Abstract
Store depletion induces STIM1 to aggregate and relocate into clusters at ER-plasma membrane junctions where it functionally interacts with and activates plasma membrane channels that mediate store-operated Ca2+ entry (SOCE). Thus, the site of peripheral STIM1 clusters is critical for the regulation of SOCE. However, what determines the location of the STIM1 clusters in the ER-PM junctional regions, and whether these represent specific sites in the cell is not yet known. Here we report that clustering of STIM1 in the subplasma membrane region of the cell and activation of TRPC1-dependent SOCE are determined by lipid raft domains (LRD). We show that store depletion increased partitioning of TRPC1 and STIM1 into plasma membrane LRD. TRPC1 and STIM1 associated with each other within the LRD, and this association was dynamically regulated by the status of the ER Ca2+ store. Peripheral STIM1 clustering was independent of TRPC1. However, sequestration of membrane cholesterol attenuated thapsigargin-induced clustering of STIM1 as well as SOCE in HSG and HEK293 cells. Recruitment and association of STIM1 and TRPC1 in LRD was also decreased. Additionally STIM1D76A, which is peripherally localized and constitutively activates SOCE in unstimulated cells, displayed a relatively higher partitioning into LRD and interaction with TRPC1, as compared with STIM1. Disruption of membrane rafts decreased peripheral STIM1D76A puncta, its association with TRPC1 and the constitutive SOCE. Together, these data demonstrate that intact LRD determine targeting of STIM1 clusters to ER-plasma membrane junctions following store depletion. This facilitates the functional interaction of STIM1 with TRPC1 and activation of SOCE.
Store-operated Ca2+ entry (SOCE)3 is a critical Ca2+ entry mechanism that is ubiquitously present in all cell types. SOCE not only determines refilling of intracellular Ca2+ stores but also regulates a wide variety of cellular functions (1–3). The molecular basis for store-operated calcium entry has long remained an enigma (3–5). Recently STIM1, an ER Ca2+ sensor protein, was suggested to be involved in coupling of ER Ca2+ store depletion to activation of plasma membrane Ca2+ entry channels (6–8). Depletion of ER Ca2+ stores results in the relocation of STIM1 into puncta in the subplasma membrane region, which has been demonstrated to be the site at which Ca2+ entry occurs (8–12). Therefore, it has been proposed that store-operated Ca2+ (SOC) channels reside in the plasma membrane juxtaposing the puncta. Indeed, Orai1 and TRPC1, which are critical components of CRAC and SOC channels, respectively, have been shown to co-localize with STIM1 puncta in stimulated cells (13–17). Thus, the site of STIM1 clusters in the peripheral ER determines recruitment and activation of plasma membrane SOCE channels. However, what determines the location of the STIM1 clusters in the ER-PM junctional regions, and whether these represent specific sites in the cell are not yet known.
Plasma membrane lipid rafts domains (LRD), which contain high concentrations of cholesterol and sphingolipids, are known to function as centers for the assembly of signaling complexes. Such assembly is suggested to facilitate both specificity and the rate of signaling events by positioning functionally associated molecules in close proximity to each other (18–21). Caveolin 1, a cholesterol-binding protein that is involved in the generation of caveolar lipid rafts, has been previously suggested to be required for SOCE (21, 22). We reported earlier that TRPC1, a core component of SOC channels (23, 24), is assembled in a signaling complex with key Ca2+-signaling proteins from both the ER and plasma membrane (25) and that intact LRD are required for activation of TRPC1-mediated SOCE. These findings have been more recently confirmed using caveolin knock-out mice (26). It has been shown that disruption of caveolar LRD or deletion of caveolin 1 results in mislocalization of TRPC1 and decreased SOCE (25–27). Here, we have examined the role of lipid raft domain in STIM1-dependent regulation of SOCE. The results presented below demonstrate that intact plasma membrane lipid rafts are required for stimulation-dependent clustering of STIM1 at the ER-plasma membrane junctional regions and STIM1-dependent regulation of SOCE. Further, LRD facilitate the store-dependent interaction of STIM1 with TRPC1 and activation of TRPC1-SOC channels.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Reagents—HSG and HEK293 cells were cultured in MEM and Dulbecco's modified Eagle's medium, respectively, supplemented with 10% fetal bovine serum and antibiotics. Cells were transfected in OptiMEM with Lipofectamine reagent 2000 (Invitrogen, Carlsbad, CA) using standard procedures and were used 36–48-h post-transfection. All reagents were of molecular biology grade obtained from Sigma Aldrich, unless mentioned otherwise.
Caveolar Raft Preparation—HSG cells were washed with phosphate-buffered saline, pH 7.4, and lysed for 30 min on ice in prechilled TNE buffer (1% v/v Triton X-100, 25 mm Tris-HCl, 150 mm NaCl, and 5 mm EDTA pH 7.5) supplemented with 1× protease and phosphatase inhibitors (Roche Applied Science). Lysates were homogenized using a dounce homogenizer followed by a brief centrifugation. 1 ml of the postnuclear supernatant (PNS) was mixed with an equal volume of 80% sucrose (w/v), and overlaid with 6 ml of 35% sucrose followed by 4 ml of 5% sucrose (in TNE buffer). Samples were centrifuged at 34,000 rpm for 18 h at 4 °C. Ten 1.2-ml fractions were collected from the top of the tube and used as required. Detergent-free fractionation of caveolar rafts was done essentially as described in Ref. 28. Cells were lysed in 500 mm sodium carbonate (pH 11.0) solution, homogenized, and centrifuged. PNS (1 ml) was adjusted to 45% sucrose by mixing with 1 ml of 90% sucrose (w/v) in MBS buffer (25 mm MES-NaOH, 150 mm NaCl, pH 6.5) and overlaid with 6.5 ml of 35% sucrose followed by 3.5 ml of 5% sucrose. Centrifugation, fraction collection, and analysis were done as described above. Detergent-resistant LRD (R) and soluble (S) fractions were isolated as described in Ref. 29. To disrupt membrane rafts by cholesterol sequestration, HSG cells were treated for 1 h with 10 mm methyl-β-cyclodextrin (MβCD) at 37 °C in serum-free MEM and washed extensively prior to stimulation. For cholesterol replenishment following initial MβCD treatment, cells were incubated with 0.5 or 1 mg/ml of water soluble cholesterol complexed with 2.5 mm MβCD (1 h, 37 °C). Total protein was estimated by the Bradford method (Bio-Rad), and total cholesterol was analyzed by a Wako cholesterol E kit per the manufacturer's instruction.
Immunoprecipitation, Western Blotting, and Antibodies— Sucrose density gradients fractions 3–5 and 8–10, corresponding to caveolar rafts or buoyant fractions (BF) and soluble or heavy fractions (HF), respectively, were pooled and adjusted to 0.25 mg/ml with radioimmune precipitation assay buffer and immunoprecipitated with anti-STIM1 or anti-caveolin1 (Cav1) antibodies. HSG cells were stimulated with 2 μm Tg or DMSO (0.1% v/v) for 5 min at 37 °C in MEM, washed with ice-cold phosphate-buffered saline, and lysed in TNE buffer. Detergent-resistant LRD were isolated as above and resuspended in 1× radioimmune precipitation assay buffer supplemented with 0.1% SDS, 1% Triton X-100, 20% glycerol, 1 mm phenylmethylsulfonyl fluoride, and 1× protease and phosphatase inhibitor. Protein concentrations were adjusted to 1 mg/ml and immunoprecipitated with anti-STIM1 or anti-TRPC1 antibodies. Immunocomplexes were separated using protein A plus-agarose beads (Pierce), eluted with 50 μl of 1× SDS dye and resolved in 4–12% NuPAGE gels (Invitrogen) followed by Western blotting as described previously (16). For store repletion experiments, cells were first stimulated for 10 min with 100 μm Carbachol (CCh) in SES buffer without CaCl2, washed thoroughly, and placed in complete MEM with 1 mm CaCl2 for another 20 min. For GM1 dot blots, a 2-μl aliquot of each fraction from the density gradients were spotted manually onto nitrocellulose membranes, blocked with 5% bovine serum albumin, and probed with horseradish peroxidase-conjugated cholera toxin subunit B. Densitometric analysis of bands was performed using the Lumi-Imager software (Roche Applied Science). A detailed list of antibodies used is provided as supplemental Table S1.
Imaging—HSG cells were grown on glass-bottomed culture dishes (MatTek, Ashland, MA) and were transiently transfected with expression plasmids for ShTRPC1, NT-ShRNA (non targeting), YFP-STIM1, or YFP-STIMD76A mutant and were used either for confocal or TIRF imaging. For confocal studies cells were stimulated with 2 μm Tg for 5 min, washed with ice-cold phosphate-buffered saline, and fixed with 4% paraformaldehyde following surface staining for the caveolar marker ganglioside GM1 with Alexa Fluor 594-conjugated cholera toxin subunit B per the manufacturer's instructions (Molecular Probes). Images were acquired using confocal laser-scanning microscope (LSM 510 Meta; Carl Zeiss, Thornwood, NY). TIRFM imaging was conducted using an Olympus IX81 motorized inverted microscope (Olympus, Centre Valley, PA), as described previously (16). Briefly, cells were bathed in a Ca2+-free or Ca2+-containing standard extracellular solution (145 mm NaCl, 5 mm KCl, 1 mm MgCl2, 10 mm HEPES, 10 mm glucose, pH 7.4 (NaOH)). Excitation light was provided by a 20-milliwatt Argon Krypton laser. The 514-nm laser was directed into an Olympus TIRF illuminator attached to the rear port of the microscope and through a 514-band pass filter (BP 10 nm) to a TIRF-optimized Olympus Plan APO ×60 (1.45 NA) oil immersion objective. Emitted light was collected through a 525-band pass filter (BP 50 nm). Images were collected every 0.5 s using a Hamamatsu EM CCD camera (Hamamatsu, Tokyo, Japan) controlled using the MetaMorph imaging software (Molecular Devices, Downington, PA).
[Ca2+]i Measurements—Cells were grown on glass bottom dishes. Treatments with 10 mm MβCD or 5 μm Filipin-III for 1 h were done in serum-free MEM at 37 °C prior to Fura2-loading. Fluorescent measurements were performed as described before (16, 24). Each fluorescence trace (340/380 nm ratio) represents an average from at least 20–30 cells.
Electrophysiology—Cells were transferred to the recording chamber and perfused with an external Ringer's solution as described in Ref. 24. All electrophysiological experiments were performed in the tight-seal whole cell configuration at room temperature using an Axopatch 200B amplifier (Molecular Devices). Development of the current was assessed by measuring the current amplitudes at a potential of -80 mV, taken from high resolution currents in response to voltage ramps ranging from -90 to 90 mV over a period of 1 s imposed every 4 s (holding potential was 0 mV) and digitized at a rate of 1 kHz. For analysis, the current recorded during the first ramp was used for leak subtraction of the subsequent current records. Tg (2 μm), dissolved in the bath solution, was used to stimulate the cells.
Statistics—Data analysis was performed using Origin 7.0 (OriginLab, Northampton, MA). Statistical comparisons were made using analysis of variance. Experimental values are expressed as means ± S.D. or S.E. Differences in the mean values were considered to be significant at p < 0.05.
RESULTS
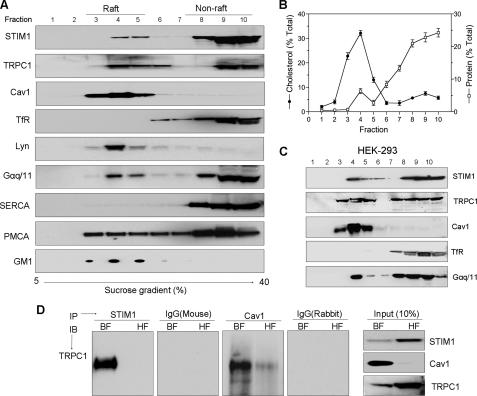
Localization of STIM1 and TRPC1 in the Lipid Raft Domain—Lipid rafts were isolated from two different cell lines (human submandibular gland, HSG, and HEK293) using density gradient ultracentrifugation. Individual fractions were collected, separated on SDS gels, and probed with the desired antibodies. Caveolin1 (Cav1), GM1, and Lyn were used as markers for LRD, while transferrin receptor (TfR), was used to identify non-raft compartments (Fig. 1, A and C). Total cholesterol and protein were also measured in these fractions to further identify the lipid raft-containing fractions (Fig. 1B). In HSG cell extracts, a fraction of endogenous STIM1 co-migrated with lipid raft markers in fractions 3–5 (low density or buoyant fractions, BF). However, a majority (75–80%) of the STIM1 was found in non-raft, heavy density fractions 8–10 (HF). Endogenous TRPC1, PMCA, and Gαq/11 were also partitioned into both lipid raft and non-raft fractions, while SERCA and TfR were detected in the heavy fractions. The BF also had relatively higher cholesterol to protein content than the HF (Fig. 1B). This further confirms that these fractions (BF) contain LRD. A similar distribution of proteins in the BF and HF fractions was observed in HEK293 cells as well (Fig. 1C). These findings were further confirmed using a detergent-free method to obtain the raft and non-raft fractions. A comparable partitioning of STIM1, TRPC1, and other proteins in BF and HF fractions was observed (supplemental Fig. S1). Thus, the distribution of proteins (STIM1, TRPC1) in raft and non-raft shown in Fig. 1 is not due to any artifacts induced by the use of Triton X-100.
FIGURE 1.
Lipid raft-associated STIM1 interacts with TRPC1. A, presence of proteins including STIM1 and TRPC1 density gradient fractions isolated from HSG cells demonstrate partitioning into raft and non-raft domains. B, total cholesterol and protein profiles in fractions shown in A. C, rafts association of STIM1 and TRPC1 in HEK293 cells. D, immunoprecipitation of endogenous TRPC1 using STIM1 and caveolin1 (Cav1) antibodies from pooled buoyant raft fractions 3–5 (BF) and from heavy non-raft fractions 8–10 (HF) of HSG cells. Respective IgGs were used as control; 10% of the inputs used for immunoprecipitation are indicated at the right.
The role of LRD in the association of TRPC1 and STIM1 (16, 17, 30) was assessed by immunoprecipitation using the raft (BF) and non-raft (HF) fractions collected from the gradient. As shown in Fig. 1D, the interaction between TRPC1 and STIM1 was primarily detected in BF fractions. Consistent with our previous results, TRPC1 is associated with Cav1 in this fraction. (Input levels of the proteins in the respective fractions are shown in Fig. 1D, adjacent panel.) Overall, these results indicate that STIM1 and TRPC1 partition into LRD and interact with each other preferentially within this domain.
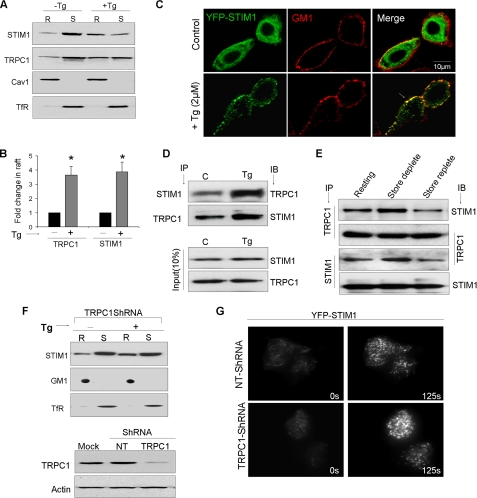
Activation-dependent Recruitment of STIM1 and TRPC1 into Lipid Raft Domains—The effect of Ca2+ store depletion on STIM1 partitioning into LRD was assessed by comparing its distribution in raft and non-raft fractions. Triton X-100 soluble (non-raft, S) and insoluble (raft, R) fractions were isolated from cells treated with 2 μm thapsigargin (+Tg) or vehicle (-Tg) and assessed for the presence of STIM1 and TRPC1. STIM1 was seen in both S and R fractions (Fig. 2A), consistent with its distribution in the sucrose density gradient described in Fig. 1A. Interestingly, as compared with the distribution in unstimulated cells, the fraction of STIM1 in rafts (R) was relatively increased (Fig. 2, A and B) in Tg-treated cells (>3-fold increase), with a concomitant decrease in the non-raft fraction. Similarly, TRPC1 also displayed stimulation-dependent increases in the raft fractions. Confocal microscopy further confirmed that co-localization of STIM1 with lipid raft marker GM1 was increased in cells stimulated with Tg (Fig. 2C and supplemental Fig. S2A) and carbachol (data not shown). Note the apparent co-localization of peripheral STIM1 puncta, but not the internal ones, with GM1.
FIGURE 2.
Store-dependent movement of STIM1 and TRPC1 into lipid raft domains. A, Western blots showing STIM1 and TRPC1 in detergent-resistant raft (R) and soluble (S) fractions obtained from control (C) or stimulated (2 μm Tg, 5 min) cells. B, bar graph summarizing optical densities (OD) obtained from five individual experiments that are plotted as mean ± S.D. ODs of untreated samples were set to 1. * indicate significant difference than control (p < 0.05). C, confocal images of HSG cells showing the co-localization of YFP-STIM1 punctae (green) with the caveolar marker ganglioside GM1 (red) in resting cells and after store depletion with Tg. D, immunoprecipitation of STIM1 and TRPC1 from raft fractions obtained from control and Tg-stimulated HSG cells. E, immunoprecipitations of STIM1 and TRPC1 using raft fractions isolated from CCh-treated cells under conditions where store is depleted and after store refilling (replete). F, Western blots showing STIM1 movement in control and TRPC1-Sh-RNA-expressing cells. Lower panel shows TRPC1 protein levels in TRPC1-ShRNA or a NT-ShRNA-expressing cells. Actin is used as a loading control. G, TIRF imaging on cells expressing either a control or TRPC1-Sh-RNA in HSG cells.
To determine whether the stimulation of cells affects the interaction between STIM1 and TRPC1 in LRD, immunoprecipitations were performed using STIM1 or TRPC1 antibodies on lipid raft preparations isolated from stimulated and unstimulated cells. Fig. 2D shows that STIM1-TRPC1 interactions were markedly increased upon stimulation. Store-dependent regulation of STIM1-TRPC1 interaction was examined by immunoprecipitations using raft preparations isolated from carbachol (CCh)-stimulated cells under conditions where ER stores were either depleted of Ca2+ or refilled. Stimulation with 100 μm CCh for 10 min in Ca2+-free medium resulted in an increase in TRPC1-STIM1 association. However, when stores were allowed to be refilled (replete) upon stimulation by washing off CCh, followed by re-addition of Ca2+ (1 mm), there was a relative decrease in the STIM1 interaction with TRPC1 (Fig. 2E). Similar findings were obtained with IP using either anti-TRPC1 or anti-STIM1 antibodies. Thus, together these data demonstrate that store depletion induces recruitment of STIM1 and TRPC1 into lipid rafts and that the status of ER Ca2+ stores predicts the magnitude of STIM1 and TRPC1 interaction.
To evaluate if TRPC1 is required for thapsigargin-mediated movement of STIM1 into lipid rafts, we silenced TRPC1 using sh-RNA as described before (16). Interestingly, thapsigargin-mediated recruitment of STIM1 into LRD was independent of TRPC1 (Fig. 2F). To demonstrate this more conclusively, we used total internal reflection fluorescence microscopy (TIRFM) and examined STIM1 translocation in thapsigargin-treated cells. Peripheral clustering of STIM1 was seen in control cells in response to thapsigargin and importantly, this movement of STIM1 was similar in cells expressing shTRPC1 (Fig. 2G). Note that this treatment decreases TRPC1 expression (Fig. 2F, lower panel) and SOCE in HSG cells (16). Overall, these data suggest that STIM1 movement and puncta formation in lipid raft domains is independent of TRPC1 expression, but is regulated by intracellular Ca2+ store depletion.
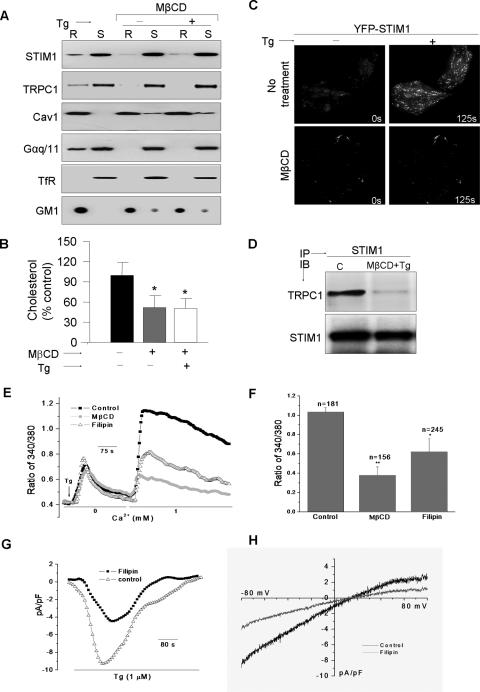
Disruption of Lipid Rafts Inhibits STIM1 Translocation and Decreases SOCE—The role of lipid rafts in targeting STIM1 clusters to the subplasma membrane region was assessed by examining the effect of LRD disruption. MβCD treatment, which depletes membrane cholesterol, decreased TRPC1 and STIM1 localization in lipid rafts (Fig. 3A and supplemental Fig. S2B) along with a ∼50% decrease in total cholesterol levels (Fig. 3B). Furthermore, it attenuated Tg-stimulated recruitment of STIM1 and TRPC1 into the raft domains (Figs. 3A and supplemental Fig. S2B). Note the relative increase of Cav1 in the non-raft fraction (internal controls for LRD disruption) as a result of reduction in the membrane cholesterol content following MβCD treatments (Fig. 3B). LRD proteins Gαq/11 and GM1 were also similarly affected while non-raft TfR was not. Importantly, replenishing of membrane cholesterol in HSG cells reversed the effect of MβCD and restored Tg-mediated STIM1 movement (supplemental Fig. S2B). Further, TIRFM demonstrated that thapsigargin-stimulated STIM1 puncta in the cell periphery was strongly decreased in cells treated with MβCD (Fig. 3C). However, the sequestration of membrane cholesterol did not have any significant effect on the internal aggregation of the STIM1 puncta, following store depletion (see epifluorescence image in supplemental Fig. S3A). Additionally, co-immunoprecipitation of TRPC1 and STIM1 from raft-fractions obtained after Tg stimulation was decreased in cells treated with MβCD prior to stimulation compared with untreated cells (Fig. 3D). These data suggest that intact LRD are required for clustering of STIM1 in the subplasma membrane region of the cells, and its increased association with TRPC1 in the ER-plasma membrane junctional region in response to Ca2+ store depletion.
FIGURE 3.
Lipid raft integrity determines STIM1 clustering and SOCE. Conditions for raft disruption by MβCD or Filipin-III are described under “Experimental Procedures.” A, Western blots were performed using individual antibodies in raft and non-raft fractions as described in supplemental Table S1. B, quantification of total cholesterol expressed as mean ± S.D. from at least three individual experiments. * denotes groups that are significantly different (using analysis of variance) from control (p < 0.05), but not from each other. C, TIRF imaging was performed on HSG cells expressing YFP-STIM1, with and without MβCD treatment; acquired images reveal STIM1 punctae at 0 or 125 s post-Tg stimulation. D, immunoprecipitation demonstrating a requirement of membrane rafts for the functional association between endogenous STIM1 and TRPC1. E, Tg-stimulated Ca2+ mobilization and G, currents were measured as described in Ref. 4. F, indicates the averaged data and the number of cells (n) imaged. * denotes values significantly different from controls, and ** indicates values significantly different from both sets (p < 0.05). H, indicates the I-V curves with and without filipin treatment.
The consequence of LRD disruption on SOCE was examined by treating HSG cells either with MβCD or filipin, both of which deplete membrane cholesterol. Both treatments decreased SOCE without significantly affecting ER-Ca2+ stores (Fig. 3E, average data are shown in F). Tg-stimulated Ca2+ currents, ISOC, measured in HSG cells were also inhibited (∼50% decreased) by treatment with either reagent, which did not alter the I-V relationship of the current (Fig. 3, G and H). Additionally we measured the effect of MβCD on SOCE in HEK293 cells. This cell line has been widely used to study the role of STIM1 in the regulation of SOCE mediated via both CRAC and SOC channels (17, 32). Activation of both types of channels has been shown to require peripheral STIM1-clustering. As seen with HSG cells (Fig. 3C), MβCD treatment of HEK293 cells also decreased Tg-stimulated peripheral clustering of STIM1 and consequently SOCE (supplemental Fig. S2C and D, respectively). These data demonstrate that lipid rafts also determine STIM1-dependent activation of SOCE in other cell types.
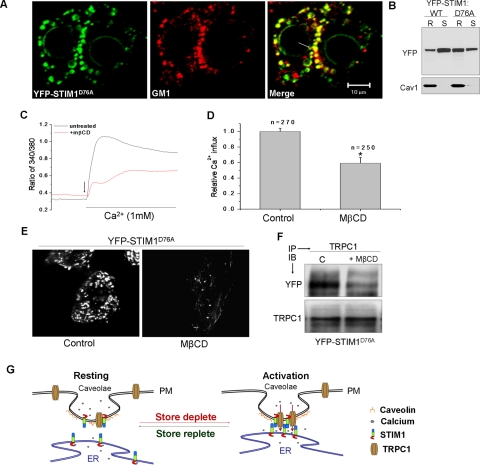
Lipid Raft Domains Are Essential for Constitutive Localization of STIM1D76A in Peripheral ER:— STIM1D76A has a mutation in the EF hand domain that renders it insensitive to ER-[Ca2+]. Hence it is predominantly aggregated in the plasma membrane region of the cell resulting in a constitutively SOCE in cells expressing this mutant protein (4, 6). It has been reported that YFP-STIM1D76A is localized in the ER from where it communicates with plasma membrane SOCE channels (10, 11). Consistent with previous reports, STIM1D76A was detected as clusters predominantly in the plasma membrane region of unstimulated cells (Fig. 4A). Notably these aggregates co-localized with the LRD marker, GM1 (see merged image). Further, a significant fraction of STIM1D76A was found to be a raft associated in unstimulated cells (Fig. 4B). Importantly, cells expressing STIM1D76A showed a high level of constitutive Ca2+ entry, which was significantly reduced (∼50%) upon treatment with MβCD (Fig. 4, C and D). Correspondingly, TIRFM demonstrated that MβCD decreased the constitutive STIM1D76A clustering in the plasma membrane region (Fig. 4E). However, STIMD76A aggregation per se was not affected by MβCD (supplemental Fig. S3B). Consistent with the constitutive Ca2+ entry in these cells, STIM1D76A showed relatively high association with TRPC1 even in unstimulated cells. Additionally STIM1D76A-TRPC1 interaction was disrupted in cells treated with MβCD (Fig. 4F). These findings further substantiate the requirement of plasma membrane LRD in targeting STIM1 to specific ER-PM junctional sites and in facilitating SOCE by compartmentalizing the functional STIM1-TRPC1 interactions.
FIGURE 4.
Lipid raft domains are essential for constitutive localization of STIM1D76A in the peripheral ER. A, confocal microscopy was performed on HSG cells expressing the D76A EF hand mutant of STIM1 (STIM1D76A). Pre-existing YFP-STIM1D76A punctae (green) co-localize with raft marker and GM1 (red), co-localization is indicated by an arrow. B represents raft and non-raft association of STIM1 and STIM1D76A. C, Ca2+ imaging was performed on cells expressing YFP-STIM1D76A treated with or without MβCD. Constitutive Ca2+ influxes were monitored by stimulating cells with the addition of 1 mm CaCl2 to the external medium (indicated by arrow). Averaged data and the number of cells (n) imaged are shown in D. * denotes values significantly different from control (p < 0.05). E, TIRFM images indicating YFP-STIM1D76A punctae sensitive to raft disruption. F, immunoprecipitations indicating dependence of membrane rafts for TRPC1 and STIM1D76A association. G, proposed model indicating raft recruitment of STIM1 as a step obligatory to SOCE.
DISCUSSION
Recent reports have established STIM1 as a critical regulatory protein for SOCE (4, 6). STIM1, a protein primarily localized in the ER, undergoes clustering and translocation to the subplasma membrane regions of the cells, where it displays punctate localization (8–10). Recent data suggest that STIM1 puncta in the peripheral region of the cells marks the location where the protein functionally interacts with plasma membrane SOCE channels and regulates Ca2+ entry. Indeed SOCE has been shown to occur at sites coincident with the STIM1 peripheral clusters (11, 12) of these puncta. STIM1-dependent clustering of CRAC channel component, Orai1, in the plasma membrane requires STIM1 puncta and is coincident with the location of the puncta (13, 14). Similarly, SOC component, TRPC1, is also co-localized with STIM1 clusters (16, 17). These results suggest that in order to mediate SOCE, STIM1 needs to be targeted to specific regions of the cell, where the likelihood of its interaction with plasma membrane SOC channel will be high. The mechanisms that determine the site of STIM1 clusters in the cell periphery as well as its functional interaction with PM and regulation of SOCE are not yet known.
Our findings provide novel data that suggest that plasma membrane LRD determine the peripheral clustering of STIM1 and regulation of SOCE. Our data demonstrate that internal Ca2+ store depletion increases the association of STIM1 with LRD. Further, this association appears to be critical for the formation of STIM1-punctae at the ER-plasma membrane junctional region of the cells. Disruption of the LRD by sequestering membrane cholesterol resulted in severe attenuation of STIM1 clustering near the plasma membrane. Furthermore, STIM1 partitioning into LRD in response to thapsigargin stimulation of cells was also inhibited by cholesterol depletion. Interestingly, the LRD association of STIM1 was restored by replenishing the depleted cholesterol. Coincident with this, raft-disrupted cells displayed reduced SOCE and ISOC. Importantly, STIM1D76A was constitutively clustered in the cell periphery and was present at relatively higher levels in LRD fractions compared with STIM1. Subplasma membrane localization of STIM1D76A was also determined by LRD integrity, because the depletion of plasma membrane cholesterol by MβCD treatment not only disrupted STIM1D76A targeting, but also its interaction with TRPC1 resulting in a significant reduction of the constitutive Ca2+ entry. Together, these data suggest a critical role for LRD in the peripheral clustering of STIM1 in the subplasma membrane region that occurs as a result of ER-Ca2+ store depletion and consequently in STIM1-dependent regulation of SOCE.
While recent studies have provided evidence that Orai proteins are core components of CRAC channels (31–33), previous studies had established a role for TRPC1 in SOCE and SOC channel function (23, 24, 34, 35). Furthermore, several recent studies show that TRPC1-dependent SOCE is regulated by STIM1 (34, 35). Additionally, there is an increase in the association of TRPC1 and STIM1 following Ca2+ store depletion (16, 17). The data presented above show that functional interaction between STIM1 and TRPC1 preferentially occurs within LRD and is dynamically regulated by ER Ca2+ store status; increases upon depletion and decreases when store is refilled. Thus, our data provide an important insight into the mechanism that is involved in the store-dependent regulation of TRPC1-SOC channels by STIM1. Based on our findings, we suggest that LRD in the plasma membrane provide a unique platform for clustering and interaction of STIM1 and TRPC1 in the ER-plasma membrane junctions (see model in Fig. 4G). While we do not know whether STIM1 is required for TRPC1 clustering and recruitment into LRD, decreasing TRPC1 expression did not affect STIM1 clustering. We propose that interaction, either direct or indirect, of STIM1 with LRD results in bringing the two membranes in close proximity to each other, which facilitates the critical association between STIM1 in the ER and TRPC1 in the plasma membrane that is involved in the activation of SOCE. Although we have not yet determined whether Orai1-STIM1 interactions also occur within LRD, previous studies have established that STIM1 puncta determine Orai1 clustering and activation in HEK293 cells (13–15). Our data show that LRD also determine STIM1 puncta in HEK293 cells and thus could also have a role in regulation of Orai1+STIM1-CRAC channels. However, further studies will be required to establish this.
The findings discussed above demonstrate that STIM1 clusters in peripheral ER that are formed in response to ER-Ca2+ store-depletion are coincident with LRD in the juxtaposed plasma membrane. We suggest that anchoring of STIM1 by plasma membrane LRD results in relatively stable ER-plasma membrane junctions that regulate SOCE. Ca2+ store depletion induces oligomerization of STIM1, which has been reported to occur prior to puncta formation in the cell periphery (8–10). The latter likely requires additional mechanisms for translocation and targeting of STIM1 oligomers to specific ER-plasma membrane junctional regions where STIM1 can interact with SOCE channels in the surface membrane (10–12). The coiled-coil domain in the C terminus of STIM1 is reported to be crucial for its aggregation while amino acids 425–671, which contain a serine-proline-rich region, appear to be important for the correct targeting of the STIM1 cluster to the cell periphery after calcium store depletion (13, 14). The polycationic region in the C-terminal tail of STIM1 also appears to help STIM1 targeting to PM region but is not essential for oligomerization after Ca2+ store depletion (12–14). Thus, aggregation of STIM1 that occurs in response to a decrease in ER-[Ca2+] and its translocation to the subplasma membrane region can be dissociated although the latter is dependent on the former. Our data demonstrate that clustering of STIM1 in the cell periphery, but not its aggregation per se, depends on plasma membrane lipid rafts. Although we have not mapped out the domain of STIM1 that is involved in its interaction with LRD, we suggest that the C terminus of STIM1 might either directly or indirectly interact with lipid or protein components of the LRD, and that this interaction serves to anchor STIM1 clusters in specific regions of the cell where it can interact with and regulate SOCE.
In conclusion, we report here that the clustering of STIM1 in the plasma membrane region, but not its aggregation per se, that occurs in response to ER Ca2+ store depletion is determined by plasma membrane LRD. We show that STIM1 association with LRD increases upon store depletion. This is further supported by our observation that the constitutively active STIM1D76A mutant exhibits increased partitioning into LRD. The disruption of LRD induced decreases in peripheral STIM1 clustering as well as SOCE. Consistent with this, disruption of LRD also attenuated recruitment of store-operated TRPC1 channels into LRD, and its functional association with STIM1. Together these data demonstrate that LRD anchors STIM1, and thus determines its localization in specific ER-plasma membrane junctions where it can functionally interact with plasma membrane channels and regulate SOCE.
Supplementary Material
Acknowledgments
We thank the confocal facility at UND for help. We thank Drs. Tobias Meyer for the STIM1 plasmids, James D. Foster (UND), and Suman R. Das (NIAID, National Institutes of Health) for reagents and discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DE017102 and 5P20RR017699 (to B. B. S.) and the Intramural Research Program of NIDCR (to I. S. A.). This work was also supported by Grant 0548733 from the National Science Foundation and a University of North Dakota Student fellowship award (to B. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
Footnotes
The abbreviations used are: SOCE, store-operated Ca2+ entry; LRD, lipid raft domains; MES, 4-morpholineethanesulfonic acid; MβCD, methyl-β-cyclodextrin; Tg, thapsigargin; ER, endoplasmic reticulum.
References
- 1.Berridge, M. J., Bootman, M. D., and Roderick, H. L. (2003) Nat. Rev. Mol. Cell Biol. 4 517-529 [DOI] [PubMed] [Google Scholar]
- 2.Parekh, A. B., and Putney, J. W. (2005) Physiol. Rev. 85 757-810 [DOI] [PubMed] [Google Scholar]
- 3.Putney, J. W., Jr. (1990) Cell Calcium 11 611-624 [DOI] [PubMed] [Google Scholar]
- 4.Putney, J. W., Jr. (2007) Cell Calcium 42 103-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatachalam, K., van Rossum, D. B., Patterson, R. L., Ma, H. T., and Gill, D. L. (2002) Nat. Cell Biol. 4 263-272 [DOI] [PubMed] [Google Scholar]
- 6.Lewis, R. S. (2007) Nature 446 284-287 [DOI] [PubMed] [Google Scholar]
- 7.Roos, J., DiGregorio, P. J., Yeromin, A. V., Ohlsen, K., Lioudyno, M., Zhang, S., Safrina, O., Kozak, J. A., Wagner, S. L., Cahalan, M. D., Velicelebi, G., and Stauderman, K. A. (2005) J. Cell Biol. 169 435-445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liou, J., Kim, M. L., Heo, W. D., Jones, J. T., Myers, J. W., Ferrell, J. E., Jr., and Meyer, T. (2005) Curr. Biol. 15 1235-1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, S. L., Yu, Y., Roos, J., Kozak, J. A., Deerinck, T. J., Ellisman, M. H., Stauderman, K. A., and Cahalan, M. D. (2005) Nature 437 902-905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liou, J., Fivaz, M., Inoue, T., and Meyer, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 29 9301-9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu, M. M., Buchanan, J., Luik, R. M., and Lewis, R. S. (2006) J. Cell Biol. 174 803-813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luik, R. M., Wu, M. M., Buchanan, J., and Lewis, R. S. (2006) J. Cell Biol. 174 815-825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, P., Lu, J., Li, Z., Yu, X., Chen, L., and Xu, T. (2006) Biochem. Biophys. Res. Commun. 350 969-976 [DOI] [PubMed] [Google Scholar]
- 14.Li, Z., Lu, J., Xu, P., Xie, X., Chen, L., and Xu, T. (2007) J. Biol. Chem. 282 29448-29456 [DOI] [PubMed] [Google Scholar]
- 15.Várnai, P., Tóth, B., Tóth, D. J., Hunyady, L., and Balla, T. (2007) J. Biol. Chem. 2007 282 29678-29690 [DOI] [PubMed] [Google Scholar]
- 16.Ong, H. L., Cheng, K. T., Liu, X., Bandyopadhyay, B. C., Paria, B. C., Soboloff, J., Pani, B., Gwack, Y., Srikanth, S., Singh, B. B., Gill, D. L., and Ambudkar, I. S. (2007) J. Biol. Chem. 282 9105-9116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, G. N., Zeng, W., Kim, J. Y., Yuan, J. P., Han, L., Muallem, S., and Worley, P. F. (2006) Nat. Cell Biol. 9 1003-1010 [DOI] [PubMed] [Google Scholar]
- 18.Galbiati, F., Razani, B., and Lisanti M. P. (2001) Cell 106 403-411 [DOI] [PubMed] [Google Scholar]
- 19.Simons, K., and Toomre, D. (2000) Nat. Rev. Mol. Cell Biol. 1 31-39 [DOI] [PubMed] [Google Scholar]
- 20.Okamoto, T., Schlegel, A., Scherer, P. E., and Lisanti, M. P. (1998) J. Biol. Chem. 273 5419-5422 [DOI] [PubMed] [Google Scholar]
- 21.Isshiki, M., and Anderson, R. G. (2003) Traffic 4 717-723 [DOI] [PubMed] [Google Scholar]
- 22.Ambudkar, I. S. (2006) Trends Pharmacol. Sci. 27 25-32 [DOI] [PubMed] [Google Scholar]
- 23.Liu, X., Singh, B. B., and Ambudkar, I. S. (2003) J. Biol. Chem. 278 11337-11343 [DOI] [PubMed] [Google Scholar]
- 24.Liu, X., Cheng, K. T., Bandyopadhyay, B. C., Pani, B., Dietrich, A., Paria, B. C., Swaim, W. D., Beech, D., Yildrim, E., Singh, B. B., Birnbaumer, L., and Ambudkar, I. S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 17542-17547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockwich, T. P., Liu, X., Singh, B. B., Jadlowiec, J., Weiland, S., and Ambudkar, I. S. (2000) J. Biol. Chem. 275 11934-11942 [DOI] [PubMed] [Google Scholar]
- 26.Murata, T., Lin, M. I., Stan, R. V., Bauer, P. M., Yu, J., and Sessa, W. C. (2007) J. Biol. Chem. 282 16631-16643 [DOI] [PubMed] [Google Scholar]
- 27.Brazer, S. C., Singh, B. B., Liu, X., Swaim, W., and Ambudkar, I. S. (2003) J. Biol. Chem. 278 27208-27215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song, K. S., Li, Shengwen., Okamoto, T., Quilliam, L. A., Sargiacomo, M., and Lisanti, M. P. (1996) J. Biol. Chem. 271 9690-9697 [DOI] [PubMed] [Google Scholar]
- 29.Legler, D. F., Micheau, O., Doucey, M. A., Tschopp, J., and Bron, C. (2003) Immunity 18 655-664 [DOI] [PubMed] [Google Scholar]
- 30.Yuan, J. P., Zeng, W., Huang, G. N., Worley, P. F., and Muallem, S. (2007) Nat. Cell Biol. 9 636-645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakriya, M., Feske, S., Gwack, Y., Srikanth, S., Rao, A., and Hogan, P. G. (2006) Nature 443 230-233 [DOI] [PubMed] [Google Scholar]
- 32.Vig, M., Beck, A., Billingsley, J. M., Lis, A., Parvez, S., Peinelt, C., Koomoa, D. L., Soboloff, J., Gill, D. L., Fleig, A., Kinet, J. P., and Penner, R. (2006) Curr. Biol. 16 2073-2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogan, P. G., and Rao, A. (2007) Trends Biochem. Sci. 32 235-245 [DOI] [PubMed] [Google Scholar]
- 34.Worley, P. F., Zeng, W., Huang, G. N., Yuan, J. P., Kim, J. Y., Lee, M. G., and Muallem, S. (2007) Cell Calcium 42 205-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ambudkar, I. S., Ong, H. L., Liu, X., Bandyopadhyay, B., and Cheng, K. T. (2007) Cell Calcium 42 213-223 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.