Abstract
Misfolding and aggregation of normally soluble proteins into amyloid fibrils and their deposition and accumulation underlies a variety of clinically significant diseases. Fibrillar aggregates with amyloid-like properties can also be generated in vitro from pure proteins and peptides, including those not known to be associated with amyloidosis. Whereas biophysical studies of amyloid-like fibrils formed in vitro have provided important insights into the molecular mechanisms of amyloid generation and the structural properties of the fibrils formed, amyloidogenic proteins are typically exposed to mild or more extreme denaturing conditions to induce rapid fibril formation in vitro. Whether the structure of the resulting assemblies is representative of their natural in vivo counterparts, thus, remains a fundamental unresolved issue. Here we show using Fourier transform infrared spectroscopy that amyloid-like fibrils formed in vitro from natively folded or unfolded β2-microglobulin (the protein associated with dialysis-related amyloidosis) adopt an identical β-sheet architecture. The same β-strand signature is observed whether fibril formation in vitro occurs spontaneously or from seeded reactions. Comparison of these spectra with those of amyloid fibrils extracted from patients with dialysis-related amyloidosis revealed an identical amide I′ absorbance maximum, suggestive of a characteristic and conserved amyloid fold. Our results endorse the relevance of biophysical studies for the investigation of the molecular mechanisms of β2-microglobulin fibrillogenesis, knowledge about which may inform understanding of the pathobiology of this protein.
Tissue deposition of amyloid fibrils, the principal component of amyloid deposits, and their persistence in vivo underlies a range of diseases known collectively as the amyloidoses (1). Amyloid deposits also universally contain sulfated glycosaminoglycans and the normal plasma glycoprotein serum amyloid P component (SAP),3 which may contribute to fibrillogenesis (2), stability (3), and persistence of amyloid (1). A striking feature of natural amyloid fibrils and amyloid-like fibrils generated in vitro is their shared global structural and morphological properties, suggesting a common underlying core conformation despite the differences in structure and function of the many unrelated precursor proteins from which they are derived. Thus, amyloid fibrils are characteristically proteinase-resistant, elongated unbranched fibrils (∼10 nm in diameter and of indefinite length), and composed of two or more twisted protofilaments as visualized using transmission electron microscopy (TEM). They exhibit birefringence, which when stained by the dye Congo red under appropriate conditions and examined by cross-polarized light microscopy gives rise to transmission of a characteristic bright green color and an x-ray fiber diffraction pattern consistent with a cross-β conformation in which the polypeptide chain forms β-sheets parallel to the fibril long axis with their constituent β-strands perpendicular to this axis (1, 4). Furthermore, all amyloid fibrils display a generic protein conformational epitope recognized by antifibril antibodies (5) and are bound with high affinity by SAP (1, 3). Nonetheless, it is clear that heterogeneity of the assembly, structure, and other properties of amyloid fibrils exists, which depends on the identity of the fibril precursor protein and the conditions under which fibrillogenesis occurs (6–11).
Use of amyloid-like fibrils as tools to probe the molecular mechanisms of protein misfolding and aggregation has enabled significant insights to be made into the ways by which soluble proteins assemble into insoluble amyloid fibrils (4, 12). Such studies have prompted development of different therapeutic strategies to inhibit fibrillogenesis (13). Although such efforts are hampered by the lack of high resolution structures of amyloid fibrils and their oligomeric intermediates, advances in solid state NMR spectroscopy and x-ray crystallography are now beginning to provide a molecular description of the specific and well-ordered packing of the β-strands within small crystallizable fragments of peptides or proteins that form amyloid-like fibrils (14, 15). Recent studies have demonstrated the apparent ubiquitous ability of polypeptides to form amyloid-like fibrils under appropriate conditions, commencing from different precursor states dependent on the solution conditions employed (16, 17). For example, amyloid-like fibrils with similar morphological and dye binding properties are formed from lysozyme at low pH and high temperature (18), after proteolysis (19) and disulfide bond reduction (20), or in the presence of denaturants such as guanidinium chloride (GuHCl) (21) or ethanol (22). However, the detailed architecture of these fibrils remains unknown. Studies on protein fragments of apolipoprotein A1 have indicated large structural differences between amyloid-like aggregates formed in vitro and ex vivo amyloid fibrils formed from the full-length precursor protein (23). Thus, while amyloid-like fibrils are useful tools for biophysical investigations of amyloid formation, how closely the fibrils that are induced to form rapidly under denaturing conditions resemble fibrils formed from natively folded proteins under physiological conditions remains an open question. Furthermore, how closely fibrils generated under artificial conditions in vitro resemble genuine amyloid fibrils formed in vivo also remains unresolved.
Here we have used β2-microglobulin (β2m), a member of the immunoglobulin superfamily with a β-sandwich structure, as a model system with which to address these questions. Deposition of β2m amyloid fibrils in vivo, predominantly in osteoarticular sites, is associated with dialysis-related amyloidosis (DRA), a complication of long-term dialysis for end stage renal failure (24). Amyloid-like fibrils generated in vitro from pure recombinant β2m in seeded or unseeded reactions, commencing from denatured or natively folded protein, were examined using Fourier transform infrared (FTIR) spectroscopy and compared with spectra of natural β2m amyloid fibrils isolated from a patient with DRA. FTIR provides a powerful means of comparing the structural properties of protein aggregates, because it is not only sensitive to the secondary structural composition of proteins, but the length and twist of the constituent β-strands also influence the resulting absorbance bands (25). Our results indicate that amyloid-like fibrils formed in vitro from distinct precursor conformations of β2m and under different assembly conditions adopt an indistinguishable β-sheet architecture that is closely similar to that found in ex vivo amyloid fibrils, highlighting a common amyloid fold, at least for fibrils formed from this protein.
EXPERIMENTAL PROCEDURES
Recombinant Proteins—Full-length wild-type β2m and a double mutant containing the amino acid substitutions P32G and I7A (P32G/I7A), were expressed in E. coli and purified as described previously (7, 26). The correct molecular mass of the resulting purified proteins was confirmed by electrospray ionization mass spectrometry (27). Protein stock solutions were freshly prepared from lyophilized β2m and filtered (0.2 μm, Minisart, Sartorius) before use.
Nuclear Magnetic Resonance (NMR) Spectroscopy—One-dimensional 1H NMR spectra were obtained using 84 μm protein in 25 mm sodium phosphate, 25 mm sodium acetate at pH 2.5 (pH 2.5 buffer), or 10 mm sodium phosphate at pH 7.0 (pH 7.0 buffer), containing 90% (v/v) H2O and 10% (v/v) D2O. Under these conditions in the absence of agitation, fibril growth is very slow, allowing analysis of the structural properties of the protein monomer under fibril-growth conditions. Spectra were recorded at 37 °C on a Varian Inova 500 MHz spectrometer using 512 transients and processed using the NMRPipe suite of programs (28).
Equilibrium Denaturation—Tryptophan fluorescence of β2m samples was recorded at 37 °C using 4.2 μm β2m in pH 2.5 or pH 7.0 buffer containing increasing concentrations of GuHCl. All samples were incubated for 1 h at 37 °C prior to data acquisition. Fluorescence measurements were performed using a Photon Technology International (PTI) C-61 spectrofluorimeter with excitation at 295 nm and emission at 340 nm using 4-nm slit widths. The data were fitted using a two-state equilibrium model (29) and are represented as fractional occupancy of the native state (fraction native) at each denaturant concentration.
Fibril Formation and Characterization—Fibrillogenesis in vitro was conducted in 1.5-ml Eppendorf tubes. Pure β2m (84 μm) was incubated in either pH 2.5 or pH 7.0 buffer, containing 0.05% (w/v) NaN3, at 37 °C with agitation (250 rpm) for varying periods of time. Control experiments demonstrated that altering the type or speed of agitation affected the rate, but not the morphology, of the fibrils formed. For selected experiments, seeds were prepared from fibrils grown for 14 days at pH 2.5 from wild-type β2m that were stabilized against depolymerization at pH 7.0 by addition of the sulfated glycosaminoglycan heparin (porcine, average mass 5 kDa, Sigma-Aldrich) (2), after which they were fragmented by freeze-thaw cycling (2) and stored at -20 °C. For seeded growth experiments, seeds were added to a final concentration of 5% (w/w) of total β2m. Fibril formation was monitored by the enhanced fluorescence intensity of Thioflavin-T (ThT) that occurs upon interaction of the dye with amyloid fibrils (30). Aliquots (10 μl) of the suspensions were withdrawn at specific times after initiation of fibrillogenesis, diluted 100-fold into freshly prepared 0.5 m Tris-HCl buffer, pH 8.5 containing 10 μm ThT, and the fluorescence emission at 480 nm (4-nm slit width) was measured using a PTI C-61 spectrofluorimeter after excitation at 444 nm (4-nm slit width). Data were highly reproducible (within ± 10%) and the average of triplicate readings for each sample was normalized to that of ThT in the starting buffer alone. Aggregates of β2m were also prepared by incubating β2m (84 μm) at (i) 37 °C in 25 mm sodium phosphate, 25 mm sodium acetate buffer at pH 3.6 containing 200 mm NaCl and 0.05% (w/v) NaN3 for 3 days without agitation to produce short (<500 nm) fibrils with a worm-like morphology (8), or (ii) 60 °C in 10 mm sodium phosphate, pH 5.0 for 1 h to yield amorphous aggregates. β2m amyloid fibrils were isolated by water extraction, after removal of endogenous SAP (31), from unfixed frozen (-80 °C) amyloidotic shoulder synovial tissue of a British patient with DRA obtained with written consent (Helsinki Declaration) and ethical approval (Ethics Committee, Royal Free Hampstead NHS Trust, London). Fractions containing the highest concentration of the ex vivo fibrils were pooled, and the purity examined by reducing SDS-PAGE using 15% homogeneous gels (31). The fibrils were characterized also by mass spectrometry and protein sequencing as described elsewhere (32). All fibrils were stored after preparation at 4 °C in the presence of 0.05–0.1% (w/v) NaN3, and their amyloid-like characteristics examined by TEM after negative staining (7), cross-polarized light microscopy after staining with alkaline alcoholic Congo red (31) and binding by 125I-labeled SAP (125I-SAP) (2, 3).
Thin Film Attenuated Total Reflectance Fourier Transform Infrared (TF-ATR-FTIR) Spectroscopy—Lyophilized monomeric β2m was dissolved directly in D2O at the appropriate pD to a concentration of ∼0.4 mm. β2m fibrils, prepared as described above, were centrifuged (20 min at 13,000 rpm) to remove any soluble material, and the pellets were washed once in D2O before resuspension in D2O at the appropriate pD to a concentration of ∼0.4 mm (2). Aliquots (∼150 μl) of monomeric β2m or fibrils were spread onto the surface of a germanium crystal ATR plate, and excess water was evaporated using a current of N2 gas to form a hydrated thin protein film ideal for the analysis of such samples (33, 34). The presence of residual solvent in the film was apparent from the absorbance band of D2Oat ∼1210 cm-1. Control experiments demonstrated that the spectra obtained were not affected by the extent of solvent evaporation. After purging the system with dry air for 15 min to remove water vapor, FTIR absorbance spectra were recorded at room temperature 2 h after suspension in D2O using a Thermo-Nicolet 560 FTIR spectrometer. For each sample, 1024 scans were accumulated at a spectral resolution of 2 cm-1. Spectra acquired of the background (ATR plate without protein sample) were subtracted from those of the samples before curve fitting the amide I′ region (1700-1600 cm-1). Second derivative and Fourier self-deconvoluted spectra (employing a resolution enhancement factor of ∼1.6 and a bandwidth of ∼16 cm-1) were used to identify peak maxima. Using this information, the raw spectra were then fitted to a series of Gaussian peaks with the identified absorbance maxima using an iterative curve-fitting procedure performed in SigmaPlot (Systat Software Inc.). The position of individual peaks was held fixed while the bandwidth (assumed to be identical for all peaks) was allowed to globally vary between 10 and 20 cm-1 during the fit. The band assignments as well as the resulting calculated FTIR spectra (33) upon solution convergence to a minimum are shown. The amide I′ band with the highest intensity is defined as the amide I′ maximum throughout this report. Replicate experiments commencing with new preparations of monomer, demonstrated both the reproducibility of fibrillogenesis and the FTIR analysis.
RESULTS
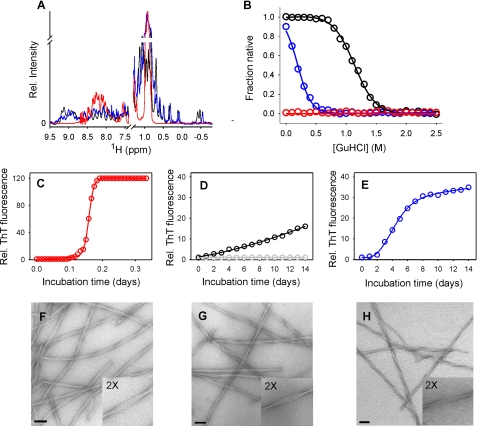
Different Precursors Facilitate Fibril Formation of β2m—The formation of amyloid-like fibrils in vitro from wild-type β2m has been extensively studied over a number of years, the results demonstrating that fibrils with a cross-β structure displaying all of the hallmarks of amyloid are generated rapidly (within hours) and spontaneously when β2m is incubated at pH 2.5 in low ionic strength (<50 mm) buffer (8, 26, 35). It has been shown previously that under these conditions, monomeric β2m is highly unfolded; the protein no longer retains β-sheet structure, and specific side-chain contacts are also absent in this highly dynamic ensemble (36, 37). Accordingly as shown here by one-dimensional 1H NMR, the spectrum of acid-unfolded β2m contains sharp resonances with little chemical shift dispersion (Fig. 1A), consistent with an unfolded state. Under these conditions the protein does not display a cooperative unfolding transition when incubated with increasing concentrations of GuHCl, a feature also characteristic of a highly unfolded state (Fig. 1B). At pH 2.5, fibril formation proceeds with nucleation-dependent polymerization kinetics (7, 35), typified by an initial lag-phase during which small oligomeric species are formed (38) prior to rapid fibril growth reflected by a substantial (∼120-fold) increase in ThT fluorescence (Fig. 1C). At the end point of incubation, amorphous aggregates are not visible by TEM (Fig. 1F), and the suspension consists predominantly of fibrils (more than 95% yield) that are ∼12 nm in diameter and have a long, straight morphology with a characteristic helical twist (7).
FIGURE 1.
Analysis of the structure and stability of different monomeric β2m precursors and the resulting amyloid-like fibrils from which they were derived. A, one-dimensional 1H NMR spectra and (B) equilibrium denaturation curves of monomeric wild-type β2m at pH 2.5 (red) and pH 7.0 (black) and P32G/I7A β2m at pH 7.0 (blue) at 37 °C. C–E, fibrillogenesis monitored by ThT fluorescence. C, unseeded fibril formation of wild-type β2m at pH 2.5. D, unseeded (gray circles) and seeded growth (black circles) of fibrils of wild-type β2m at pH 7.0. Fragmented fibrils from C stabilized by the presence of heparin were used as seeds (see “Experimental Procedures”). E, amyloid-like fibril formation of P32G/I7A β2m at pH 7.0 in the absence of added seeds. F–H, representative negative stain TEM images of amyloid-like fibrils formed from wild-type β2m at pH 2.5 (F) and at pH 7.0 upon addition of seeds (G), and from P32G/I7A β2m in unseeded growth at pH 7.0 (H). Scale bars represent 100 nm.
By contrast with the behavior of acid-unfolded β2m, incubation of β2m in its native state in low ionic strength buffer at neutral pH does not result in an increase in ThT fluorescence (Fig. 1D, gray circles), and fibrils are not observed even after incubation for months at 37 °C (2, 39, 40). Instead, the protein retains its native structure as demonstrated by the well-dispersed resonances in the one-dimensional 1H NMR spectrum of this state (Fig. 1A) and its cooperative unfolding transition when incubated with GuHCl (Fig. 1B).
To generate fibrils from native β2m at neutral pH, a procedure was employed in which fibrils formed by spontaneous (unseeded) growth at pH 2.5 were first stabilized by the addition of low molecular weight heparin (see “Experimental Procedures”). These fibrils were subsequently fragmented and used to seed fibril growth from the native wild-type protein (2). Consistent with previous results (2), the addition of these so-called heparin-stabilized seeds to monomeric β2m at pH 7.0 abolished the lag phase and resulted in a small, but significant (∼18-fold) increase in ThT fluorescence after incubation for 14 days (Fig. 1D). At the endpoint of this incubation, fibrils that are morphologically indistinguishable from those produced spontaneously from the acid-unfolded protein were formed (Fig. 1G).
To form amyloid-like fibrils from native β2m at neutral pH in the absence of additives or seeds, new variants of the protein were created, building on our previous observations that single amino acid substitutions in the N- or C-terminal regions of the polypeptide chain confer enhanced amyloidogenic potential at neutral pH, as does the variant in which the single cis Pro-32 is replaced with trans Gly (40, 41). Accordingly, the double mutant P32G/I7A was selected for further analysis. At 37 °C at neutral pH, P32G/I7A β2m is natively folded as judged by the linewidth and chemical shift dispersion of its one-dimensional 1H NMR spectrum, but is significantly destabilized compared with wild-type β2m(ΔΔGun = 14 kJ mol-1) (Fig. 1, A and B). Importantly, P32G/I7A is able to self-assemble in a nucleation-dependent manner to produce amyloid-like fibrils without the need for denaturant, heparin, other additives, or seeds, which give rise to a moderately (∼30-fold) enhanced ThT fluorescence signal (Fig. 1E). TEM reveals abundant fibrils ∼12 nm in diameter that morphologically resemble those generated de novo from acid-unfolded wild-type β2m and by seeded growth of native β2m at pH 7.0 (Fig. 1, F and H).
The amyloid-like nature of the fibrils produced in vitro using each of these conditions was confirmed by their ability to bind Congo red under selective staining conditions and produce green birefringence when viewed in cross-polarized light microscopy. In addition, the wild-type fibrils displayed specific, concentration-dependent binding by 125I-SAP and gave rise to a classical cross-β diffraction pattern after examination by x-ray fiber diffraction (Ref. 2 and results not shown), confirming their identity as genuine amyloid. Using these protein variants and incubation conditions, therefore, amyloid-like fibrils can be produced from acid-unfolded β2m, as well as from the native protein in seeded and unseeded reactions at pH 7.0, allowing a direct comparison of the fibrils formed by different mechanisms, under different assembly conditions, and from highly distinct initial precursor conformations.
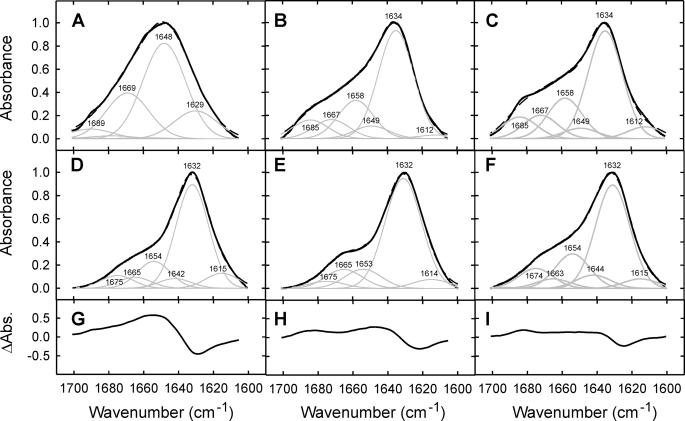
The β2m Amyloid Fold Is Precursor-independent—To determine the similarity in the content and organization of secondary structure in each of the fibril samples formed from β2m under different conditions, the fibrils and their precursors were examined by ATR-FTIR spectroscopy (Fig. 2). This technique can distinguish between the ordered β-sheet conformations in native proteins and insoluble ones such as amyloid fibrils (25). The amide I′ band that absorbs between the wavelengths 1600 and 1700 cm-1 is a widely used and sensitive probe for characterizing the secondary structure components of proteins (42, 43). The FTIR spectra of acid-unfolded and native β2m are highly distinct (Fig. 2, A–C). The spectrum of the acid-unfolded protein at pH 2.5 contains a broad amide I′ peak with an absorbance maximum determined by second derivative and Fourier self-deconvoluted spectra (see “Experimental Procedures”), of 1648 cm-1 (Fig. 2A). These data are consistent with an unordered polypeptide chain, in accord with results obtained by one-dimensional 1H NMR and equilibrium denaturation (Fig. 1, A and B). The spectrum of wild-type β2minits native conformation at pH 7.0 differs markedly from that of the unfolded state, showing a relatively narrow amide I′ absorbance maximum at 1634 cm-1 (Fig. 2B), characteristic of a folded immunoglobulin domain (44). The spectrum of P32G/I7A β2m is indistinguishable from that of native wild-typeβ2m (Fig. 2C), indicating that despite being significantly destabilized, this variant also adopts a native immunoglobulin fold at neutral pH, consistent with the 1H NMR spectrum of this protein (Fig. 1A).
FIGURE 2.
Analysis of the secondary structure of monomeric and fibrillarβ2m by ATR-FTIR spectroscopy. FTIR spectra of the amide I′ region of deuterated monomeric (A–C) and fibrillar (D–F) β2m(solid black lines) showing the component bands (gray lines). G–I, difference spectra illustrating the structural changes between monomeric and fibrillar species for each growth condition. A, D, G, wild-type β2m at pH 2.5 (unseeded growth); B, E, H, wild-type β2m at pH 7.0 (seeded growth); and C, F, I, P32G/I7A β2m at pH 7.0 (unseeded growth). In A–F, the sum (black dashed lines) of individual spectral components (gray lines) after Fourier self-deconvolution closely matches the experimental data (solid black lines).
FTIR spectra of the amyloid-like fibrils of β2m formed under the three different conditions studied are shown in Fig. 2, D–F. Strikingly, and by contrast with the very different FTIR spectra of their native and denatured monomeric precursors, the FTIR spectra of all three amyloid-like fibrils are identical, each showing a distinct absorbance maximum at 1632 cm-1, suggesting that they possess a common β-sheet architecture. Analysis of the difference spectra between each monomeric precursor and its corresponding fibrillar species reveals distinct structural transitions upon fibril formation (Fig. 2, G–I). A large shift in the absorbance maximum is observed upon fibril formation at pH 2.5 (Fig. 2G), consistent with the generation of β-sheet structure from the highly unfolded monomeric precursor. By contrast, only a small shift in the absorbance maximum (from 1634 to 1632 cm-1) occurs when fibrils are formed from the native protein at neutral pH using either seeded or unseeded growth (Fig. 2, H and I). The identity of the FTIR spectra of the fibrils formed in vitro from different precursors is remarkable given the high sensitivity of FTIR spectroscopy to the length and twist of β-strands, as well as the number of β-strands per sheet (25), and considering the very different growth conditions, assembly mechanisms (nucleated and templated growth), and timescales of fibril formation (hours to weeks) of each sample.
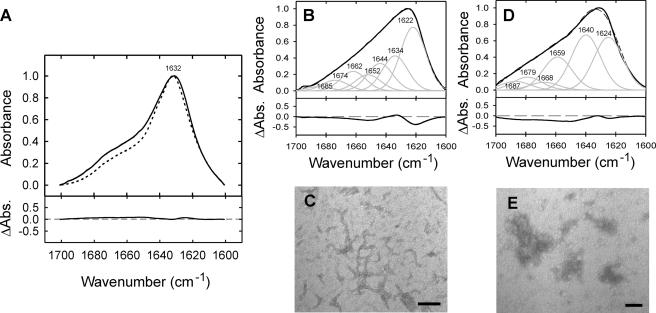
As a further demonstration of the sensitivity of FTIR spectroscopy to the conformational state of β2m, different aggregated states of the wild-type protein were generated, and the resulting spectra compared with those obtained for β2m amyloid-like fibrils. Thus, worm-like fibrils formed by the assembly of partially folded monomers at pH 3.6 (8) and amorphous aggregates formed by heating the protein to 60 °C close to its pI (see “Experimental Procedures”) were generated (Fig. 3). Although the worm-like fibrils have been shown previously to bind ThT and give rise to an x-ray fiber diffraction pattern consistent with a cross-β structure, they do not exhibit clear green birefringence after staining by Congo red and display reactivity to anti-amyloid antibodies distinct from the long straight amyloid-like fibrils analyzed here, suggesting that their structure is distinct from that typical of amyloid (8, 26). Importantly, the worm-like fibrils and amorphous aggregates give rise to FTIR spectra with amide I′ absorbance maxima at different frequencies from those of the amyloid-like fibrils of β2m (Fig. 3, B and C), confirming that the amide I′ maximum at 1632 cm-1 is specific and characteristic of the cross-β structure of β2m amyloid.
FIGURE 3.
Comparison of ATR-FTIR spectra from different β2m aggregates. A, overlay FTIR spectra of amyloid-like fibrils formed in vitro from wild-type β2m at pH 2.5 (dotted line) and P32G/I7A β2m fibrils formed at pH 7.0 (solid line), indicating their common amide I′ absorbance maximum at 1632 cm-1. The difference spectrum is shown below. FTIR spectra (B, D) and TEM analyses (C, E) of non-amyloid short fibrils with a worm-like morphology formed by wild-type β2m at pH 3.6 (B, C) and amorphous non-amyloid aggregates formed by heating wild-type β2m to 60 °C at pH 5.0 (D, E). Difference spectra of each aggregate type compared with amyloid-like fibrils formed from wild-type β2m at pH 2.5 are shown below each spectrum. Scale bars in C and E represent 200 nm.
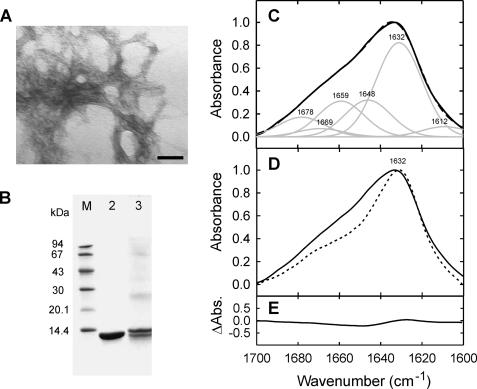
The Structure of Amyloid-like Fibrils of β2m Generated in Vitro Resembles Their ex Vivo Counterparts—To determine how closely β2m amyloid-like fibrils resemble their natural in vivo counterparts, amyloid fibrils were isolated under non-denaturing conditions and without additional purification from the tissue of a patient with DRA. These fibrils appear in negative stain TEM as predominantly dense aggregates in which adjacent fibrils are often intertwined and interconnected (Fig. 4A), a feature typical of ex vivo amyloid fibril preparations (31). SDS-PAGE analysis (Fig. 4B) revealed that the fibrils consisted predominantly of proteins that migrated as a ∼12-kDa doublet, and which corresponded, as identified by proteomics and described elsewhere (32), to intact full-length β2m (∼75% abundance) and a truncated species (∼25% abundance) lacking the N-terminal 6 amino acids, which is consistently found in ex vivo β2m fibril isolates (32, 45, 46). FTIR spectroscopy of the ex vivo fibrils enabled a detailed comparison of the structural properties of the β-sheets of natural amyloid with their in vitro counterparts. Importantly, the FTIR spectrum of ex vivo β2m amyloid exhibits an amide I′ maximum at 1632 cm-1 (Fig. 4C), precisely as observed for amyloid-like fibrils formed in vitro by wild-type β2m (Fig. 2, D and E) and the variant P32G/I7A (Figs. 2F and 4D). Although the ex vivo preparation consists predominantly of amyloid fibrils, it may also contain traces of co-isolated lipids, glycans, non-fibrillar extracellular matrix proteins, and possibly also unfolded proteins resulting from the extraction procedure, the latter of which may give rise to the increased intensity of the 1648 cm-1 band observed in its FTIR spectrum (Fig. 2A). Alternatively, the increased intensity of this band could reflect differences in the structure of non β-sheet components of the fibrils formed in vivo compared with those generated in vitro. Despite these minor differences, the distinct amide I′ maximum at 1632 cm-1 for all β2m amyloid samples examined here demonstrates, for the first time, the structural similarity in the β-sheet architecture of amyloid-like fibrils formed in vitro from pure recombinant β2m and their in vivo counterparts (Fig. 4, D and E).
FIGURE 4.
Analysis of ex vivo β2m amyloid fibrils. A, negative stain TEM micrograph of ex vivo β2m amyloid fibrils (scale bar represents 200 nm). B, reduced denatured SDS-PAGE stained with Coomassie Brilliant Blue R-350, of pure recombinant wild-type β2m(lane 2) and isolated pooled ex vivo β2m amyloid fibrils (lane 3). The relative molecular mass of standard marker proteins (M) is indicated in kDa on the left. C, amide I′ region of the ATR-FTIR spectrum of ex vivo β2m amyloid fibrils (black lines). Spectral components (gray lines) are shown after deconvolution, with individual band assignments and their sum (black dotted line). D, overlay FTIR spectra of amyloid-like fibrils formed in vitro from P32G/I7A β2m at pH 7.0 (dotted line) and isolated ex vivo β2m amyloid fibrils (solid line), indicating the common β-sheet component at 1632 cm-1. E, FTIR difference spectrum between the fibrils formed from P32G/I7A β2m in vitro and the ex vivo amyloid fibrils shown in D indicates their close structural identity.
DISCUSSION
In this study, we have exploited the sensitivity of FTIR spectroscopy to the secondary structural content, as well as the length and twist of β-strands in soluble and insoluble proteins (25, 47), to demonstrate the similarity in β-sheet structure of amyloid-like fibrils generated from β2m in vitro under an array of different conditions with their ex vivo counterparts. While subtle differences in the fibril architecture (for example in the precise stacking of the β-strands, the organization of non-β-sheet components, and the arrangement of the side chains) cannot be discerned by FTIR spectroscopy and would require more detailed elucidation, for example, by solid state NMR spectroscopy (48), the ability of FTIR spectroscopy to distinguish the structures of long straight versus worm-like β2m fibrils and different types of aggregates (amyloid-like fibrils versus amorphous aggregates) supports the concept of a common β-sheet organization for the β2m amyloid fibrils studied here. These results are consistent with previous findings whereby limited proteolysis revealed a common structural core of ex vivo and in vitro β2m amyloid involving residues ∼10–99 (27, 49, 50).
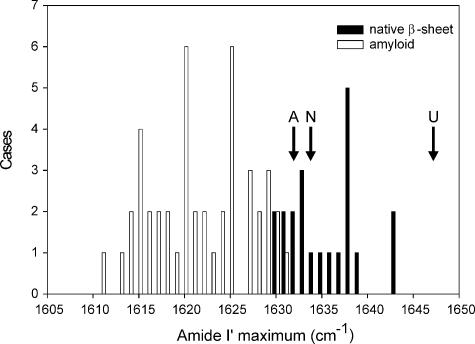
The structural similarity between amyloid fibrils formed from different precursor states of β2m under different growth conditions has several implications. First, the FTIR amide I′ absorbance maximum of β2m amyloid fibrils at 1632 cm-1 is highly distinct compared with the FTIR absorbance maxima of amyloid-like fibrils generated in vitro from other proteins and peptides that typically yield a maximum below 1625 cm-1 (25) (Fig. 5 and supplemental data). As noted by Fändrich and co-workers (25) and shown here in Fig. 5, a clear difference is observed in the frequency distribution of the amide I′ absorbance maxima of native β-sheet proteins and that of amyloid-like fibrils, consistent with significant differences in the twist and/or length of their constituent β-sheets. Interestingly, while the FTIR absorbance maximum of native β2m monomers lies within the expected range for native β-sheet proteins, β2m amyloid-like fibrils exhibit an amide I′ maximum at the edge of the range reported to date for amyloid-like fibrils formed in vitro from many other different proteins (Fig. 5). While a higher frequency at 1632 cm-1, obtained by synchrotron FTIR microspectroscopy of Aβ amyloid deposits in situ in sections of brain tissue of patients with Alzheimer disease, was previously attributed to the non-fibrillar components of amyloid (51), such an explanation can be ruled out for the β2m fibrils examined here based on their purity, high yield, and the fact that the amide I′ absorbance maximum does not change upon limited proteolysis.4 Our results here indicate, therefore, an amide I′ absorbance maximum for β2m amyloid that is highly distinct from that (1618 cm-1) suggested to be typical for other amyloid-like fibrils (52).
FIGURE 5.
Comparison of the amide I′ absorbance maxima in the ATR-FTIR spectra of different native β-sheet proteins and amyloid-like fibrils formed in vitro from various different proteins and peptides. The amide I′ maximum of native β-sheet proteins is shown as black bars and that of fibrils is shown as gray bars. This figure updates that published previously by Zandomeneghi et al. (25). A clear difference in the frequency distribution of the amide I′ absorbance maxima of native β-sheet proteins and that of fibrils is observed, indicating differences in their β-sheet architecture. Importantly, while the amide I′ absorbance maximum of native β2m(N) falls within the expected range for native β-sheet proteins, the amyloid-like fibrils formed from β2m in vitro and in vivo (A) result in an amide I′ maximum at the edge of the range reported previously for all amyloid-like fibrils reported to date. Interestingly, this band also falls within the distribution of native β-sheet proteins. The absorbance maximum of acid-unfolded β2m is also indicated (U). The proteins used for this analysis are listed in supplemental Tables S1 and S2.
The finding that an amyloid fold with a common β-sheet architecture results from the incubation of β2m under a wide range of conditions: including different growth rates, in seeded and unseeded reactions, from precursors with very different initial conformations, and in the absence or presence of additional biological factors suggests that the aggregation free energy landscape of β2m possesses a distinct global minimum that can be accessed readily irrespective of the conditions employed (53, 54). This result contrasts markedly with amyloid-like fibrils formed from the human prion protein, for example, wherein different protein sequences and seeding conditions result in amyloid-like fibrils with different structural properties resolvable by FTIR spectroscopy (55, 56). Establishing the underlying shape of the aggregation free energy landscape for different protein sequences and how this responds to changes in the growth conditions, therefore, will be needed to determine the full repertoire of amyloid structures (15, 57) and to reveal how the aggregation landscapes lead to functional amyloid or amyloid assembly routes and protein depositions associated with disease (54, 58).
Finally, our results provide the first direct evidence that the organization of β-sheets in β2m amyloid-like fibrils formed in vitro in a highly acidic environment, as well as at neutral pH, is indistinguishable from that of fibrils formed from the same protein in vivo under pathophysiological conditions, at least as determined by FTIR spectroscopy. This study therefore validates structural and mechanistic analyses of fibrillogenesis in vitro to provide insights into the formation of β2m amyloid fibrils and their pathological consequences and confirm the validity of these amyloid-like fibrils as mimics of natural β2m amyloid fibrils. Interestingly, a recent analysis of Aβ42 fibrils formed on a synthetic surface containing immobilized Aβ42 oligomers in the presence of Fe3+ also resulted in an FTIR spectrum with an absorbance maximum at 1632 cm-1, identical to that observed here for amyloid fibrils of β2m (59). Further comparative studies of amyloid-like fibrils derived in vitro from amyloidogenic precursor proteins associated with other types of amyloidosis and their in vivo counterparts will now be needed to determine whether the FTIR amide I′ absorbance maximum at 1632 cm-1 is a feature shared by genuine amyloid-like fibrils and genuine amyloid fibrils of other chemical types.
Supplementary Material
Acknowledgments
We thank Alan Berry and the S. E. Radford group for valuable discussions, Geoff Platt for providing data on the effect of proteolysis on the FTIR spectrum of β2m, Louise Serpell for performing x-ray fiber diffraction experiments, and Professor M. B. Pepys FRS for the DRA amyloid tissue.
This work was supported by the Wellcome Trust. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and additional references.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: SAP, serum amyloid P component; ATR, attenuated total reflectance; β2m, β2-microglobulin; DRA, dialysis-related amyloidosis; FTIR, Fourier transform infrared; NMR, nuclear magnetic resonance; TEM, transmission electron microscopy; ThT, thioflavin-T; GuHCl, guanidinium chloride.
G. W. Platt and S. E. Radford, unpublished results.
References
- 1.Pepys, M. B. (2006) Annu. Rev. Med. 57 223-241 [DOI] [PubMed] [Google Scholar]
- 2.Myers, S. L., Jones, S., Jahn, T. R., Morten, I. J., Tennent, G. A., Hewitt, E. W., and Radford, S. E. (2006) Biochemistry 45 2311-2321 [DOI] [PubMed] [Google Scholar]
- 3.Tennent, G. A., Lovat, L. B., and Pepys, M. B. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 4299-4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiti, F., and Dobson, C. M. (2006) Annu. Rev. Biochem. 75 333-366 [DOI] [PubMed] [Google Scholar]
- 5.O'Nuallain, B., and Wetzel, R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1485-1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez, J. L., Tennent, G., Pepys, M., and Saibil, H. R. (2001) J. Mol. Biol. 311 241-247 [DOI] [PubMed] [Google Scholar]
- 7.Kad, N. M., Myers, S. L., Smith, D. P., Smith, D. A., Radford, S. E., and Thomson, N. H. (2003) J. Mol. Biol. 330 785-797 [DOI] [PubMed] [Google Scholar]
- 8.Gosal, W. S., Morten, I. J., Hewitt, E. W., Smith, D. A., Thomson, N. H., and Radford, S. E. (2005) J. Mol. Biol. 351 850-864 [DOI] [PubMed] [Google Scholar]
- 9.van der Wel, P. C., Lewandowski, J. R., and Griffin, R. G. (2007) J. Am. Chem. Soc. 129 5117-5130 [DOI] [PubMed] [Google Scholar]
- 10.Sawaya, M. R., Sambashivan, S., Nelson, R., Ivanova, M. I., Sievers, S. A., Apostol, M. I., Thompson, M. J., Balbirnie, M., Wiltzius, J. J., McFarlane, H. T., Madsen, A. O., Riekel, C., and Eisenberg, D. (2007) Nature 447 453-457 [DOI] [PubMed] [Google Scholar]
- 11.Luca, S., Yau, W. M., Leapman, R., and Tycko, R. (2007) Biochemistry 46 13505-13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodali, R., and Wetzel, R. (2007) Curr. Opin. Struct. Biol. 17 48-57 [DOI] [PubMed] [Google Scholar]
- 13.Sciarretta, K. L., Gordon, D. J., and Meredith, S. C. (2006) Methods Enzymol. 413 273-312 [DOI] [PubMed] [Google Scholar]
- 14.Tycko, R. (2006) Q. Rev. Biophys. 39 1-55 [DOI] [PubMed] [Google Scholar]
- 15.Nelson, R., and Eisenberg, D. (2006) Curr. Opin. Struct. Biol. 16 260-265 [DOI] [PubMed] [Google Scholar]
- 16.Uversky, V. N., and Fink, A. L. (2004) Biochim. Biophys. Acta 1698 131-153 [DOI] [PubMed] [Google Scholar]
- 17.Calamai, M., Chiti, F., and Dobson, C. M. (2005) Biophys. J. 89 4201-4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krebs, M. R., Wilkins, D. K., Chung, E. W., Pitkeathly, M. C., Chamberlain, A. K., Zurdo, J., Robinson, C. V., and Dobson, C. M. (2000) J. Mol. Biol. 300 541-549 [DOI] [PubMed] [Google Scholar]
- 19.Frare, E., Polverino De Laureto, P., Zurdo, J., Dobson, C. M., and Fontana, A. (2004) J. Mol. Biol. 340 1153-1165 [DOI] [PubMed] [Google Scholar]
- 20.Cao, A., Hu, D., and Lai, L. (2004) Protein Sci. 13 319-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernaglia, B. A., Huang, J., and Clark, E. D. (2004) Biomacromolecules 5 1362-1370 [DOI] [PubMed] [Google Scholar]
- 22.Goda, S., Takano, K., Yamagata, Y., Nagata, R., Akutsu, H., Maki, S., Namba, K., and Yutani, K. (2000) Protein Sci. 9 369-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreola, A., Bellotti, V., Giorgetti, S., Mangione, P., Obici, L., Stoppini, M., Torres, J., Monzani, E., Merlini, G., and Sunde, M. (2003) J. Biol. Chem. 278 2444-2451 [DOI] [PubMed] [Google Scholar]
- 24.Jahn, T. R., and Radford, S. E. (2005) in Amyloid Proteins: The Beta Sheet Conformation and Diseases (Sipe, J. D., ed), Vol. 2, pp. 667-695, Wiley-VCH, Weinheim [Google Scholar]
- 25.Zandomeneghi, G., Krebs, M. R., McCammon, M. G., and Fandrich, M. (2004) Protein Sci. 13 3314-3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McParland, V. J., Kad, N. M., Kalverda, A. P., Brown, A., Kirwin-Jones, P., Hunter, M. G., Sunde, M., and Radford, S. E. (2000) Biochemistry 39 8735-8746 [DOI] [PubMed] [Google Scholar]
- 27.Myers, S. L., Thomson, N. H., Radford, S. E., and Ashcroft, A. E. (2006) Rapid Commun. Mass Spectrom. 20 1628-1636 [DOI] [PubMed] [Google Scholar]
- 28.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 29.Santoro, M. M., and Bolen, D. W. (1988) Biochemistry 27 8063-8068 [DOI] [PubMed] [Google Scholar]
- 30.Groenning, M., Olsen, L., van de Weert, M., Flink, J. M., Frokjaer, S., and Jorgensen, F. S. (2007) J. Struct. Biol. 158 358-369 [DOI] [PubMed] [Google Scholar]
- 31.Tennent, G. A. (1999) Methods Enzymol. 309 26-47 [DOI] [PubMed] [Google Scholar]
- 32.Giorgetti, S., Stoppini, M., Tennent, G. A., Relini, A., Marchese, L., Raimondi, S., Monti, M., Marini, S., Ostergaard, O., Heegaard, N. H., Pucci, P., Esposito, G., Merlini, G., and Bellotti, V. (2007) Protein Sci. 16 343-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberg, K. A., and Fink, A. L. (1998) Anal. Biochem. 256 92-106 [DOI] [PubMed] [Google Scholar]
- 34.Goormaghtigh, E., Cabiaux, V., and Ruysschaert, J. M. (1990) Eur. J. Biochem. 193 409-420 [DOI] [PubMed] [Google Scholar]
- 35.Naiki, H., Hashimoto, N., Suzuki, S., Kimura, H., Nakakuki, K., and Gejyo, F. (1997) Amyloid 4 223-232 [Google Scholar]
- 36.Katou, H., Kanno, T., Hoshino, M., Hagihara, Y., Tanaka, H., Kawai, T., Hasegawa, K., Naiki, H., and Goto, Y. (2002) Protein Sci. 11 2218-2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platt, G. W., McParland, V. J., Kalverda, A. P., Homans, S. W., and Radford, S. E. (2005) J. Mol. Biol. 346 279-294 [DOI] [PubMed] [Google Scholar]
- 38.Smith, A. M., Jahn, T. R., Ashcroft, A. E., and Radford, S. E. (2006) J. Mol. Biol. 364 9-19 [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto, S., Hasegawa, K., Yamaguchi, I., Tsutsumi, S., Kardos, J., Goto, Y., Gejyo, F., and Naiki, H. (2004) Biochemistry 43 11075-11082 [DOI] [PubMed] [Google Scholar]
- 40.Jahn, T. R., Parker, M. J., Homans, S. W., and Radford, S. E. (2006) Nat. Struct. Mol. Biol. 13 195-201 [DOI] [PubMed] [Google Scholar]
- 41.Jones, S., Smith, D. P., and Radford, S. E. (2003) J. Mol. Biol. 330 935-941 [DOI] [PubMed] [Google Scholar]
- 42.Surewicz, W. K., and Mantsch, H. H. (1988) Biochim. Biophys. Acta 952 115-130 [DOI] [PubMed] [Google Scholar]
- 43.Dong, A., Huang, P., and Caughey, W. S. (1990) Biochemistry 29 3303-3308 [DOI] [PubMed] [Google Scholar]
- 44.Souillac, P. O., Uversky, V. N., and Fink, A. L. (2003) Biochemistry 42 8094-8104 [DOI] [PubMed] [Google Scholar]
- 45.Linke, R. P., Hampl, H., Lobeck, H., Ritz, E., Bommer, J., Waldherr, R., and Eulitz, M. (1989) Kidney Int. 36 675-681 [DOI] [PubMed] [Google Scholar]
- 46.Stoppini, M., Mangione, P., Monti, M., Giorgetti, S., Marchese, L., Arcidiaco, P., Verga, L., Segagni, S., Pucci, P., Merlini, G., and Bellotti, V. (2005) Biochim. Biophys. Acta 1753 23-33 [DOI] [PubMed] [Google Scholar]
- 47.Seshadri, S., Khurana, R., and Fink, A. L. (1999) Methods Enzymol. 309 559-576 [DOI] [PubMed] [Google Scholar]
- 48.Makin, O. S., and Serpell, L. C. (2005) FEBS J. 272 5950-5961 [DOI] [PubMed] [Google Scholar]
- 49.Monti, M., Amoresano, A., Giorgetti, S., Bellotti, V., and Pucci, P. (2005) Biochim. Biophys. Acta 1753 44-50 [DOI] [PubMed] [Google Scholar]
- 50.Relini, A., De Stefano, S., Torrassa, S., Cavalleri, O., Rolandi, R., Gliozzi, A., Giorgetti, S., Raimondi, S., Marchese, L., Verga, L., Rossi, A., Stoppini, M., and Bellotti, V. (2007) J. Biol. Chem. 283 4912-4920 [DOI] [PubMed] [Google Scholar]
- 51.Choo, L. P., Wetzel, D. L., Halliday, W. C., Jackson, M., LeVine, S. M., and Mantsch, H. H. (1996) Biophys. J. 71 1672-1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zurdo, J., Guijarro, J. I., and Dobson, C. M. (2001) J. Am. Chem. Soc. 123 8141-8142 [DOI] [PubMed] [Google Scholar]
- 53.Jahn, T. R., and Radford, S. E. (2005) FEBS J. 272 5962-5970 [DOI] [PubMed] [Google Scholar]
- 54.Jahn, T. R., and Radford, S. E. (2008) Arch. Biochem. Biophys. 469 100-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones, E. M., and Surewicz, W. K. (2005) Cell 121 63-72 [DOI] [PubMed] [Google Scholar]
- 56.Surewicz, W. K., Jones, E. M., and Apetri, A. C. (2006) Acc. Chem. Res. 39 654-662 [DOI] [PubMed] [Google Scholar]
- 57.Mukhopadhyay, S., Krishnan, R., Lemke, E. A., Lindquist, S., and Deniz, A. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2649-2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fowler, D. M., Koulov, A. V., Balch, W. E., and Kelly, J. W. (2007) Trends Biochem. Sci. 32 217-224 [DOI] [PubMed] [Google Scholar]
- 59.Ha, C., Ryu, J., and Park, C. B. (2007) Biochemistry 46 6118-6125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.