25 217–232 [DOI] [PubMed] [Google Scholar]
- 21.Kazlauskaite, J., Sanghera, N., Sylvester, I., Venien-Bryan, C., and Pinheiro, T. J. (2003) Biochemistry 42 3295–3304 [DOI] [PubMed] [Google Scholar]
- 22.Matsuzaki, K., and Horikiri, C. (1999) Biochemistry 38 4137–4142 [DOI] [PubMed] [Google Scholar]
- 23.Davidson, W. S., Jonas, A., Clayton, D. F., and George, J. M. (1998) J. Biol. Chem. 273 9443–9449 [DOI] [PubMed] [Google Scholar]
- 24.Knight, J. D., and Miranker, A. D. (2004) J. Mol. Biol. 341 1175–1187 [DOI] [PubMed] [Google Scholar]
- 25.Bokvist, M., Lindstrom, F., Watts, A., and Grobner, G. (2004) J. Mol. Biol. 335 1039–1049 [DOI] [PubMed] [Google Scholar]
- 26.Chirita, C. N., Necula, M., and Kuret, J. (2003) J. Biol. Chem. 278 25644–25650 [DOI] [PubMed] [Google Scholar]
- 27.Necula, M., Chirita, C. N., and Kuret, J. (2003) J. Biol. Chem. 278 46674–46680 [DOI] [PubMed] [Google Scholar]
- 28.Lee, H. J., Choi, C., and Lee, S. J. (2002) J. Biol. Chem. 277 671–678 [DOI] [PubMed] [Google Scholar]
- 29.Jayasinghe, S. A., and Langen, R. (2005) Biochemistry 44 12113–12119 [DOI] [PubMed] [Google Scholar]
- 30.Knight, J. D., Hebda, J. A., and Miranker, A. D. (2006) Biochemistry 45 9496–9508 [DOI] [PubMed] [Google Scholar]
- 31.Jayasinghe, S. A., and Langen, R. (2004) J. Biol. Chem. 279 48420–48425 [DOI] [PubMed] [Google Scholar]
- 32.Altenbach, C., Greenhalgh, D. A., Khorana, H. G., and Hubbell, W. L. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 1667–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbell, W. L., Gross, A., Langen, R., and Lietzow, M. A. (1998) Curr. Opin. Struct. Biol. 8 649–656 [DOI] [PubMed] [Google Scholar]
- 34.Langen, R., Oh, K. J., Cascio, D., and Hubbell, W. L. (2000) Biochemistry 39 8396–8405 [DOI] [PubMed] [Google Scholar]
- 35.White, S. H., and Wimley, W. C. (1998) Biochim. Biophys. Acta 1376 339–352 [DOI] [PubMed] [Google Scholar]
- 36.Williamson, J. A., and Miranker, A. D. (2007) Protein Sci. 16 110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriarty, D. F., and Raleigh, D. P. (1999) Biochemistry 38 1811–1818 [DOI] [PubMed] [Google Scholar]
- 38.Tenidis, K., Waldner, M., Bernhagen, J., Fischle, W., Bergmann, M., Weber, M., Merkle, M. L., Voelter, W., Brunner, H., and Kapurniotu, A. (2000) J. Mol. Biol. 295 1055–1071 [DOI] [PubMed] [Google Scholar]
- 39.Westermark, P., Engstrèom, U., Johnson, K. H., Westermark, G. T., and Betsholtz, C. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 5036–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wimley, W. C., Hristova, K., Ladokhin, A. S., Silvestro, L., Axelsen, P. H., and White, S. H. (1998) J. Mol. Biol. 277 1091–1110 [DOI] [PubMed] [Google Scholar]
- 41.Luca, S., Yau, W. M., Leapman, R., and Tycko, R. (2007) Biochemistry 46 13505–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Koning, E. J., Morris, E. R., Hofhuis, F. M., Posthuma, G., Hoppener, J. W., Morris, J. F., Capel, P. J., Clark, A., and Verbeek, J. S. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McHaourab, H. S., Lietzow, M. A., Hideg, K., and Hubbell, W. L. (1996) Biochemistry 35 7692–7704 [DOI] [PubMed] [Google Scholar]
- 44.Margittai, M., Fasshauer, D., Pabst, S., Jahn, R., and Langen, R. (2001) J. Biol. Chem. 276 13169–13177 [DOI] [PubMed] [Google Scholar]
- 45.Isas, J. M., Langen, R., Haigler, H. T., and Hubbell, W. L. (2002) Biochemistry 41 1464–1473 [DOI] [PubMed] [Google Scholar]
- 46.Perozo, E., Cortes, D. M., and Cuello, L. G. (1998) Nat. Struct. Biol. 5 459–469 [DOI] [PubMed] [Google Scholar]
- 47.Gross, A., Columbus, L., Hideg, K., Altenbach, C., and Hubbell, W. L. (1999) Biochemistry 38 10324–10335 [DOI] [PubMed] [Google Scholar]
- 48.Rost, B., Yachdav, G., and Liu, J. (2004) Nucleic Acids Res. 32 W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breeze, A. L., Harvey, T. S., Bazzo, R., and Campbell, I. D. (1991) Biochemistry 30 575–582 [DOI] [PubMed] [Google Scholar]
- 50.Motta, A., Andreotti, G., Amodeo, P., Strazzullo, G., and Castiglione Morelli, M. A. (1998) Proteins 32 314–323 [PubMed] [Google Scholar]