Abstract
The M protein of Streptococcus equi subsp. equi known as fibrinogen-binding protein (FgBP) is a cell wall-associated protein with antiphagocytic activity that binds IgG. Recombinant versions of the seven equine IgG subclasses were used to investigate the subclass specificity of FgBP. FgBP bound predominantly to equine IgG4 and IgG7, with little or no binding to the other subclasses. Competitive binding experiments revealed that FgBP could inhibit the binding of staphylococcal protein A and streptococcal protein G to both IgG4 and IgG7, implicating the Fc interdomain region in binding to FgBP. To identify which of the two IgG Fc domains contributed to the interaction with FgBP, we tested two human IgG1/IgA1 domain swap mutants and found that both domains are required for full binding, with the CH3 domain playing a critical role. The binding site for FgBP was further localized using recombinant equine IgG7 antibodies with single or double point mutations to residues lying at the CH2-CH3 interface. We found that interaction of FgBP with equine IgG4 and IgG7 was able to disrupt C1q binding and antibody-mediated activation of the classical complement pathway, demonstrating an effective means by which S. equi may evade the immune response. The mode of interaction of FgBP with IgG fits a common theme for bacterial Ig-binding proteins. Remarkably, for those interactions studied in detail, it emerges that all the Ig-binding proteins target the CH2-CH3 domain interface, regardless of specificity for IgG or IgA, streptococcal or staphylococcal origin, or host species (equine or human).
Strangles is a highly contagious upper respiratory tract disease of the horse and is caused by Streptococcus equi subsp. equi, a Lancefield group C streptococcus. The disease is characterized by fever, mucopurulent nasal discharge, and the formation of abscesses in lymph nodes of the head and neck. It is one of the most frequently reported equine clinical problems worldwide and is associated with serious economic cost, because severe outbreaks may last for months or years (1-3). A proportion of horses that recover from strangles become persistent carriers of S. equi, capable of infecting naïve horses and continuing the spread of the disease (4). Complications can occur in up to 20% of strangles cases and include formation of abscesses in other body organs (“bastard strangles”) and purpura hemorrhagica, an immune complex-mediated vasculitis (3).
The S. equi M protein, also known as fibrinogen-binding protein (FgBP),2 is a major cell wall-associated protein of S. equi subsp. equi (5-7). It is 534 amino acids in length, contains two blocks of degenerate repeated sequences, and, in common with M-proteins of other Gram-positive streptococci, is predicted to possess a non-helical N terminus and extensive regions of α-helical coiled-coil structure throughout the rest of the extracellular portion of the protein (7, 8). FgBP plays a significant role in the resistance of S. equi to phagocytosis and contributes substantially to its virulence (9). The N-terminal region of FgBP is able to bind strongly to fibrinogen, and this appears to play an important part in the antiphagocytic effect of the protein and survival of S. equi in the host (6, 7, 10-12).
In addition to the binding of host fibrinogen, FgBP has been demonstrated to bind to the Fc region of horse IgG, as well as IgG from several other species (human, rabbit, and cat but not mouse, rat, goat, sheep, cow, or chicken) (9). Hence FgBP represents another example of a bacterial Ig-binding protein. A number of such proteins, produced by pathogenic staphylococcal and streptococcal strains and including the widely employed staphylococcal protein A and streptococcal protein G, have been described. The region of FgBP that binds to IgG Fc is distinct from that responsible for fibrinogen binding and involves the central part of the protein (8, 9). The development of specific mucosal and systemic antibody responses is considered crucial for combating infection with S. equi and for protection from future disease (2). Significantly, a specific IgG response has been shown to contribute to this protective humoral immunity (13). The horse has seven IgG subclasses (14, 15), and to date there is no information with regard to the subclass specificity of FgBP and whether the interaction of FgBP with IgG Fc contributes to subversion of the immune response by S. equi. Here, we demonstrate that the binding of FgBP to horse IgG is highly subclass specific and that the binding site for FgBP on horse IgG Fc is located in the interdomain region and overlaps with that of protein A and G. Furthermore, binding of FgBP to horse IgG is able to disrupt antibody-mediated complement activation suggesting another mechanism by which FgBP can mediate resistance to phagocytosis and compromise the host immune response to S. equi subsp. equi.
EXPERIMENTAL PROCEDURES
Production of FgBPs—FgBP1, a recombinant form of FgBP that lacks the wall/membrane anchor domain, was produced as described previously (7). For generation of FgBPM-, a recombinant form of FgBP that lacks the central domain between the A and B repeats (amino acids 279-348) and does not bind to equine IgG,3 FgBP gene sequences encoding amino acids 37-278 and 349-470 were amplified and juxtaposed in the vector pQE30. Transformation of Escherichia coli XL-1Blue with pQE30 and purification of the recombinant protein was carried out as previously described (7, 11).
Analysis of the Interaction of FgBP with Recombinant Equine Igs by ELISA—Interaction of FgBP1 with recombinant forms of the seven equine IgG (reqIgG) subclasses, produced and purified as described previously (16), was analyzed by ELISA. FgBP1 (10 μg/ml in carbonate buffer) was coated overnight at 4 °C onto 96-well Maxisorp immunoplates (Nunc, Roskilde, Denmark). Control wells were coated with 10 μg/ml FgBPM-. Plates were washed in Milli-Q water and then blocked for 1 h at room temperature with 5% nonfat milk powder in PBS containing 0.5% Tween 20 (PBS-T). Plates were then incubated with serial dilutions (0-20 μg/ml in PBS-T) of one of the reqIgG subclasses or recombinant equine monomeric equine IgA4 (eqIgA) for 1 h at room temperature. Binding of reqIgs to FgBP was detected by incubation for 1 h at room temperature with HRP-conjugated goat anti-mouse λ light chain antibody (0.2 μg/ml in PBS-T) (Bethyl Laboratories Inc., Montgomery, TX). Goat IgG does not bind to FgBP (9) and so was suitable as the detection reagent in these experiments. Plates were developed as previously described (17). The pH dependence of the FgBP-IgG interaction was analyzed using the same ELISA except that reqIgGs (serial dilutions of 0-10 μg/ml) were diluted in PBS-T at either pH 5.5, pH 7.4, or pH 8.5, and FgBP1 and FgBPM- were used at 5 μg/ml.
Biosensor Analysis of FgBP1 Binding to reqIgG—Biosensor experiments were carried out using a BIAcore X instrument (BIAcore AB, Uppsala, Sweden). Recombinant FgBP1 (20 μm in 10 mm sodium acetate, pH 5.5) was immobilized on a CM5 biosensor chip (BIAcore) using the manufacturer's instructions for amine coupling chemistry. Recombinant FgBPM- was coupled under the same conditions to the reference surface of the chip. Recombinant eqIgGs were diluted in BIAcore HBS-EP buffer (0.01 m HEPES, 0.15 m NaCl, 3 mm EDTA, 0.005% surfactant P20) across a concentration range of 0-150 μg/ml and were independently injected over the chip (60 μl and 30 μl/min). After each injection the sensor chip was regenerated with 20 μl of 10 mm glycine, pH 3, followed by a 10-μl injection of 10 mm glycine, pH 2, if required. Biosensor analysis of recombinant human IgG1 (18) binding to FgBP1 was also carried out as described above. BIAevaluation 3.2 software was used for data analysis.
Protein G and Protein A Competition ELISAs—Plates were coated with 2.5 μg/ml of either reqIgG4 or reqIgG7 and then washed and blocked as described above. Plates were placed on ice, and 100 μl/well ice-cold HRP-conjugated protein G (0.25 μg/ml in PBS-T) (Sigma), and serial dilutions of either FgBP1 or FgBPM- (both 0-50μg/ml in PBS-T) were added simultaneously to each well. Plates were incubated for 2 h at room temperature and then developed as described above. A similar ELISA was used to assess competition between HRP-conjugated protein A (Sigma) (20μg/ml) and FgBP1 or FgBPM- (serial dilutions of 0-50μg/ml) using plates coated with 5 μg/ml reqIgG4 or reqIgG7. It was necessary to use different concentrations of HRP-conjugated protein G and protein A in the assay because of the different binding affinities of the two proteins for IgG.
Binding of Recombinant Human Igs to FgBP—Two previously described recombinant human IgG1/IgA1 domain-swap mutants (18) were assessed for binding to FgBP1. In the first mutant (γ1γ2α3), the CH3 domain of human IgG1 is replaced by the CH3 domain of human IgA1, whereas in the second (α1α2γ3), the CH3 domain of human IgA1 is replaced by the CH3 domain of human IgG1. Binding of FgBP1 to a recombinant version of the human IgG3 allotype, IGHG3*01 or IgG3m(5) (19), purified from transfectant supernatant by affinity chromatography on 3-nitro-4-hydroxy-5-iodophenylacetate-Sepharose, was also investigated. This allotype is known to lack the ability to bind protein A (20, 21). Binding analysis, using recombinant human IgG1 and human IgA1, produced as described previously (17), as positive and negative controls, respectively, was carried out by ELISA as described above using 5 μg/ml FgBP1 to coat and serial dilutions (up to 10 μg/ml) of recombinant Igs.
Site-directed Mutagenesis of eqIgG7 Heavy (H) Chain—Single or double point mutations were introduced into the CH2 or CH3 domains of the eqIgG7 H-chain using a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA), following the manufacturer's instructions. Residues mutated in eqIgG7 corresponded to those in human IgG1 responsible for binding protein A or protein G (22, 23). Selected residues in eqIgG7 were exchanged for the amino acid(s) present in the non-FgBP-binding eqIgG subclasses, at positions where a non-conservative amino acid alteration was found. The five mutants generated are described in Table 1. Mutant antibodies were purified and analyzed by SDS-PAGE and Western blotting as previously described (16). The FgBP1 binding capacities of mutant reqIgG7s were compared with that of wild-type reqIgG7 using biosensor analysis as described above except that 100-μl injections of antibody were used.
TABLE 1.
Names and description of reqIgG7 mutants
| Mutant | Descriptiona | eqIgG subclasses carrying the same residues as those substituted |
|---|---|---|
| M252K | Met252 in CH2 domain substituted with Lys | IgG5 |
| K382Q | Lys382 in CH3 domain substituted with Gln | IgG1, IgG2, IgG3, IgG5, IgG6 |
| HN433/434EH | His433 in CH3 domain substituted with Glu, Asn434 in CH3 domain substituted with His | Glu433 in IgG5 |
| H433R | His433 in CH3 domain substituted with Arg | IgG6 |
| HY435/436TV | His435 in CH3 domain substituted with Thr, Tyr436 in CH3 domain substituted with Val | Thr435 in IgG6 |
| Val436 in IgG3 |
Numbering corresponds to human IgG1 numbering.
Analysis of the Consequences of FgBP-IgG Binding on Complement Activation—FgBP-mediated inhibition of C1q binding and activation of the classic complement pathway by equine IgG was assessed by ELISA using reqIgG3, reqIgG4, and reqIgG7 (10 μg/ml), as described previously (16), with the following modifications. After coating of wells with 3-nitro-4-hydroxy-5-iodophenylacetate-bovine serum albumin and subsequent incubation with recombinant Igs, plates were incubated with serial dilutions of FgBP1 or FgBPM- (0-10 μg/ml in PBS-T containing 0.5% bovine serum albumin) for 1 h at room temperature. Plates were incubated then with either human C1q protein (0.5 μg/ml, Calbiochem) or human serum. In the former case, binding of C1q to IgG was detected using sheep anti-human C1q-HRP, as previously described, whereas in the latter, activation of the classic complement pathway was detected using a 1/500 dilution of biotin-conjugated mouse anti-human C4c (Quidel, Santa Clara, CA) followed by streptavidin-HRP (1/500, Dako, Ely, Cambs, UK). C4c generation is a specific marker for activation of the classic complement pathway (24). Human C1q and human serum (as a source of complement) were used rather than their equine equivalents because of the availability of detection reagents for the human but not equine proteins. Moreover, cross-species reactivity of IgG and complement components has been well demonstrated (25).
RESULTS
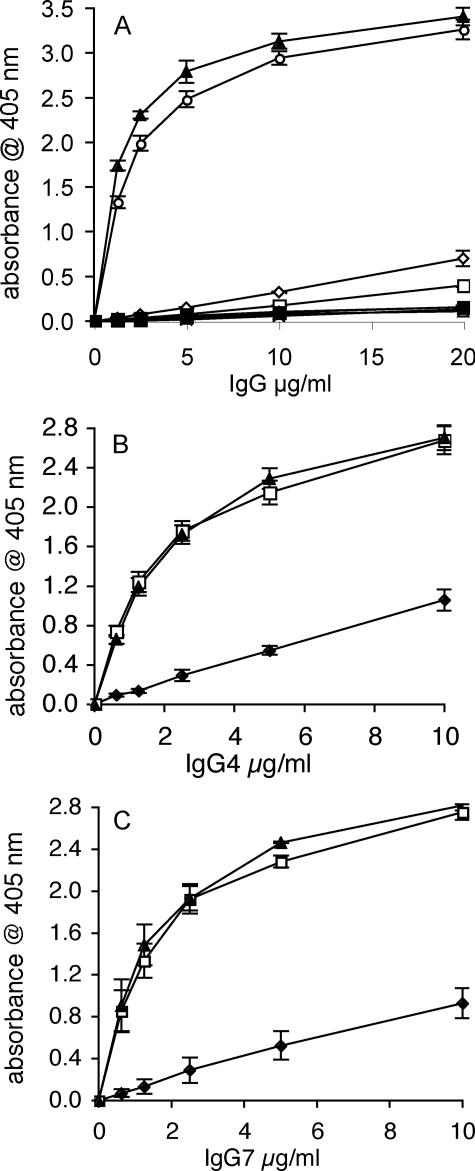
Analysis of FgBP-IgG Binding—Binding of recombinant FgBP1 to the seven reqIgG subclasses and monomeric reqIgA was first analyzed by ELISA. FgBP1 bound strongly to reqIgG4 and reqIgG7, with little or no binding to the remaining five reqIgG subclasses or to reqIgA (Fig. 1A). Because IgG binding by the bacterial IgG-binding proteins staphylococcal protein A and streptococcal protein G is known to be pH-dependent (26), we tested the effect of pH variation on the FgBP1-IgG interaction (Fig. 1, B and C). An increase in pH from 7.4 to pH 8.5 had little effect on binding, whereas a decrease in pH to 5.5 reduced binding to roughly a third of that seen at physiological pH. Hence, the pH dependence of IgG binding by FgBP is more similar to that of protein A (strongest binding at pH 8) than to that of protein G (strongest binding at pH 4-5) (26).
FIGURE 1.
ELISA analysis of the interaction of recombinant equine Igs with FgBP1. A, subclass specificity of the interaction. FgBP1 bound strongly to IgG4 and IgG7, but the remaining five IgG subclasses and IgA showed little or no binding. Open diamond, reqIgG1; cross, reqIgG2; open square, reqIgG3; open circle, reqIgG4; open triangle, reqIgG5; plus sign, reqIgG6; closed triangle, reqIgG7; closed square, reqIgA. B and C, pH dependence of the binding to FgBP1. For both equine IgG4 (B) and IgG7 (C), binding to FgBP1 was most efficient at pH 7.4-8.5. Closed diamond, pH 5.0; open square, pH 7.4; closed triangle, pH 8.5. Binding of reqIgGs is expressed as absorbance obtained for the binding to FgBP1 minus absorbance for binding to the FgBPM- control (binding to the latter was typically <0.15 absorbance units). The mean (±S.E.) of three independent experiments is shown.
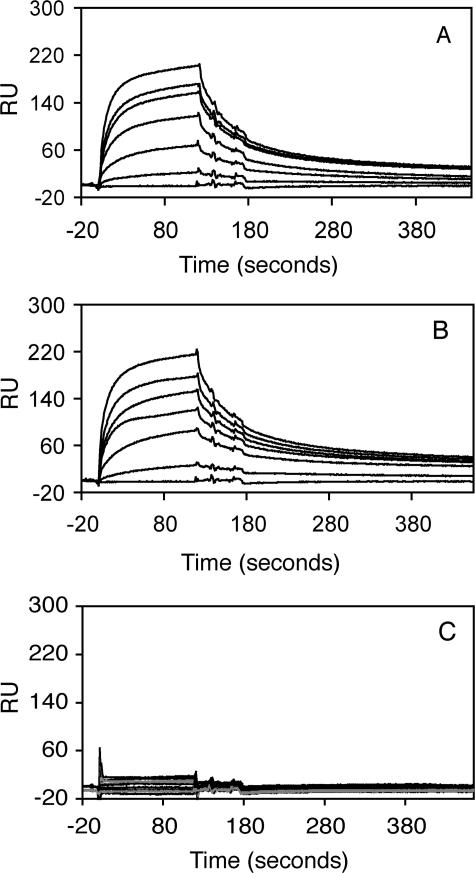
To establish the affinity of the FgBP-IgG interaction and confirm the equine IgG subclass specificity we used surface plasmon resonance measurements. These experiments gave identical results with regard to subclass specificity to those obtained by ELISA, confirming that reqIgG4 and reqIgG7 are the predominant subclasses capable of binding FgBP1 (Fig. 2). These subclasses share similar affinity in the micromolar range (Ka values of 6.96 × 106 m-1 for reqIgG7 and 6.79 × 106 m-1 for reqIgG4). Hence, binding affinity is much lower than that of protein G for IgG (26) and closer to that of fragment B of protein A (27).
FIGURE 2.
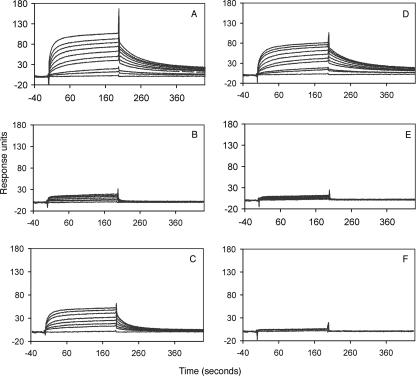
Biosensor analysis of reqIgG binding to immobilized FgBP1. Measurements were carried out at different concentrations (0, 35, 50, 75, 100, 125, and 150 μg/ml) for the soluble analyte equine IgG4 (A) and equine IgG7 (B), respectively. Measurements for the remaining five subclasses were carried out at 150 μg/ml only (C). The response curves shown represent specific binding to FgBP1 (following subtraction of any nonspecific binding to the FgBPM- control). Only IgG4 and IgG7 showed concentration-dependent interaction with immobilized FgBP1. RU, response units.
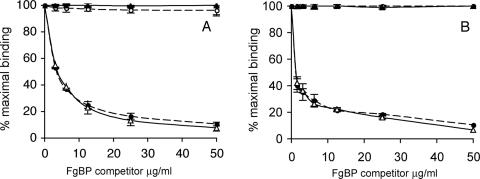
Localization of the FgBP Binding Site on Equine IgG—Protein A and protein G are known to bind in the interdomain region of IgG Fc (22, 23) and have both been shown to interact with reqIgG4 and reqIgG7, although the binding to protein A is relatively weak (16). Therefore, we used a competition ELISA to see if FgBP could inhibit binding of proteins A and G to reqIgG4 and reqIgG7. We found that FgBP1, but not the non-IgG-binding control protein FgBPM-, was able to compete with both protein A and protein G for binding to reqIgG4 and reqIgG7 (Fig. 3). This inhibition was concentration-dependent, and binding of both protein A and protein G was negligible at the maximum concentration of FgBP1 (50 μg/ml). These findings suggest that the FgBP binding site(s) on reqIgG4 and reqIgG7 overlap with those of protein A and protein G in the Fc interdomain region.
FIGURE 3.
ELISA analysis of competition for binding to reqIgG4 and reqIgG7 between FgBP proteins and protein A and protein G. A, competition of binding of HRP-conjugated protein G (0.25 μg/ml) to reqIgG4 and reqIgG7 coated at 2.5 μg/ml. B, competition of binding of HRP-conjugated protein A (20 μg/ml) to reqIgG4 and reqIgG7 coated at 5 μg/ml. The figures shows mean (±S.E.) of three independent experiments. reqIgG4 with FgBP1 competitor (closed circle) and FgBPM- competitor (open circle); reqIgG7 with FgBP1 competitor (open triangle) and FGBPM- competitor (closed triangle).
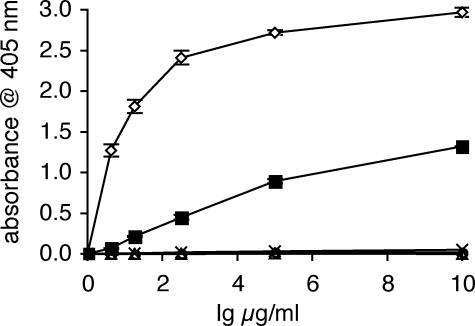
To determine whether one or both of the IgG Fc domains contribute to FgBP binding, we utilized recombinant human IgG1 and two human IgG1/IgA1 domain-swap mutants. Using biosensor analysis (data not shown), we found that human IgG1 interacted with FgBP1 with an affinity (Ka of 5.71 × 106 m-1) only slightly lower than that of reqIgG4 and reqIgG7. Like eqIgA, human IgA1 was found not to bind FgBP1 (Fig. 4). A domain-swap mutant lacking the CH2 domain but retaining the CH3 domain of IgG1 (α1α2γ3) was able to bind to FgBP1, but at a much reduced level compared with that of wild-type human IgG1 (Fig. 4). However, FgBP1 binding was completely absent for a mutant retaining the CH2 domain but lacking the CH3 domain of IgG1 (γ1γ2α3) (Fig. 4). Hence, both Fc domains are required for maximal binding, but the CH3 domain appears to be the most critical.
FIGURE 4.
ELISA analysis of FgBP1 binding to human IgG1, human IgG1/IgA1 domain-swap mutants and human IgG3 allotype IGHG3*01. Binding of Igs is expressed as absorbance obtained for the binding to FgBP1 minus absorbance for binding to the FgBPM- control (binding to the latter was typically <0.15 absorbance units). Fig. shows mean (±S.E.) of three independent experiments. Open diamond, human IgG1; open triangle, human IgA1; closed square, α1α2γ3 mutant; open circle, γ1γ2α3 mutant; cross, human IgG3 IGHG3*01 allotype.
In addition, we included in this analysis a recombinant human IgG3 antibody of the IGHG3*01 allotype. IgG3 allotypes capable of binding protein A possess a His residue at position 435 within the interaction site for protein A. However, the IGHG3*01 allotype carries an Arg at this critical CH3 domain residue and is unable to bind protein A (20, 21). We found that the IGHG3*01 allotype did not bind FgBP1, indicating that residue 435 is critical also for interaction with FgBP (Fig. 4).
To further localize the FgBP binding site on equine IgG, we generated five IgG7 mutants with single or double point mutations in the regions corresponding to the protein A/protein G binding sites (Table 1). Analysis of the IgG7 mutants by SDS-PAGE and Western blotting revealed that all the mutant reqIgG7s assembled like wild-type reqIgG7 and retained the anticipated reactivity with antigen and both polyclonal and monoclonal anti-equine IgGb (i.e. anti-eqIgG4 and -eqIgG7) reagents (Fig. 5). These test outcomes and the fact that the substitutions were based on residues present in other equine IgG isotypes suggest that the likelihood of unintended conformational disruption in the mutants is extremely low, allowing us to draw meaningful conclusions on the relative contributions of the mutated residues to FgBP binding. Binding of the mutant reqIgG7s to FgBP1 was analyzed using surface plasmon resonance experiments (Fig. 6). Mutants M252K and K382Q both showed decreased FgBP1 binding compared with wild-type reqIgG7. However, the 45-fold decrease in FgBP1 binding by M252K (Ka of 1.53 × 105 m-1) was more dramatic than the 2.4-fold decrease shown by K382Q (Ka of 2.91 × 106 m-1). Mutant H433R showed a small (1.7-fold, Ka of 4 × 106 m-1) reduction in FgBP1 binding capacity compared with wild-type reqIgG7. However, the double point mutants HN433/434EH and HY435/436TV showed drastically impaired abilities to bind FgBP1, retaining <5% of the binding capacity of wild-type reqIgG7. The FgBP1 binding affinities and interdomain site sequence alignment of the tested IgGs are summarized in Fig. 7. Taken together these results suggest that the binding site on IgG for FgBP encompasses some of the same interdomain residues as those involved in interaction with protein A and protein G. Residues in the Leu251-Ser254 and the Leu432-Tyr436 loops are particularly implicated. Comparison of the binding affinities of mutants H433R and HN433/434EH suggests that either His433 is not critical for FgBP binding, or that alteration of this residue to a positively charged amino acid such as arginine (as in H433R) can be tolerated without significant impact on the affinity of the interaction. In contrast, residues His435 and/or Tyr436 appear to be critical for the interaction, because in both instances where these residues are substituted (with Arg435 and Phe436 in human IgG3 allotype IGHG3*01 and with Thr435 and Val436 in reqIgG7 mutant HY435/436TV) binding to FgBP1 is ablated. Overall, we can conclude that the binding site for FgBP on reqIgG Fc appears to center on the interdomain region, utilizing residues common to both the protein A and protein G binding sites.
FIGURE 5.
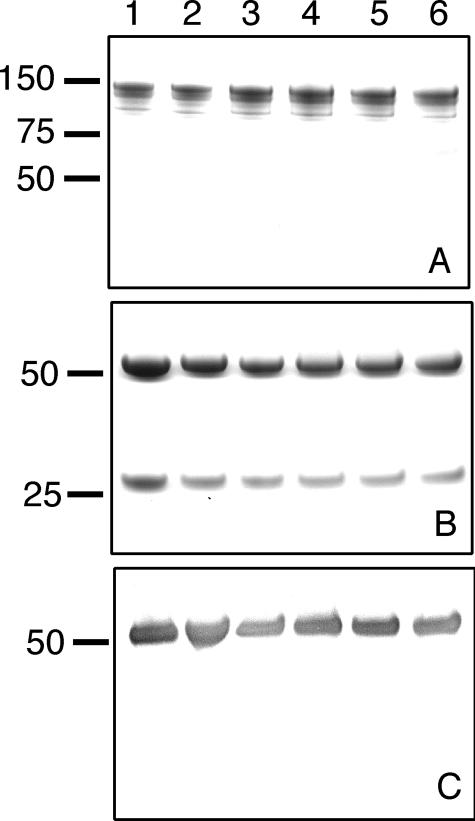
Analysis of reqIgG7 mutants by SDS-PAGE and Western blotting. Proteins were run under non-reducing (A) and reducing (B and C) conditions and stained with Coomassie Brilliant blue (A and B) or immunoblotted and probed with polyclonal goat anti-horse IgGb which recognizes reqIgG4 and reqIgG7 (C). Lane 1, wild-type reqIgG7; lane 2, M252K; lane 3, K382Q; lane 4, H433R; lane 5, HN433/434EH; lane 6, HY435/436TV.
FIGURE 6.
Biosensor analysis of the binding of FgBP1 to reqIgG7 mutants. Injections of 100 μl (30 μl/min) of reqIgG7s at 0, 5, 10, 25, 35, 50, 75, 100, 125, and 150μg/ml are shown. A, wild-type reqIgG7; B, M252K; C, K382Q; D, H433R; E, HN433/434EH; F, HY435/436TV. The response curves shown represent specific binding to FgBP1 (following subtraction of any nonspecific binding to the FgBPM- control).
FIGURE 7.
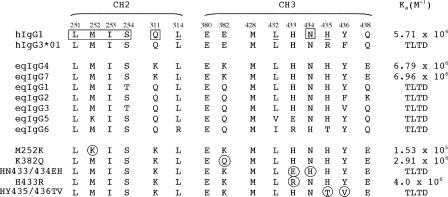
Amino acid alignments of reqIgG subclasses and reqIgG7 mutants showing only CH2 and CH3 residues critical for interaction with protein A and protein G. Residue numbers (shown above sequences) correspond to human IgG1 numbering. In the corresponding hIgG1 sequence (shown for comparison at the top), residues implicated in interactions are indicated as follows: protein G, normal text; protein A, underlined; both protein A and protein G, box. The corresponding sequence for human IgG3 allotype IGHG3*01 (hIgG3*01) is also shown. In the middle section, the FgBP-binding subclasses eqIgG4 and eqIgG7 are shown above the non-binding subclasses. reqIgG7 mutants are shown in the lower part of the figure, with substitutions encircled. Ka values for interaction with FgBP1 are shown on the right. TLTD, too low to detect.
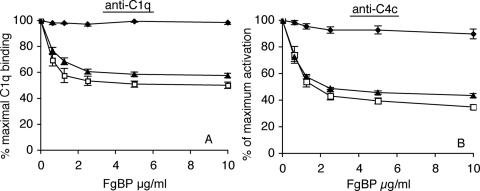
Impact of FgBP Binding to IgG on Complement Activation—We have previously demonstrated that reqIgG1, reqIgG3, reqIgG4, and reqIgG7 bind strongly to C1q and activate the classical complement pathway (16). To gain an insight into the physiological consequences of FgBP binding to eqIgG4 and eqIgG7, we investigated whether FgBP1 could inhibit their ability to bind C1q and activate the classical complement pathway. We found that FgBP1 was able to disrupt C1q binding and complement activation by reqIgG4 and reqIgG7 (Fig. 8). In contrast, FgBP1 was unable to disrupt C1q binding and complement activation by reqIgG3, which we have shown binds poorly or not at all to FgBP1 (Fig. 8). The control protein, FgBPM-, had no effect on C1q binding or complement activation. With the highest concentration of FgBP1 used (10 μg/ml), C1q binding and complement activation by reqIgG4 and reqIgG7 decreased to 50 and 40%, respectively, of that seen in the absence of FgBP1. This partial rather than complete inhibition of antibody-mediated complement activation may be a consequence of using soluble rather than cell-bound FgBP (see “Discussion” below).
FIGURE 8.
Disruption of antibody-mediated complement activation by FgBP1. Increasing concentrations of FgBP1 were able to disrupt C1q binding, detected using anti-C1q antibody (A), and activation of the classical complement pathway, detected using anti-C4c antibody (B), by reqIgG4 and reqIgG7 but not by reqIgG3, which binds poorly to FgBP1. Closed diamond, reqIgG3; open square, reqIgG4; closed triangle, reqIgG7. In each case, C1q binding/activation are shown as a percentage of the binding/activation seen in the absence of FgBP1. The figure shows mean (±S.E.) of three independent experiments.
DISCUSSION
Ig-binding proteins have been identified in a range of bacteria, and their diversity in Ig binding characteristics in terms of Ig isotype, subclass, and species has long been recognized. The horse has an unusually large number of IgG subclasses, which vary in their structural and functional attributes (14-16). In this study we addressed the eqIgG subclass specificity of FgBP and found that this bacterial protein is highly specific for only two (eqIgG4 and eqIgG7) of the seven eqIgG subclasses. eqIgG4 and eqIgG7 are 97% identical at the amino acid level, and prior to genomic identification were together thought to comprise a single subclass termed IgGb (15, 28). For all the binding characteristics and effector functions thus far tested, these two subclasses share common features, presumably due to their high sequence similarity (16).
We have demonstrated that FgBP binds to the interdomain region of IgG Fc and that both of the Fc domains are required for full binding. The residues implicated in our mutagenesis studies as critical for binding to FgBP are known to mediate interaction with staphylococcal protein A and streptococcal protein G also (22, 23). Indeed, there is a striking overlap of the regions recognized by these three distinct bacterial proteins (Fig. 9), despite the fact that they derive from very different streptococcal and staphylococcal strains and these bacteria are pathogenic in different mammalian species.
FIGURE 9.
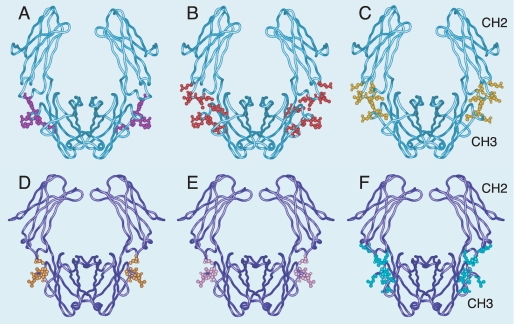
Interaction sites for streptococcal and staphylococcal Ig-binding proteins on IgG and IgA Fc regions. The CH2 and CH3 domains of the heavy chains of IgG (A-C) (PDB accession: 1FC1) and IgA (D-F) (PDB accession: 1OW0) are shown in light blue and dark blue, respectively. Residues shown to be critical for interaction by mutagenesis studies and/or by x-ray crystallography of complexes are highlighted by spheres. A, FgBP from S. equi in purple; B, protein G from group C and group G streptococci in red; C, protein A from S. aureus in yellow; D, Sir22 from S. pyogenes in orange; E,β protein from Group B streptococcus in pink; F, SSL7 from S. aureus in green. Remarkably, all the Ig-binding proteins target the CH2-CH3 domain interface, regardless of specificity for IgG or IgA, streptococcal or staphylococcal origin, or host species (equine or human).
Even more remarkably, the fact that these unrelated IgG-binding proteins bind to similar sites in the IgG interdomain region parallels the situation for the unrelated bacterial IgA binding proteins Sir22 from S. pyogenes, β protein from group B streptococcus, and SSL7 from Staphylococcus aureus, which all bind to the Fc domain interface in human IgA (29-31) (Fig. 9). Although a different immunoglobulin class is involved, again we see the equivalent region being targeted by proteins produced by very different bacterial pathogens. Thus, it seems that convergent evolution may have favored the appearance of bacterial proteins that bind to the CH2/CH3 interface in IgG and IgA. This interdomain region, in IgG at least, has been recognized as one of only a limited number of regions on the Ig surface that is particularly suited to protein-protein interaction (32, 33).
The evolutionary reasons why such sites of relative vulnerability have been retained on the surface of Ig Fc regions are likely to be complex (34) but most probably relate to their roles as interaction sites for key host receptors. In IgG, for example, the Fc interdomain region forms the interaction site for FcRn, the so-called neonatal Fc receptor that mediates a number of processes fundamental to IgG function, including regulation of IgG turnover and transepithelial transfer of IgG (35). For IgA, the CH2/CH3 interdomain region serves as the interaction site for its specific Fc receptor, FcαRI, which triggers efficient elimination mechanisms against IgA-coated pathogens (18, 36).
Despite the marked overlap in their binding sites on IgG, the eqIgG subclass specificities for FgBP, protein G, and protein A are distinct. Although FgBP binds almost exclusively to eqIgG4 and eqIgG7, protein G binds strongly to eqIgG1, eqIgG4, and eqIgG7 and shows intermediate binding to eqIgG3, low binding to eqIgG2 and eqIgG6, and no binding to eqIgG5 (16). Binding of eqIgGs to protein A is generally quite low with the subclasses binding in the following order eqIgG1 > eqIgG3 = eqIgG4 = eqIgG7 > eqIgG2 = eqIgG5 > eqIgG6 (16). His435 in the CH3 domain of IgG, which is critical for binding to protein A, either alone or together with its neighbor Tyr436, also appears to be a requirement for binding to FgBP. Thus deviations from the His435-Tyr436 motif may account, at least in part, for the inability of eqIgG6 and eqIgG2 to bind FgBP. Although our mutagenesis experiments have pinpointed amino acid differences that may account for the deficiency of some of the eqIgG subclasses in FgBP binding, we predict that there may be additional residues in the eqIgG Fc outside the protein A and protein G binding sites that make unique contributions to the IgG-FgBP interaction.
Together eqIgG4 and eqIgG7 are the most prevalent forms of IgG in serum and are also found in mucosal secretions (37). Furthermore, both subclasses are able to stimulate a strong respiratory burst from equine peripheral blood leukocytes and effectively activate complement via the classical pathway (16). Accumulated evidence has highlighted that antibodies, in particular those that recognize FgBP, play a vital role in protection against strangles (38-40). Furthermore, FgBP-specific IgGb (i.e. eqIgG4/eqIgG7) has been recognized as the predominant IgG subclass present in the sera of acute and convalescent strangles cases and in the nasal mucosa during the acute response (13). The non-immune binding of antibodies by bacterial immunoglobulin binding proteins is presumed to subvert the host immune response. Functionally, this could be through prevention of initial antibody binding to bacterial epitopes, evasion of detection by other components of the immune response by coating of bacteria with host antibody, or by blocking of Fc-mediated effector functions such as complement activation or Fc-receptor binding. Several studies have investigated the ability of bacterial Ig binding proteins to interfere with the effector functions of IgG. The majority of available evidence appears to argue against the ability of such proteins to inhibit the binding of IgG to FcγR on phagocytic cells (41). However, protein A has been shown to be capable of inhibiting C1q binding to surface-immobilized IgG, thereby preventing complement activation (42). Similarly, the binding of protein H from S. pyogenes to surface-immobilized IgG can inhibit C1q binding and partially block complement-mediated lysis (42). In the current study, binding of FgBP to surface-immobilized eqIgG4 and eqIgG7 was clearly able to disrupt the binding of C1q and prevent subsequent activation of the classical complement pathway. Our results indicate that the observed inhibition of C1q binding and complement activation depends on the binding of FgBP to eqIgG, because when an eqIgG subclass that is unable to bind FgBP is used (eqIgG3), no inhibition is seen. Because specific antibody and complement have both been shown necessary to combat S. equi (43), such targeting of the major complement-activating eqIgG subclasses may afford the bacterium an effective means to disrupt antibody-mediated elimination.
The observed FgBP-mediated inhibition of both C1q binding to IgG and complement activation, although substantial, was not complete. Our use of a soluble version of FgBP may have underestimated the potency, in this regard, of FgBP, which in its natural form is an abundant cell wall-associated protein (5-7). One might speculate that anchoring of IgG close to the bacterial surface by cell wall-bound FgBP is likely to increase the chance of steric hindrance. Thus the approach and binding of such a large molecule as C1q would be compromised, resulting in more complete inhibition of the complement pathway. Support for this notion comes from related studies with protein G. Although one study found soluble streptococcal protein G unable to block both C1q binding to IgG and complement-mediated lysis of IgG-sensitized red blood cells (42), another demonstrated that, when IgG is bound by bacterial cell-surface protein G, interaction with C1q is completely blocked (44). The authors of the second study proposed that the presence of protein G in a defined orientation and spacing on the streptococcal cell surface facilitates inhibition of the interaction of C1q with protein G-bound IgG. In a similar fashion, one might argue that the particular spatial arrangement of FgBP on the surface of S. equi cells may effectively prevent the approach and interaction of bulky C1q molecules with any IgG that is bound to FgBP. Thus we consider that there are good reasons to believe that the FgBP-IgG interaction is likely to make an important contribution to the pathophysiology of strangles, at least in part through inhibition of complement-mediated clearance mechanisms.
Under the physiological conditions of plasma, one might expect binding of both fibrinogen and IgG to FgBP, assuming access/steric hindrance is not an issue. Further experiments will be necessary to assess the impact that fibrinogen binding might have on the interaction of IgG with FgBP and on the prevalent mechanism of inhibition of complement activation (45).
S. equi has evolved multiple secreted and cell surface-associated factors that interact with host proteins and promote adherence and/or help to evade immune responses (1). These include another immunoglobulin-binding protein, the α2-macroglobulin, albumin, and IgG-binding protein or EAG (46). The equine IgG subclass specificity and the biological effects of IgG binding by EAG have not been elucidated and would make an informative comparison with FgBP, particularly because this protein is the subject of vaccine studies (47-49). Interestingly, protein ZAG, the homologue of EAG present in S. equi subsp. zooepidemicus, is reported to bind IgG from a broader range of species than FgBP and to bind horse IgG (subclass not specified) with a much higher affinity (Ka of 1 × 1010 m-1) (50).
In conclusion, we have shown that the Fc-mediated binding of equine IgG by S. equi FgBP is highly specific for the eqIgG4 and eqIgG7 subclasses. The FgBP-IgG interaction provides an effective means by which S. equi subsp. equi can circumvent antibody effector function. In common with several other streptococcal and staphylococcal IgG- and IgA-specific Ig-binding proteins, FgBP binds to the domain interface of the Ig Fc region, illustrating a common mode of interaction that has been adopted by these important virulence factors.
Acknowledgments
We are grateful to Adele Page for technical assistance with BIAcore experiments.
Author's Choice—Final version full access.
This work was supported by the Wellcome Trust (Grant 074863) and the Privy Purse Charitable Trust. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FgBP, fibrinogen-binding protein; reqIgG, recombinant equine IgG; reqIgA, recombinant equine IgA; PBS, phosphate-buffered saline; HRP, horseradish peroxidase; H-chain, heavy chain; EAG, α2-macroglobulin, albumin, and IgG-binding protein; ELISA, enzyme-linked immunosorbent assay.
M. Meehan and P. Owen, unpublished observations.
M. J. Lewis, B. Wagner, R. Irvine, and J. M. Woof, manuscript in preparation.
References
- 1.Harrington, D. J., Sutcliffe, I. C., and Chanter, N. (2002) Microb. Infect. 4 501-510 [DOI] [PubMed] [Google Scholar]
- 2.Timoney, J. F. (2004) Vet. Res. 35 397-409 [DOI] [PubMed] [Google Scholar]
- 3.Taylor, S. D., and Wilson, W. D. (2006) Clin. Tech. Equine Pract. 5 211-217 [Google Scholar]
- 4.Newton, J. R., Verheyen, K., Talbot, N. C., Timoney, J. F., Wood, J. L. N., Lakhani, K. H., and Chanter, N. (2000) Equine Vet. J. 32 515-526 [DOI] [PubMed] [Google Scholar]
- 5.Galan, J. E., and Timoney, J. F. (1987) Infect. Immun. 55 3181-3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timoney, J. F., Artiushin, S. C., and Boschwitz, J. F. (1997) Infect. Immun. 65 3600-3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meehan, M., Nowlan, P., and Owen, P. (1998) Microbiology 144 993-1003 [DOI] [PubMed] [Google Scholar]
- 8.Meehan, M., Kelly, S. M., Price, N. C., and Owen, P. (2002) FEMS Microbiol. Lett. 206 81-86 [DOI] [PubMed] [Google Scholar]
- 9.Meehan, M., Lynagh, Y., Woods, C., and Owen, P. (2001) Microbiology 147 3311-3322 [DOI] [PubMed] [Google Scholar]
- 10.Boschwitz, J. S., and Timoney, J. F. (1994) Microb. Pathog. 17 121-129 [DOI] [PubMed] [Google Scholar]
- 11.Meehan, M., Muldowney, D. A., Watkins, N. J., and Owen, P. (2000) Microbiology 146 1187-1194 [DOI] [PubMed] [Google Scholar]
- 12.Meehan, M., Muldowney, D. A., O'Meara, F., and Owen, P. (2000) FEMS Microbiol. Lett. 190 317-321 [DOI] [PubMed] [Google Scholar]
- 13.Sheoran, A. S., Sponseller, B. T., Holmes, M. A., and Timoney, J. F. (1997) Vet. Immunol. Immunopathol. 59 239-251 [DOI] [PubMed] [Google Scholar]
- 14.Wagner, B., Greiser-Wilke, I., Wege, A. K., Radbruch, A., and Leibold, W. (2002) Immunogenetics 54 353-364 [DOI] [PubMed] [Google Scholar]
- 15.Wagner, B., Miller, D. C., Lear, T. L., and Antczak, D. F. (2004) J. Immunol. 173 3230-3242 [DOI] [PubMed] [Google Scholar]
- 16.Lewis, M. J., Wagner, B., and Woof, J. M. (2008) Mol. Immunol. 45 818-827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morton, H. C., Atkin, J. D., Owens, R. J., and Woof, J. M. (1993) J. Immunol. 151 4743-4752 [PubMed] [Google Scholar]
- 18.Pleass, R. J., Dunlop, J. I., Anderson, C. M., and Woof, J. M. (1999) J. Biol. Chem. 274 23508-23514 [DOI] [PubMed] [Google Scholar]
- 19.Bruggemann, M., Williams, G. T., Bindon, C. I., Clark, M. R., Walker, M. R., Jefferis, R., Waldmann, H., and Neuberger, M. (1987) J. Exp. Med. 166 1351-1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Recht, B., Frangione, B., Franklin, E., and van Loghem, E. (1982) J. Immunol. 127 917-923 [PubMed] [Google Scholar]
- 21.van Loghem, E., Frangione, B., Recht, B., and Franklin, E. C. (1982) Scand. J. Immunol. 15 275-278 [DOI] [PubMed] [Google Scholar]
- 22.Deisenhofer, J. (1981) Biochemistry 20 2361-2370 [PubMed] [Google Scholar]
- 23.Sauer-Eriksson, A. E., Kleywegt, G. J., Uhlen, M., and Jones, T. A. (1995) Structure 3 265-278 [DOI] [PubMed] [Google Scholar]
- 24.Wolbink, G. J., Bollen, J., Baars, J. W., ten Berge, R. J. M., Swaak, A. J. G., Paardekooper, J., and Hack, C. E. (1993) J. Immunol. Meth. 163 67-76 [DOI] [PubMed] [Google Scholar]
- 25.Burton, D. R., and Woof, J. M. (1992) Adv. Immunol. 51 1-84 [DOI] [PubMed] [Google Scholar]
- 26.Åkerström, B., and Björck, L. (1986) J. Biol. Chem. 261 10240-10247 [PubMed] [Google Scholar]
- 27.Lancet, D., Isenman, D., Sjödahl, J., Sjöquist, J., and Pecht, I. (1978) Biochem. Biophys. Res. Commun. 85 608-614 [DOI] [PubMed] [Google Scholar]
- 28.Wagner, B. (2006) Dev. Comp. Immunol. 30 155-164 [DOI] [PubMed] [Google Scholar]
- 29.Pleass, R. J., Areschoug, T., Lindahl, G., and Woof, J. M. (2001) J. Biol. Chem. 276 8197-8204 [DOI] [PubMed] [Google Scholar]
- 30.Wines, B. D., Willoughby, N., Fraser, J. D., and Hogarth, P. M. (2006) J. Biol. Chem. 281 1389-1393 [DOI] [PubMed] [Google Scholar]
- 31.Ramsland, P. A., Willoughby, N., Trist, H. M., Farrugia, W., Hogarth, P. M., Fraser, J. D., and Wines, B. D. (2007) Proc. Natl. Aacd. Sci. U. S. A. 104 15051-15056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burton, D. R. (1985) Mol. Immunol. 22 161-206 [DOI] [PubMed] [Google Scholar]
- 33.DeLano, W. L., Ultsch, M. H., De Vos, A. M., and Wells, J. A. (2000) Science 287 1279-1283 [DOI] [PubMed] [Google Scholar]
- 34.Abi-Rached, L., Dorighi, K., Norman, P. J., Yawata, M., and Parham, P. (2007) J. Immunol. 178 7943-7954 [DOI] [PubMed] [Google Scholar]
- 35.Ghetie, V., and Ward, E. S. (2000) Annu. Rev. Immunol. 18 739-766 [DOI] [PubMed] [Google Scholar]
- 36.Herr, A. B., Ballister, E. R., and Bjorkman, P. J. (2003) Nature 423 614-620 [DOI] [PubMed] [Google Scholar]
- 37.Sheoran, A. S., Timoney, J. F., Holmes, M. A., Karzenski, S. S., and Crisman, M. V. (2000) Am. J. Vet. Res. 61 1099-1105 [DOI] [PubMed] [Google Scholar]
- 38.Timoney, J. F., and Trachman, J. (1985) Infect. Immun. 48 29-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galan, J. E., and Timoney, J. F. (1985) Infect. Immun. 47 623-628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galan, J. E., Timoney, J. F., and Lengemann, F. W. (1986) Infect. Immun. 54 202-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wines, B. D., Powell, M. S., Parren, P. W., Barnes, N., and Hogarth, P. M. (2000) J. Immunol. 164 5313-5318 [DOI] [PubMed] [Google Scholar]
- 42.Berge, A., Kihlberg, B.-M., Sjoholm, A. G., and Björck, L. (1997) J. Biol. Chem. 272 20774-20781 [DOI] [PubMed] [Google Scholar]
- 43.Boschwitz, J. S., and Timoney, J. F. (1994) Infect. Immun. 62 3515-3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nitsche-Schmitz, D. P., Johansson, H. M., Sastalla, I., Reissmann, S., Frick, I.-M., and Chhatwal, G. S. (2007) J. Biol. Chem. 282 17530-17536 [DOI] [PubMed] [Google Scholar]
- 45.Carlsson, F., Sandin, S., and Lindahl, G. (2005) Mol. Microbiol. 56 28-39 [DOI] [PubMed] [Google Scholar]
- 46.Lindmark, H., Jonsson, P., Engvall, E. O., and Guss, B. (1999) Res. Vet. Sci. 66 93-99 [DOI] [PubMed] [Google Scholar]
- 47.Flock, M., Jacobsson, K., Frykberg, L., Hirst, T. R., Franklin, A., Guss, B., and Flock, J.-I. (2004) Infect. Immun. 72 3228-3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flock, M., Karlström, Å., Lannergård, J., Guss, B., and Flock, J.-I. (2006) Vaccine 24 4144-4151 [DOI] [PubMed] [Google Scholar]
- 49.Waller, A., Flock, M., Smith, K., Robinson, C., Mitchell, Z., Karlström, Å., Lannergård, J., Bergman, R., Guss, B., and Flock, J.-I. (2007) Vaccine 25 3629-3635 [DOI] [PubMed] [Google Scholar]
- 50.Jonsson, H., Lindmark, H., and Guss, B. (1995) Infect. Immun. 63 2968-2975 [DOI] [PMC free article] [PubMed] [Google Scholar]