Linking the phagocyte defect in patients with chronic granulomatous disease (CGD)2 with the biochemical basis for oxygen consumption by stimulated neutrophils (1) represented a seminal advance in understanding the molecular basis for a key component of innate immunity. In subsequent decades, the catalytic and regulatory elements of the “respiratory burst oxidase” were elucidated, tacitly assuming all along that the NADPH-dependent oxidase under study represented a system uniquely expressed in phagocytic cells and dedicated to generating relatively large amounts of reactive oxygen species (ROS) destined to destroy invading microbes. Development of more sensitive analytical systems for ROS detection revealed that some physiological and pathophysiological events in non-phagocytic cells were associated with ROS generation, although the subcellular source of oxidants remained uncertain. With the identification of mox1 in 1999 (2) as a homolog of gp91phox, the catalytic component of the phagocyte oxidase, came the birth of the NADPH oxidase (NOX) protein family and the eventual identification of its seven members. With remarkable rapidity, many features of the structure, activity, cell biology, and physiology of the NOX proteins have been described, as summarized in several excellent and comprehensive recent reviews (3–5). This more circumscribed minireview provides an overview of the organizing features of the protein family, a summary of the physiology and pathophysiology in which NOX proteins participate (or might participate), and identification of some of the remaining unanswered questions in the field.
The Phagocyte Paradigm
The patriarch of the NOX protein family is gp91phox (NOX2), the heavy subunit of flavocytochrome b558, the catalytic component of the phagocyte NADPH oxidase (6). Associated with p22phox in plasma membrane and membranes of selected intracellular compartments, gp91phox operates as an electron transferase, shuttling electrons from NADPH in the phagocyte cytoplasm, across two nonequivalent hemes (7) buried in the membrane, to O2, the electron acceptor, thereby generating superoxide anion. Substantial evidence suggests a stepwise flow of electrons during oxidase activity: a single electron transfers sequentially from cytosolic NADPH to NOX2-associated FAD, to the more proximal heme in the membrane (Em7 = –225 mV) (7), to the distal heme (Em7 = –265 mV), and finally to molecular O2. The univalent nature of the reaction with O2 produces superoxide anion as the immediate product of the oxidase, with hydrogen peroxide subsequently generated by dismutation of the superoxide. As discussed below, some of the non-phagocyte NOX proteins display unanticipated and unexplained reactivity.
Although the crystal structure of flavocytochrome b558 has not been solved, mutagenesis data suggest that the NOX2 subunit contains both hemes, where they are bishistidine-ligated in parallel transmembrane helices, coordinated with His101– His115 and His209–His222, respectively (8). The currently accepted model resembles that suggested for FRE1, the iron reductase of Saccharomyces cerevisiae (9), with the paired hemes stacked between two α-helices, perpendicular to the plane of the membranes, and coordinated with histidines 12–13 amino acids apart in the linear sequence (10).
Although gp91phox is the catalytic component of flavocytochrome b558, its partner, p22phox, is essential for optimal activity. In myeloid cells, coexpression of p22phox with gp91phox and subsequent heterodimer formation are prerequisites for egress from the endoplasmic reticulum and proper localization in the plasma membrane. Expression of either subunit alone results in retention of the expressed protein in the endoplasmic reticulum, with subsequent degradation, in part mediated by the proteasome (11). Consistent with the requirement of heterodimer formation for stability of the individual subunits, neutrophils from patients with genetic defects in either gp91phox or p22phox lack both proteins (12). When expressed heterologously in COS-7 cells, gp91phox alone reaches the cell surface (albeit less efficiently), exhibits the same spectral properties as flavocytochrome b558, and contains hemes with mid-potentials of –264 and –233 mV, nearly the same as the native protein. However, membrane-dependent generation of superoxide in the cell-free system from the COS-7 system absolutely requires coexpression of both subunits (13).
The phagocyte oxidase is agonist-dependent, exhibiting no detectable constitutive activity, and transfers electrons and consumes oxygen nearly instantaneously when stimulated. Characterization of NOX2 benefited not only from seminal studies of patients with CGD, but also from the development of the broken cell superoxide-generating system. First described by Bromberg and Pick (14) using macrophages and by several other laboratories (15, 16) using neutrophil subcellular components, the broken cell system combines membrane and cytosol from resting phagocytes, which lack oxidase activity, in the presence of anionic amphiphiles to generate superoxide anion in an NADPH-dependent fashion. Availability of the broken cell system was essential for the identification of p47phox, p67phox, and Rac as cytosolic components required for phagocyte oxidase activity (17–19) and has been a powerful vehicle for applying a reductionist approach to dissecting the biochemistry of this oxidase.
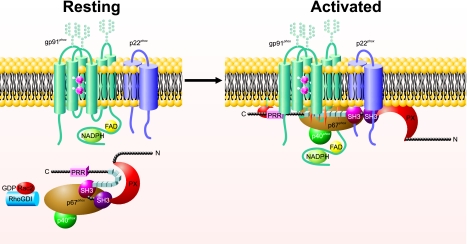
Regulation of the phagocyte system relies on spatial segregation of its essential components (Fig. 1); upon stimulation, regulatory components residing in the cytoplasm of resting phagocytes translocate to the target membranes and associate there with flavocytochrome b558 to assemble into a functional NADPH oxidase (20). In the cytoplasm of resting cells are two protein complexes, one composed of p47phox, p67phox, and p40phox and the other containing Rac and RhoGDI, the GDP dissociation inhibitor for Rho. Activation-dependent phosphorylation of p47phox and RhoGDI triggers conformational changes in both targets, thereby exposing otherwise cryptic sites in p47phox that bind p22phox, membrane phospholipids, and perhaps other sites and generating Rac-GTP that subsequently translocates to the membrane. Although the multiple intermolecular interactions that mediate oxidase assembly have been extensively studied, it suffices for the purpose of this minireview to recognize that regulation of the phagocyte oxidase depends on compartmentalization of its components in the resting cell and agonist-dependent assembly at the integral membrane protein, flavocytochrome b558.
FIGURE 1.
Assembly and activation of the phagocyte NADPH oxidase. In resting phagocytes, heterodimeric gp91phox-p22phox resides in the membrane, whereas the complex of p47phox-p67phox-p40phox is cytosolic. Agonist-triggered phosphorylation of the autoinhibitory domain of p47phox (series of small boxes) releases a conformational restriction, making interactive protein motifs, including the PX domain, Src homology 3 regions (SH3), and proline-rich regions (PRR), in p47phox accessible to associate at the membrane and to mediate oxidase assembly. See text for additional details.
Biochemical studies using the broken cell system have demonstrated convincingly that p47phox serves as an organizing adaptor protein that lacks intrinsic catalytic activity. On the other hand, p67phox represents an essential activating cofactor, possessing a domain that regulates the reduction of FAD by NADPH (21). Whereas p47phox and p67phox translocate to the membrane in a complex with p40phox, Rac2 translocates in its GTP-bound form independent of p47phox and p67phox (22). Rac2 participates in the catalytic activity of the phagocyte oxidase directly, via its interactions with p67phox, or both (23, 24). Additional factors modulate the stability and activity of the assembled complex, although the extent of their contribution to oxidase function is incompletely understood. In summary, the assembled and active phagocyte oxidase includes an electron transferase and associated organizing, activating, and stabilizing elements.
Features of the NOX Protein Family Members
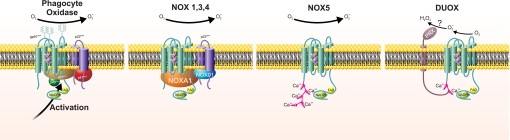
The seven-member NOX protein family is defined by its shared sequence homology with gp91phox: all members have six predicted transmembrane domains, motifs for NADPH and FAD binding, and conserved paired histidines that could ligate heme groups. NOX5, Duox1, and Duox2 have additional structural features not shared by the other NOX proteins. All three have cytosolic EF-hands (four in NOX5 and two in Duox proteins) that subserve calcium-dependent regulation of their oxidase activity. In addition, Duox has a long (>550 amino acids) N-terminal extracellular region with sequence homology to the animal peroxidases (Fig. 2). Although there is ∼25% amino acid identity between this N-terminal region and myeloperoxidase, the invariant histidines that provide the proximal and distal axial bonds with heme iron in animal peroxidases (25) are not conserved in Duox but are replaced by serine residues. This observation is not evidence against the region being a functional peroxidase domain, as alternative residues could form the heme pocket in Duox. However, the lack of homology in residues that characterize the animal peroxidases serves as a caveat to extrapolation from sequence similarity. Furthermore, were the terminal region in Duox capable of peroxidase activity, one would anticipate that it would support normal thyroid hormone synthesis in those patients in whom defective thyroid peroxidase activity causes hypothyroidism (26). However, thyroid hormone synthesis requires functional thyroid peroxidase, suggesting that Duox cannot mediate iodination of thyroid hormone. As iodination ranks low in the hierarchy of halogenation reactions mediated by animal peroxidases, the absence of a detectable contribution to thyroid synthesis suggests that Duox lacks peroxidase activity in this particular in vivo setting (27). Interferon-γ-induced augmentation of peroxidase activity by cultured human bronchial epithelial cells is inhibited by the stable expression of short hairpin RNA and parallels the decrease in Duox2 mRNA, consistent with Duox2 exhibiting peroxidase activity in this experimental setting. In the absence of spectral data or rigorous characterization of the enzymology of the purified protein or putative peroxidase domain, the function of this structural feature in Duox remains undefined.
FIGURE 2.
Structural organization of NOX protein family members. There are four different formats for the structural organization of members of the NOX protein family depending on the nature of their activity (constitutive versus agonist-dependent), the requirement of p22phox, dependence on cytosolic cofactors, the presence of EF-hands, and dependence on calcium. Duox alone has an additional transmembrane domain with an extracellular domain with limited sequence homology to animal peroxidases (PRX).
Whereas correct cellular localization and optimal function of flavocytochrome b558 require NOX2 to associate with p22phox, this organizational feature is not shared by all NOX protein family members; only NOX1, NOX3, and NOX4 also form heterodimers with p22phox. As with NOX2, the structural bases and the functional consequences of the association of NOX with p22phoxare not completely delineated. It is noteworthy that the EF-hand-containing, calcium-activated NOX proteins, specifically NOX5, Duox1, and Duox2, are those that function without an association with p22phox, perhaps hinting of a specific functional requirement fulfilled by p22phox or simply marking concurrent modifications during the evolution of the protein family (28).
Just as the phagocyte oxidase has components in addition to the membrane electron transferase, so too do related NOX proteins have associated factors. Homologs of p47phox and p67phox, named NOX organizing (NOXO1) and activating (NOXA1) proteins to reflect their presumed function, have been identified (29–31), isoforms of NOXO1 described (32, 33), and each implicated in the activity of NOX1–3. Of special note, dependence of NOX3 on NOXA1 exhibits species specificity; whereas murine NOX3 requires both NOXO1 and NOXA1 for maximal activity, optimal activity of human NOX3 depends on NOXO1 alone (30, 34). Human and murine NOXO1 are 67% identical at the amino acid level, with most differences at the C terminus. Human NOXO1 is 21 amino acids longer than its murine homolog and includes a proline-rich nonomer (PSQATAPPP) at the C terminus. Although the basis for this striking species difference is not understood, it raises important questions about the precise regulation of NOX3 activity by cytosolic factors. There are conflicting data regarding the contribution of Rac to NOX3 activity, and there is no evidence that Rac figures in the activities of NOX4, NOX5, or Duox proteins. For NOX5 and the Duox proteins, calcium serves as the definitive regulator, with phosphorylation of NOX5 providing a mechanism for regulating its sensitivity to ambient calcium concentrations (35). Taken together, the NOX proteins exhibit a full range of dependence on subunits (supplemental Table S1), from NOX2, which depends on multiple factors in the membrane and recruited from the cytoplasm, to Duox, operating apparently as an independent membrane protein.
Physiology
ROS are generated in a wide variety of biological contexts, both physiological and pathophysiological, and in nearly all tissues and organs of multicellular organisms, including animals, plants, and even microbes (36). Although NOX proteins represent the major non-mitochondrial sources of ROS, identification of the specific NOX protein(s) responsible for ROS production in a particular tissue or organ system has been undermined by limited availability of reliable tools for robust analysis. For example, whereas several well characterized antibodies against NOX2 are widely used, no such immunochemical probes exist at this time for NOX3. Antibodies for other non-phagocyte NOX proteins are variably or incompletely characterized, are nonspecific (i.e. recognize more than one NOX protein), or exhibit properties that dampen enthusiasm for their use (e.g. recognize protein in mice in which the gene encoding the NOX protein is not expressed). By necessity, tissue localization studies have relied on detection of mRNA and not protein, or the identification of p22phox, as a surrogate marker for NOX1–4. It is in this context, using the limited tools available, that NOX proteins have been identified in nearly every tissue and organ examined, often with more than one isoform implicated in a given biological activity. In some sites, identification of multiple isoforms likely reflects genuine isoform-specific subcellular locations or dedicated functions. For example, a cell might have a constitutively active NOX at the plasma membrane, providing a constant supply of oxidants at the cell surface to serve a specific extracellular purpose, whereas another NOX isoform in endosomes serves as an agonist-dependent source of oxidants as second messengers for intracellular signal transduction. When better analytical tools are applied to examine tissue expression of NOX proteins in situ, it will be possible to clarify ambiguities and to distinguish the apparent redundancy that results in organellar specialization of specific isotypes from misinformation that inadvertently arises from the use of nonspecific tools.
With the exception of the contribution of NOX2 to phagocyte-mediated antimicrobial activity, there are few unambiguous functional assignments of NOX proteins in specific tissues. NOX3-p22phox and NOXO1 are essential for normal otoconium formation in the inner ear, and the absence of any of the three components results in vestibular dysfunction (34, 37, 51). Duox serves as the H2O2 source in the thyroid that supports thyroid peroxidase-dependent iodination of thyroid hormone (38). Although most cases of congenital hypothyroidism reported to date reflect mutations in thyroid peroxidase, inherited defects in Duox are responsible in some patients (39). Duox also is implicated in mucosal host defense by airway epithelium (40), providing oxidants to support the lactoperoxidase-H2O2-thiocyanate antimicrobial system (41, 42). Defective thiocyanate transport in cystic fibrosis subverts optimal activity of this system and contributes in part to the frequent respiratory infections seen in patients with this disorder.
A comprehensive body of work from several disciplines links NOX activity to a wide range of physiological functions. This is especially true in cardiovascular biology (43), where NOX1, NOX2, and NOX4 have been localized in many tissues, including vascular smooth muscle (NOX1, NOX2, and NOX4), endothelium (NOX2 and NOX4), cardiomyocytes (NOX2 and NOX4), and vascular adventitium (NOX4 > NOX2 = 1). NOX isoforms contribute to maintenance of vascular tone, surveillance of ambient oxygen tension, and promotion of vascular cell growth and angiogenesis. The signaling pathways that regulate vascular tone reflect the complex interplay between ROS from various sources, including NOX proteins and reactive nitrogen species produced by endothelial nitric-oxide synthase. The interactions of these different reactive species can synergize or antagonize in producing varied biological effects and can obfuscate the precise tissue localization of their cellular origin. Aberrant NOX activity can precipitate endothelial cell dysfunction and contribute to atherosclerosis, hypertension, congestive heart failure, ischemia-reperfusion injury, and the gamut of vasculopathies that complicate diabetes mellitus. As predicted from the identification of NOX expression in nearly all tissues (supplemental Table S2), the biological processes in which NOX proteins are implicated are equally broad, including diseases of the renal, endocrine, pulmonary, gastrointestinal, hepatic, and central nervous systems as well as carcinogenesis, aging, and a host of degenerative diseases (44, 45).
Unresolved Issues Meriting Further Exploration
To put into perspective the unresolved issues in NOX biology that merit further study and the attendant challenges that face investigators in the field, it helps to reiterate the confluence of factors that promoted elucidation of the phagocyte oxidase. First, the phagocyte oxidase exhibits robust enzymatic activity; stimulated human neutrophils generate ∼10 nmol of superoxide/min/106 cells, which requires the transfer of ∼108 electrons/s/cell or a current of ∼16 pA (46). Quantitative methods such as superoxide dismutase-inhibitable reduction of ferricytochrome c and oxygen consumption readily detect oxidase activity. Second, the tissue source of the phagocyte oxidase is easily accessible, and large numbers of neutrophils (2–4 × 106/ml of blood) can be obtained in high purity, thus providing for study relatively large amounts of NOX2 (∼6–10 pmol of heme/106 cells). There is no evidence that phagocytes express any NOX isoform other than NOX2, as there is no spectral evidence for a flavocytochrome or oxidase activity of neutrophil plasma membranes isolated from patients with X-linked CGD, and there is no increase in mRNA for any of the NOX isoforms in neutrophils of patients with X-CGD.3 Consequently, any agonist-triggered, NADPH-dependent oxidant production reflects the activity of NOX2. Third, CGD provides a natural context by which to gauge speculations and to test hypotheses related to the function, regulation, and physiology of the phagocyte oxidase.
In contrast, the biological context for the non-phagocyte oxidase presents very different challenges. The non-phagocyte oxidases are expressed at low levels in complex tissue compartments composed of multiple cell types and different NOX isoforms. These complexities, coupled with limited analytical tools, provide an added challenge to investigators in the field and, by necessity, have left many fundamental features of the NOX proteins undefined.
What Are the Biochemical Features of NOX Proteins?
No data describe the enzymology of the non-phagocyte NOX proteins; no studies reporting the Km, Vmax, or enzyme turnover have been published. Unexamined as well is the possibility that substrates other than oxygen may be the target of electron transfer by the NOX proteins in tissue. Study of spectral properties is limited to a single report of heterologously expressed NOX3 (47), and only one publication reports the activity of isolated membranes in a broken cell system, the latter describing the preference of NOX4 for NADPH over NADH (48). Application of such analytical approaches may elucidate the biochemical basis for the peculiar observation regarding the most proximal detectable product generated by some of the NOX family members. Whereas nearly all NOX protein members generate superoxide anion by virtue of single electron reduction of molecular oxygen, only H2O2 has been detected after activation of Duox. Whether the extracellular domain peculiar to Duox serves as a superoxide dismutase, thus converting the superoxide anion generated by the oxidase to the H2O2 detected (Fig. 2), or the unexpected activity reflects unique chemistry is currently not known. Studies of NOX4 report the NADPH-dependent production of H2O2, not superoxide anion (49), and NOX4 membranes produce H2O2, as judged by NADPH-dependent oxidation of Amplex Red, although NOX4 transfectants reduce nitro blue tetrazolium, which indicates superoxide anion generation (unless the construct exhibits direct diaphorase activity) (48). NOX4 endogenously expressed in mesangial cells generates superoxide in response to angiotensin II, hinting that some of the confusing observations may reflect peculiar activities, subcellular location, or both related to overexpression systems.
What Are the Biochemical Properties of the Non-phagocyte NOX Proteins Expressed in Situ?
Much of the understanding of the biochemistry of the non-phagocyte NOX proteins derives from studies of their overexpression in transfected heterologous systems. Although adaptor and organizing factors interact under such conditions, the specific cofactors operating in situ have not been clearly defined in all tissues. In many cases, neither NOXO1 nor NOXA1 has been identified in tissues where the implicated NOX protein would require these cofactors for activity. For example, with the exception of rat basilar artery endothelial cells (50), vascular tissue expression of NOXO1 or NOXA1 has not been fully characterized. Not only are the essential cofactors operative in situ unidentified in many settings, but the physiological and pathophysiological agents that modulate activity, both as agonists and antagonists, are incompletely characterized. In tissues expressing multiple NOX proteins, better definition of the interplay of particular isoforms and the contribution of their subcellular compartmentalization to the overall hierarchy of oxidant generation awaits further study.
Closing Comments
The prominence of ROS in biological settings as disparate as cellular signaling and microbial killing provides compelling incentive to characterize fully the NOX proteins, both in isolation and in their physiological settings. Although the phagocyte paradigm has yielded informative hints, many biological features of the NOX protein family members will likely be novel, reflecting specialized needs for their site of expression, and thus provide exhilarating challenges to the diverse group of investigators engaged in their study. As evidenced by the energy exhibited at the last NOX protein Gordon Research Conference, these are exciting times in the oxidase world!
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI34879, HL53592, and AI44642. This work was also supported by a Veterans Affairs merit review. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
Footnotes
The abbreviations used are: CGD, chronic granulomatous disease; ROS, reactive oxygen species; NOX, NADPH oxidase.
F. R. DeLeo, personal communication.
References
- 1.Baehner, R. L., and Nathan, D. G. (1967) Science 155 835–836 [DOI] [PubMed] [Google Scholar]
- 2.Suh, Y.-A., Arnold, R. S., Lassegue, B., Shi, J., Xu, X., Sorescu, D., Chung, A. B., Griendling, K. K., and Lambeth, J. D. (1999) Nature 401 79–82 [DOI] [PubMed] [Google Scholar]
- 3.Lambeth, J. D., Kawahara, T., and Diebold, B. (2007) Free Radic. Biol. Med. 43 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geiszt, M. (2006) Cardiovasc. Res. 71 289–299 [DOI] [PubMed] [Google Scholar]
- 5.Bedard, K., and Krause, K.-H. (2007) Physiol. Rev. 87 245–313 [DOI] [PubMed] [Google Scholar]
- 6.Babior, B. M. (2004) Curr. Opin. Immunol. 16 42–47 [DOI] [PubMed] [Google Scholar]
- 7.Cross, A. R., Rae, J., and Curnutte, J. T. (1995) J. Biol. Chem. 270 17075–17077 [DOI] [PubMed] [Google Scholar]
- 8.Biberstine-Kinkade, K. J., DeLeo, F. R., Epstein, R. I., LeRoy, B. A., Nauseef, W. M., and Dinauer, M. C. (2001) J. Biol. Chem. 276 31105–31112 [DOI] [PubMed] [Google Scholar]
- 9.Finegold, A. A., Shatwell, K. P., Segal, A. W., Klausner, R. D., and Dancis, A. (1996) J. Biol. Chem. 271 31021–31024 [DOI] [PubMed] [Google Scholar]
- 10.Taylor, R. M., Baniulis, D., Burritt, J. B., Gripentrog, J. M., Lord, C. I., Riesselman, M. H., Maaty, W., Bothner, B. P., Angel, T. E., Dratz, E. A., Linton, G. F., Malech, H. L., and Jesaitis, A. J. (2006) J. Biol. Chem. 281 37045–37056 [DOI] [PubMed] [Google Scholar]
- 11.DeLeo, F. R., Burritt, J. B., Yu, L., Jesaitis, A. J., Dinauer, M. C., and Nauseef, W. M. (2000) J. Biol. Chem. 275 13986–13993 [DOI] [PubMed] [Google Scholar]
- 12.Heyworth, P. G., Cross, A. R., and Curnutte, J. T. (2003) Curr. Opin. Immunol. 15 578–584 [DOI] [PubMed] [Google Scholar]
- 13.Yu, L., Quinn, M. T., Cross, A. R., and Dinauer, M. C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7993–7998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bromberg, Y., and Pick, E. (1984) Cell. Immunol. 88 213–221 [DOI] [PubMed] [Google Scholar]
- 15.McPhail, L. C., Shirley, P. S., Clayton, C. C., and Snyderman, R. (1985) J. Clin. Investig. 75 1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curnutte, J. T. (1985) J. Clin. Investig. 75 1740–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunoi, H., Rotrosen, D., Gallin, J. I., and Malech, H. L. (1988) Science 242 1298–1301 [DOI] [PubMed] [Google Scholar]
- 18.Volpp, B. D., Nauseef, W. M., and Clark, R. A. (1988) Science 242 1295–1298 [DOI] [PubMed] [Google Scholar]
- 19.Abo, A., Pick, E., Hall, A., Totty, N., Teahan, C. G., and Segal, A. W. (1991) Nature 353 668–670 [DOI] [PubMed] [Google Scholar]
- 20.Nauseef, W. M. (2004) Histochem. Cell Biol. 122 277–291 [DOI] [PubMed] [Google Scholar]
- 21.Nisimoto, Y., Motalebi, S., Han, C. H., and Lambeth, J. D. (1999) J. Biol. Chem. 274 22999–23005 [DOI] [PubMed] [Google Scholar]
- 22.Heyworth, P. G., Bohl, B. P., Bokoch, G. M., and Curnutte, J. T. (1994) J. Biol. Chem. 269 30749–30752 [PubMed] [Google Scholar]
- 23.Bokoch, G. M., and Diebold, B. A. (2002) Blood 100 2692–2696 [DOI] [PubMed] [Google Scholar]
- 24.Sarfstein, R., Gorzalczany, Y., Mizrahi, A., Berdichevsky, Y., Molshanski-Mor, S., Weinbaum, C., Hirshberg, M., Dagher, M. C., and Pick, E. (2004) J. Biol. Chem. 279 16007–16016 [DOI] [PubMed] [Google Scholar]
- 25.Fiedler, T. J., Davey, C. A., and Fenna, R. E. (2000) J. Biol. Chem. 275 11964–11971 [DOI] [PubMed] [Google Scholar]
- 26.Park, S. M., and Chatterjee, V. K. (2004) J. Med. Genet 42 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harper, R. W., Xu, C., McManus, M., Heidersbach, A., and Eiserich, J. P. (2006) FEBS Lett. 580 5150–5154 [DOI] [PubMed] [Google Scholar]
- 28.Kawahara, T., Quinn, M. T., and Lambeth, J. D. (2007) BMC Evol. Biol. 7 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bánfi, B., Clark, R. A., Steger, K., and Krause, K.-H. (2003) J. Biol. Chem. 278 3510–3513 [DOI] [PubMed] [Google Scholar]
- 30.Cheng, G., Ritsick, D., and Lambeth, J. D. (2004) J. Biol. Chem. 279 34250–34255 [DOI] [PubMed] [Google Scholar]
- 31.Geiszt, M., Lekstrom, K., Witta, J., and Leto, T. L. (2003) J. Biol. Chem. 278 20006–20012 [DOI] [PubMed] [Google Scholar]
- 32.Takeya, R., and Sumimoto, H. (2006) Antioxid. Redox Signal. 8 1523–1532 [DOI] [PubMed] [Google Scholar]
- 33.Ueyama, T., Lekstrom, K., Tsujibe, S., Saito, N., and Leto, T. L. (2007) Free Radic. Biol. Med. 42 180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiss, P. J., Knisz, J., Zhang, Y., Baltrusaitis, J., Sigmund, C. D., Thalmann, R., Smith, R. J., Verpy, E., and Bánfi, B. (2006) Curr. Biol. 16 208–213 [DOI] [PubMed] [Google Scholar]
- 35.Jagnandan, D., Church, J. E., Bánfi, B., Stuehr, D. J., Marrero, M. B., and Fulton, D. J. R. (2007) J. Biol. Chem. 282 6494–6507 [DOI] [PubMed] [Google Scholar]
- 36.Bedard, K., Lardy, B., and Krause, K.-H. (2007) Biochimie (Paris) 89 1107–1112 [DOI] [PubMed] [Google Scholar]
- 37.Paffenholz, R., Bergstrom, R. A., Pasutto, F., Wabnitz, P., Munroe, R. J., Jagla, W., Heinzmann, Y., Marquardt, A., Bareiss, A., Laufs, J., Russ, A., Stumm, G., Schimenti, J. C., and Bergstrom, D. E. (2004) Genes Dev. 18 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupuy, C., Virion, A., Ohayon, R., Kaniewski, J., Deme, D., and Pommier, J. (1991) J. Biol. Chem. 266 3739–3743 [PubMed] [Google Scholar]
- 39.Moreno, J. C., and Visser, T. J. (2007) Endocr. Dev. 10 99–117 [DOI] [PubMed] [Google Scholar]
- 40.Geiszt, M., Witta, J., Baffi, J., Lekstrom, K., and Leto, T. L. (2003) FASEB J. 17 1502–1504 [DOI] [PubMed] [Google Scholar]
- 41.Fragoso, M. A., Fernandez, V., Forteza, R., Randell, S. H., Salathe, M., and Conner, G. E. (2004) J. Physiol. (Lond.) 561 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moskwa, P., Lorentzen, D., Excoffon, K. J., Zabner, J., McCray, P. B., Jr., Nauseef, W. M., Dubuy, C., and Bánfi, B. (2007) Am. J. Respir. Crit. Care Med. 175 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cave, A. C., Brewer, A. C., Narayanapanicker, A., Ray, R., Grieve, D. J., Walker, S., and Shah, A. M. (2006) Antioxid. Redox Signal. 8 691–728 [DOI] [PubMed] [Google Scholar]
- 44.Lambeth, J. D. (2007) Free Radic. Biol. Med. 43 332–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krause, K.-H. (2007) Exp. Gerontol. 42 256–262 [DOI] [PubMed] [Google Scholar]
- 46.Murphy, R., and DeCoursey, T. E. (2006) Biochim. Biophys. Acta 1757 996–1011 [DOI] [PubMed] [Google Scholar]
- 47.Nakano, Y., Bánfi, B., Jesaitis, A. J., Dinauer, M. C., Allen, L. A., and Nauseef, W. M. (2007) Biochem. J. 403 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serrander, L., Cartier, L., Bedard, K., Bánfi, B., Lardy, B., Plastre, O., Sienkiewicz, A., Fórró, L., Schlegel, W., and Krause, K.-H. (2007) Biochem. J. 406 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martyn, K. D., Frederick, L. M., von Loehneysen, K., Dinauer, M. C., and Knaus, U. G. (2006) Cell. Signal. 18 69–82 [DOI] [PubMed] [Google Scholar]
- 50.Ago, T., Kitazono, T., Kuroda, J., Kumai, Y., Kamouchi, M., Ooboshi, H., Wakisaka, M., Kawahara, T., Rokutan, K., Ibayashi, S., and Iida, M. (2005) Stroke 36 1040–1046 [DOI] [PubMed] [Google Scholar]
- 51.Nakano, Y., Longo-Guess, C. M., Bergstrom, D. E., Nauseef, W. M., Jones, S. M., and Bánfi, B. (2008) J. Clin. Investig. 118 1176–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.