Abstract
Adenoviruses use the short noncoding RNA transcript virus-associated (VA) RNAI to counteract two critical elements of the host cell defense system, innate cellular immunity and RNA interference, mediated by the double-stranded RNA-activated protein kinase (PKR) and Dicer/RNA-induced silencing complex, respectively. We progressively shortened the VA RNAI terminal stem to examine its necessity for inhibition of PKR. Each deletion, up to 15 bp into the terminal stem, resulted in a cumulative decrease in PKR inhibitory activity. Remarkably, however, despite significant apparent destabilization of the RNA structure, the final RNA mutant that lacked the entire terminal stem (TSΔ21 RNA) efficiently bound PKR and exhibited wild-type inhibitory activity. TSΔ21 RNA stability was strongly influenced by solution pH, indicating the involvement of a protonated base within the VA RNAI central domain tertiary structure. Gel filtration chromatography and isothermal titration calorimetry analysis indicated that wild-type VA RNAI and TSΔ21 RNA form similar 1:1 complexes with PKR but that the latter lacks secondary binding site(s) that might be provided by the terminal stem. Although TSΔ21 RNA bound PKR with wild-type Kd, and overall change in free energy (ΔG), the thermodynamics of binding (ΔH and ΔS) were significantly altered. These results demonstrate that the VA RNAI terminal stem is entirely dispensable for inhibition of PKR. Potentially, VA RNAI is therefore a truly bi-functional RNA; Dicer processing of the VA RNAI terminal stem saturates the RNA interference system while generating a “mini-VA RNAI” molecule that remains fully active against PKR.
The interferon-induced double-stranded RNA (dsRNA)4-activated protein kinase (PKR) is a key component of the innate immune response that forms the first line of intracellular defense against viral infection (1, 2). PKR regulates translation initiation by phosphorylating the eukaryotic initiation factor 2 (eIF2) α-subunit at serine 51. The large increase in affinity of the phosphorylated form for its guanosine exchange factor (eIF2B) results in competitive inhibition and the reduction in available eIF2·GTP·Met-tRNAMeti ternary complex leads to a sharp reduction in both cellular and viral protein expression (3–5). Viruses devote large portions of their genomes to evading such host defenses and have evolved many different strategies to counter the PKR-mediated response (6). For example, Epstein-Barr virus and adenovirus produce large quantities of short noncoding RNA transcripts, EBER (7, 8) and VA RNAs, respectively (9, 10), that bind directly to PKR but inhibit rather than activate the kinase activity.
All adenoviruses encode at least one VA RNA sequence (VA RNAI) of ∼160 nucleotides that is transcribed by the host RNA polymerase III and accumulates to very high concentrations in the late stages of infection (11, 12). Although VA RNAI sequences from different virus serotypes vary considerably, all can be drawn in a similar extended structure consisting of three major domains (13, 14) as follows: the terminal stem (including the paired 5′ and 3′ ends), a central domain, and the apical stem capped by a loop structure (Fig. 1). The apical stem and central domain are responsible for binding and inhibition of PKR, respectively, and the structural requirements for these roles have been thoroughly dissected (13–24). Remarkably, this functional division (22) is mirrored by a structural division between the apical stem and the central domain, which are essentially independent domains within the VA RNAI global architecture (25).
FIGURE 1.
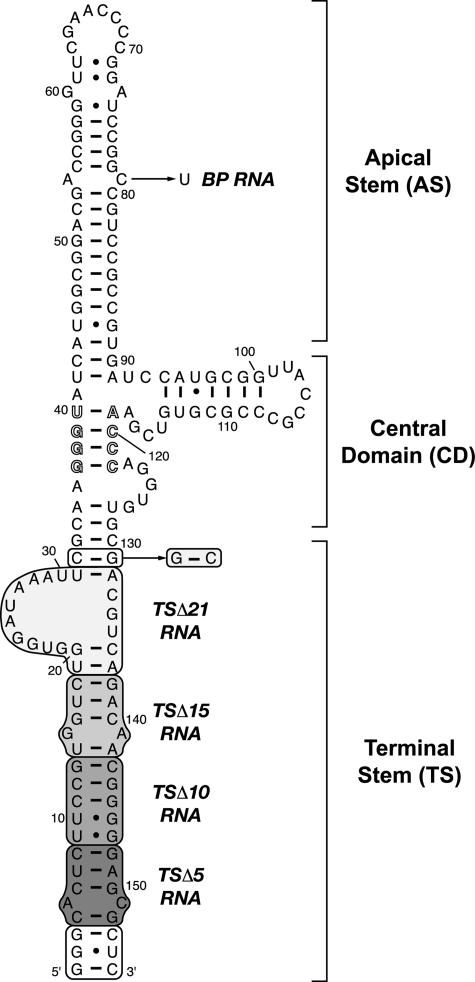
VA RNAI secondary structure and terminal stem deletion mutants. Proposed secondary structure for Ad2 VA RNAI (16), including a highly conserved pair of complementary tetranucleotide sequences (outline typeface). Shaded boxed regions indicate helical segments progressively deleted from the terminal stem to create RNAs: TSΔ5 (nts 4–8 and 148–152), TSΔ10 (nts 4–13 and 143–152), TSΔ15 (nts 4–18 and 138–152), and TSΔ21 (nts 1–32 and 131–155). The white boxed region (nts 1–3 and 153–155) was retained in TSΔ5, TSΔ10, and TSΔ15 RNAs to maintain a strong promoter for in vitro transcription by T7 RNAP; in TSΔ21, base pair C32–G131 was reversed for this purpose, as indicated. A final point mutation, 79C→U, was made to create an apical stem mutant RNA (bp RNA) with a Watson-Crick A54–U79 base pair.
In contrast, relatively few studies have examined the involvement of the terminal stem in the folding and function of VA RNAI. The terminal stem sequence is highly conserved as it contains essential transcription signals and, where differences are observed between RNAs from different serotypes, compensatory changes maintain base pairing (21). Whether the terminal stem plays any further critical role(s) in the RNA structure or function remains an open question. Although the end of the stem can be altered without reducing RNA function (13), it is possible that an intact terminal stem may be required to act as a clamp stabilizing the functional central domain or simply help protect the critical regions of the molecule from exonuclease activity in vivo (21). An intriguing additional role was more recently uncovered when sequences derived from the terminal stem were shown to be incorporated into RISC complexes following VA RNAI processing by Dicer (26). This suggests that VA RNAI suppresses both the RNA interference and PKR-mediated cellular defenses during viral infection.
To examine the contribution of the terminal stem to VA RNAI global structure, stability, and activity, we devised a mutagenesis strategy to systematically shorten the stem with the aim of determining the maximum deletion that could be accommodated without significant loss of PKR inhibition. Remarkably, despite the expected progressive reduction in inhibitory activity with shortening of the stem, a deletion of the entire terminal stem resulted in an inhibitor with wild-type level activity. Here we describe the characterization of this “mini-VA RNAI” and its interaction with PKR and discuss the implications of these results for studies of PKR-RNA interaction and PKR inhibition by viral RNA transcripts.
EXPERIMENTAL PROCEDURES
Mutagenesis and Preparation of RNA in Vitro Transcripts— A plasmid encoding adenovirus type 2 (Ad2) VA RNAI was created with a 5′-T7 RNA polymerase (T7 RNAP) promoter and 3′-hepatitis delta virus ribozyme sequence followed by a DraI restriction site for run-off transcription, as described previously (27). VA RNAI mutants were generated in this plasmid by QuikChange site-directed mutagenesis (Stratagene) and confirmed by automated DNA sequencing. The VA RNAI terminal stem (TS) was first deleted in three successive segments of 5 bp each to create the mutants TSΔ5, TSΔ10, and TSΔ15. For these RNAs, the 3 terminal bp of the wild-type sequence were maintained to provide a strong promoter for T7 RNA polymerase (T7 RNAP). A final deletion of the entire terminal stem, including the asymmetric bulge of nucleotides 21–30, created the construct TSΔ21. In TSΔ21 the T7 RNAP promoter was generated by reversal of the new terminal base pair, G32–C131. A final mutation in the apical stem of full-length VA RNAI, 78C→U, generated base pair RNA. The VA RNAI sequence and proposed secondary structure is shown in Fig. 1 with each of these mutations marked.
DraI-linearized plasmid DNA templates (100 μg/ml) were used in run-off transcription reactions (0.5 or 1.0 ml volume) under optimal conditions for VA RNAI (28) using T7 RNAP expressed from plasmid pT7-911 and purified by Ni2+ affinity chromatography (29). RNA transcripts were purified by preparative denaturing PAGE with gels containing 50% urea and 8% acrylamide. VA RNAI bands were identified by UV shadowing, excised from the gel, and eluted using a Biotrap device (Schleicher & Schuell) before ethanol precipitation and resuspension in TE buffer.
RNA UV Melting Analysis—Samples contained 20–25 μg of RNA in a solution containing 10 mm MOPS buffer, pH 7.0, and 50 mm KCl. For experiments with VA RNAI (TSΔ21) at different pH values, the following buffers were used in otherwise identical conditions: MES (pH 5.5, 6.0, and 6.5), MOPS (6.5, 7.0, and 7.5), and HEPES (7.5 and 8.0). Experiments performed with different buffers at the same pH gave identical results. UV melting curves were collected on a Varian Cary 400 UV-visible spectrophotometer with a 6-cell multichanger, running in dual beam mode. Up to five melting curves were collected in each experiment with the sixth cell containing only buffer and fitted with an in-sample temperature probe. First derivatives of the melting curves (Fig. 3A), referred to as “melting profiles,” were calculated using a Savitsky-Golay algorithm as implemented in the program OD Deriv.
FIGURE 3.
RNA UV melting analysis of VA RNAI terminal stem deletion mutants. A, UV melting curves (dashed lines) and melting profiles (first derivatives of the melting curves; solid lines) for wild-type VA RNAI collected at 260 (blue) and 280 nm (black). The assignment shown of RNA domain unfolding to each region of the melting profile was determined using compensatory base pair changes in each helical region of the RNA (25). B, melting profiles of wild-type VA RNAI (black), TSΔ5(blue), TSΔ10 (green), TSΔ15 (orange), and TSΔ21 (red) at 280 nm. All terminal stem mutations affect only the first apparent transition (Tm ∼ 60 °C) as expected. C, dependence of TSΔ21 RNA unfolding apparent Tm values upon pH. TSΔ21 RNA unfolds in three apparent transitions with the first two corresponding to the remaining central domain structure. The first central domain apparent unfolding transition is dramatically stabilized by low pH (ΔTm ∼ 11 °C between pH 5.5 and 7.5), whereas the second apparent transition is unaffected (ΔTm ∼ 0). The final apparent transition (Tm ∼ 85 °C) corresponds to the apical stem and shows a small increase in stability at lower pH (ΔTm ∼ 4 °C). D, comparison of UV melting profiles at 280 nm for full-length wild-type VA RNAI and bp RNA. As for TSΔ21 RNA, both the central domain (Tm ∼ 60 °C) and apical stem (Tm ∼ 85 °C) unfolding exhibit dependence upon pH in the context of the full-length wild-type RNA, with greater stability at lower pH. In contrast, the pH dependence is reversed for bp RNA, which lacks the (A·C)+ mismatch pair (Fig. 1).
PKR Protein Expression and Purification—PKR was expressed in Escherichia coli in a nonphosphorylated form using plasmids encoding both the kinase and λ-protein phosphatase (30, 31). Purification was accomplished using established procedures as follows: chitin affinity chromatography for PKR expressed from plasmid pTYB2-PKR(λ-PP) (30) or sequential purification by heparin affinity, poly(I)·poly(C) RNA-Sepharose affinity, and gel filtration chromatographies on an ÄKTApurifier 100 system for untagged PKR expressed from pPET-PKR/PPase (31). Poly(I)·poly(C) RNA-Sepharose was prepared using single-stranded poly(C) (4 mg/ml; GE Healthcare) and poly(I) (4 mg/ml; Sigma) RNAs coupled to CNBr-activated Sepharose 4B FF (GE Healthcare) as described by Wagner et al. (32). PKR produced by either method gave identical results in the kinase inhibition assays. PKR produced from pPET-PKR/protein phosphatase was used for all other experiments.
PKR Autophosphorylation Inhibition Assays—Purified PKR was dialyzed into 2× reaction buffer (100 mm Tris, pH 7.8, 100 mm KCl, 10% glycerol, 5 mm dithiothreitol). Wild-type VA RNAI and each terminal stem deletion mutant RNA were diluted into the same buffer to generate a 5× stock for each point in the inhibition assay (see Fig. 3 legend for details). PKR (2 μl; ∼0.1 μg) was preincubated with the VA RNAI (3 μl) at room temperature for 5 min. An equal volume of 2× PKR/activation reaction mixture was added, and each sample was incubated at room temperature for a further 10 min. The 2× PKR/activation reaction mixture contained 0.6 μg/ml poly(I)·poly(C), 4 μm MgCl2, 40 μm ATP, and 0.2 mCi/ml [32P]ATP (6000 Ci/mmol, 10 mCi/ml; PerkinElmer Life Sciences). Reactions were stopped by the addition of 0.5 volumes of 3× SDS loading dye. Samples were heated at 90 °C for 2–5 min and fractionated by 10% acrylamide SDS-PAGE at 150–200 V for ∼1 h. Gels were fixed, dried, and exposed to an imaging plate (GE Healthcare) and viewed using Typhoon 8600 PhosphorImager. Quantitation of PKR phosphorylation was done with ImageJ software (www.rsb.info.nih.gov). Control experiments without poly(I)·poly(C)-activating RNA showed that none of the VA RNAI samples activated PKR. All assays were conducted at least three times.
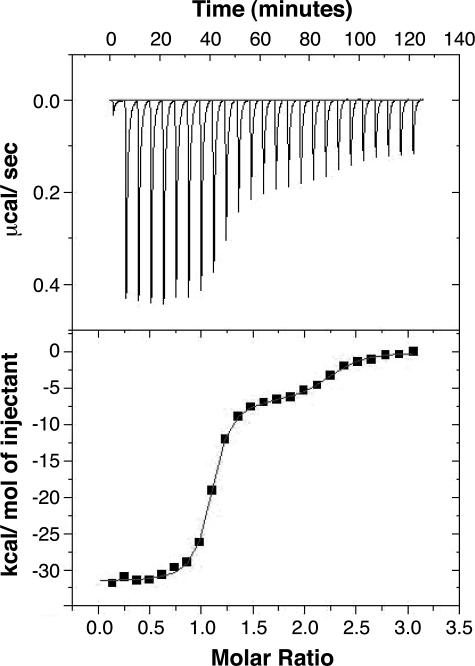
Isothermal Titration Calorimetry (ITC)—The thermodynamics of VA RNAI-PKR interactions were measured using a VP-ITC microcalorimeter following established procedures for VA RNAI (33, 34). RNA and protein were dialyzed exhaustively into 10 mm sodium phosphate, pH 6.5, 100 mm NaCl, and 5 mm β-mercaptoethanol. The sample cell contained TSΔ21 RNA at a concentration of 3 μm, and the injection syringe contained PKR at 60 μm. Titration experiments were performed at 30 °C and involved a single 2-μl injection followed by 28 × 10-μl injections of 24 s duration with 360 s spacing. Titration curves were fit by a nonlinear least squares method in Microcal Origin software using a model for one or two binding sites. A model with two binding sites was found to give the optimal fit and was used to extract thermodynamic parameters Kd, ΔH, ΔS, and N
(Table 1).
TABLE 1.
Thermodynamic parameters of PKR-RNA interaction
| RNA | Kd | N | ΔH | ΔS | ΔG |
|---|---|---|---|---|---|
| nm | kcal/mol | cal/mol·K | kcal/mol | ||
| TSΔ21 | 83 ± 20.1 | 1.16 | –6.7 | 10.4 | –9.8 |
| VA RNA1a | 79 ± 9 | NRb | –11.3 | –4.8 | –9.8 |
Data are from Ref. 38
NR indicates not reported
Gel Filtration Chromatography—Gel filtration experiments were performed using a Superdex™ 10/300 GL column (GE Healthcare) attached to an ÄKTApurifier 100 system and equilibrated with 10 mm sodium phosphate buffer, pH 6.5, and 100 mm NaCl. All samples were loaded in a total volume of 0.5 ml. The PKR concentration was 3 μm for all experiments that contained protein, whereas the RNA concentration was 0.75 μm (4:1 protein to RNA ratio), 1.5 μm (2:1), 3 μm (RNA alone and 1:1), or 6 μm (1:2).
RESULTS
To examine its role in creating or maintaining a functional VA RNAI structure that efficiently inhibits PKR, the Ad2 VA RNAI terminal stem (TS) was progressively shortened. Three successive deletions of 5 bp each and a final deletion of the entire terminal stem, including the asymmetric bulge of nucleotides 21–30, were made to create TSΔ5, TSΔ10, TSΔ15, and TSΔ21 RNAs, respectively (Fig. 1).
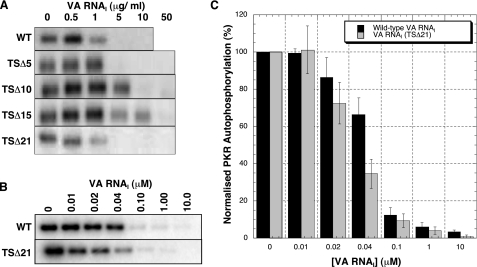
PKR Kinase Inhibition Assays—PKR autophosphorylation correlates well with subsequent activity against its primary cellular target eIF2 (35) and was used as a convenient assay of efficacy of our mutant VA RNAI inhibitors. We anticipated that the activity of VA RNAI would decrease with each successive deletion to the terminal stem. The ability of each mutant RNA to inhibit PKR autophosphorylation upon addition of dsRNA activator RNA was therefore measured and compared with wild-type VA RNAI (Fig. 2A). Wild-type VA RNAI inhibited PKR completely at a concentration of <5 μg/ml, in agreement with previous studies (23, 36). As expected, the mutant RNAs showed a decrease in activity for each successive deletion of 5 bp from the terminal stem, with ∼5, 10, and 50 μg/ml RNA required for full PKR inhibition by the TSΔ5, TSΔ10, and TSΔ15 RNAs, respectively (Fig. 2A). Remarkably, however, this trend was sharply reversed for the TSΔ21. This most severely deleted RNA displayed wild-type activity, completely inhibiting PKR autophosphorylation at a concentration of <5 μg/ml. To confirm this, the experiment was repeated at equimolar concentrations of wild-type and TSΔ21 RNAs (because for an equal mass of RNA in each reaction, the shorter VA RNAI (TSΔ21) would contain ∼50% more molecules). Quantitation of these assays confirmed that VA RNAI (TSΔ21) inhibits PKR autophosphorylation at least as efficiently as wild-type RNA despite lacking the entire terminal stem (Fig. 2, B and C).
FIGURE 2.
VA RNAI TSΔ21 is an active inhibitor of PKR autophosphorylation. A, assays of PKR autophosphorylation inhibition by wild-type VA RNAI and terminal stem deletion mutants TSΔ5, TSΔ10, TSΔ15, and TSΔ21. PKR (0.1 μg) was preincubated with 0, 0.5, 1, 5, 10, and 50 μg/ml of VA RNA prior to activation by poly(I)·poly(C) RNA (0.3 μg/ml). A progressive reduction in inhibitory activity is observed as the terminal stem is shortened, but this trend is sharply reversed for the largest deletion TSΔ21. B, assays of PKR autophosphorylation inhibition by wild-type (WT) VA RNAI and TSΔ21 RNA performed as in A but using equimolar RNA concentrations (0, 0.01, 0.02, 0.04, 0.1, 1.0, and 10 μm). C, quantification of the data shown in B. Each experiment in A and B was repeated in at least three independent experiments. Error bars show S.E.
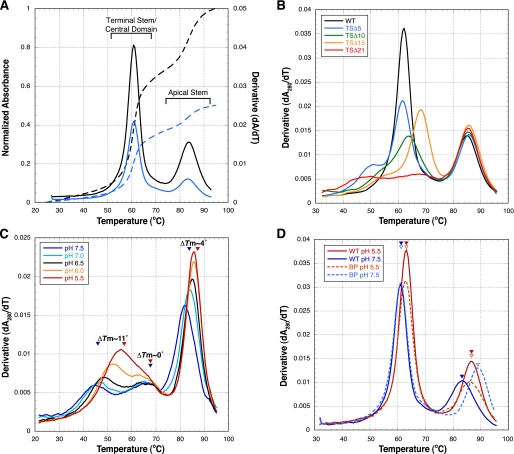
Impact of Terminal Stem Deletions on VA RNAI Structure— RNA UV melting analysis can be used to assess the impact of mutations upon the folding and stability of an RNA molecule. The unfolding of large RNAs can, however, be a complex process that occurs in a number of overlapping transitions (37). We therefore describe unfolding transitions in the melting profile, the first derivative of the UV melting curve, as “apparent transitions,” each with an associated apparent melting temperature (Tm). We have shown that wild-type VA RNAI unfolds in two independent apparent transitions (Fig. 3A) and assigned the lower (∼60 °C) and higher (∼85 °C) temperature apparent transitions to the unfolding of the terminal stem/central domain and apical stem, respectively (25). This assignment is further confirmed by the present data as deletions in the terminal stem affect only the lower temperature apparent transition (Fig. 3B). The second unfolding transition, corresponding to the apical stem unfolding, is identical to that of wild-type VA RNAI in each case.
The unfolding of the wild-type VA RNAI terminal stem and central domain are coupled and highly cooperative and result in the sharp apparent transition in the melting profile at 61 °C (Fig. 2A). Our previous studies demonstrated that even very subtle alterations in sequence, such as compensatory base pair exchanges, can be sufficient to uncouple these unfolding events into two or more apparent transitions (25). Such uncoupling of the first apparent transitions is observed for all the terminal stem deletion RNA mutants (Fig. 3B). The TSΔ5 RNA terminal stem and central domain unfold as two apparent transitions, one at the original apparent Tm (∼60 °C) and one at lower temperature (Tm ∼ 50 °C). Both TSΔ10 and TSΔ15 RNA have a similar profile shape but, surprisingly, in addition to uncoupling the unfolding process, the apparent Tm of each new apparent transition is also increased with the highest stability of the remaining terminal stem/central domain observed for TSΔ15 RNA.
The melting profile for TSΔ21 mutant is the most remarkable and distinct from wild-type VA RNAI. Deletion of the terminal stem creates an RNA with a very “loose” central domain structure that unfolds in two very broad apparent transitions centered around ∼45 and 65 °C (Fig. 3B). A range of UV melting experiments conducted in the presence of various concentrations and types of monovalent cation or with Mg2+ did not indicate any specific stabilization of any domain of the RNA (data not shown). However, significant stabilization of the first broad apparent transition (Tm ∼ 45 °C) is observed at low pH; over the range 7.5 to 5.5 a stabilization of ∼11 °C is observed (Fig. 3C). In sharp contrast, the second broad transition (Tm ∼ 65 °C) corresponding to the remaining central domain structure is unaffected by changes in pH. Equivalent melting data for full-length VA RNAI (Fig. 3D) show that the central domain stability is also dependent upon pH in this context, although the ΔTm is smaller, presumably as the structure is already stabilized close to the maximum extent possible by the intact terminal stem. Specific protonation-dependent stabilization within the central domain most likely arises from a component of tertiary structure within the RNA (see “Discussion”). The apical stem is also moderately stabilized at lower pH with a ΔTm ∼ 4 °C over the same range for both TSΔ21 and wild-type VA RNAI (Fig. 3, C and D). We hypothesized that the pH dependence of apical stem unfolding might arise from the presence of an (A·C)+ mismatch pair predicted from the current secondary structure model (Fig. 1). This was confirmed using a 79C→U mutation to create a Watson-Crick A-U base pair at this position (“bp RNA”), removing the potential protonation site. This resulted in an apical stem structure that was most stable at neutral pH (Fig. 3D) and lacked the pH dependence exhibited by the wild-type sequence.
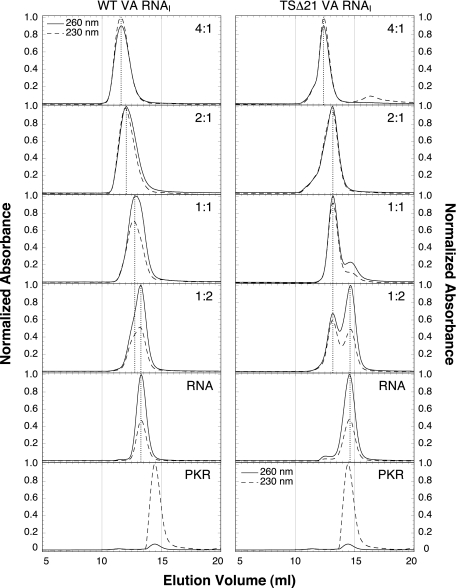
Analysis of Wild-type and TSΔ21 VA RNAI Interaction with PKR—We next examined the interaction of TSΔ21 RNA with PKR to explore potential differences in molecular recognition between this new “mini-VA RNAI” and the wild-type sequence. Analytical gel filtration chromatography of PKR, VA RNAI, and PKR-VA RNAI complexes at various molar ratios was used to investigate the nature and stoichiometry of binding (Fig. 4). Wild-type VA RNAI eluted considerably earlier from the column than TSΔ21 RNA as expected based on the molecular size (Fig. 4, RNA). PKR and both RNAs in isolation showed a single major peak in the chromatogram characterized by strong 230 nm/no 260 nm and strong 260 nm/weak 230 nm absorbances, respectively. The protein and RNA content of each peak for various complex ratios could therefore be determined using the relative absorbances.
FIGURE 4.
Gel filtration chromatographic analysis of VA RNAI-PKR complexes. Elution of wild-type (WT) VA RNAI (left column) and TSΔ21 RNA (right column) from a gel filtration column, alone and with PKR at various molar ratios (indicated in the top right of each panel as input protein:RNA ratio). UV absorbance was measured at both 260 and 230 nm to allow the relative protein and RNA content of each peak to be determined. Each experiment was normalized to the peak absorbance measurement at either wavelength.
At an input molar ratio of 1:2 PKR to TSΔ21 RNA, a single peak corresponding to a complex of 1:1 stoichiometry is observed well resolved from a second later eluting peak corresponding to free RNA. Wild-type VA RNAI exhibits the same behavior, but the peaks are not well resolved because of the larger size of the RNA. At an input ratio of 1:1, for both RNAs a single major peak is observed that elutes at the volume corresponding to the 1:1 complex. In contrast, at higher protein to RNA ratios a difference in binding behavior is observed. For TSΔ21, although some higher molecular weight species are visible as a leading shoulder, a significant portion of the protein-RNA complex still elutes at the volume corresponding to the complex of 1:1 stoichiometry (Fig. 4, marked by dotted vertical line between 1:1 and 2:1 panels in the right column). In contrast, with a 2:1 input ratio, the PKR-wild-type VA RNAI complex elutes significantly earlier, corresponding to higher molecular weight complex, with no 1:1 stoichiometry complex remaining. For both RNAs, a further increase in protein to RNA input ratio decreases the elution volume for both complexes (or mixtures of complexes), but at no concentration is any free protein observed.
Finally, the thermodynamics of TSΔ21-PKR interaction were measured by ITC (Fig. 5) under conditions used previously to characterize the binding of various RNAs to PKR (34, 38). In line with previous observations, the titration curve for TSΔ21 RNA was best fit using a model for two binding sites, and the values derived for parameters associated with the “high affinity” binding site are given in Table 1. Most strikingly, although the enthalpic and entropic contributions to binding differ dramatically for TSΔ21 and wild-type VA RNAI, the resulting binding affinity and overall free energy change are identical.
FIGURE 5.
Analysis of TSΔ21 RNA·PKR complex stability. ITC analysis of RNA-protein interaction for titration of PKR (60 μm) into TSΔ21RNA (3 μm). The titration heats measured on binding (top panel) were integrated to produce the binding isotherm (lower panel), and this fit to a model for two binding sites using the Origin software package. The thermodynamic parameters determined from the fit for the first, high affinity, binding site are shown in Table 1.
DISCUSSION
VA RNAI has long been recognized as a potent inhibitor of the cellular anti-viral defenses mediated by PKR. A large body of evidence has identified the apical stem of VA RNAI as the primary binding site and its complex central domain as the major determinant of inhibitory activity (13–24). Although far less extensively examined, prior mutagenesis suggested that an intact terminal stem structure, particularly adjacent to the central domain, might also contribute to inhibition of PKR (21). Our previous studies of VA RNAI unfolding supported this idea because the central domain stability was found to be coupled to and dependent upon that of the terminal stem (25). Thus, in addition to carrying promoter sequences, the terminal stem might play an indirect role in VA RNAI activity by acting as a structural clamp to stabilize the functional RNA tertiary fold of the central domain or to protect against RNA unwinding or nuclease attack.
To directly assess its contribution to VA RNAI structure and PKR inhibition, we used site-directed mutagenesis to systematically shorten the terminal stem helix. In doing so, we also hoped to identify an RNA with a significantly shortened terminal stem that might be beneficial for in vitro biochemical and biophysical analyses of PKR-RNA interactions. For example, deletion of a significant length of the helix would remove the potential for the terminal stem to provide secondary, or “nonspecific,” binding site(s) that might complicate such analyses.
Creation of a Minimal VA RNAI Inhibitor of PKR—The terminal stem of VA RNAI was deleted in three sequential steps of 5 bp and a final deletion of the entire helix (Fig. 1). We anticipated that a progressive reduction in PKR inhibitory activity would be observed but that it might be possible to identify a point at which the terminal stem could be deleted without significant loss of function. For the first three RNAs, TSΔ5, TSΔ10, and TSΔ15, this expectation was met, with each requiring more inhibitor RNA to provide complete inhibition of PKR. Remarkably, however, the trend was completely reversed for TSΔ21 RNA, which exhibited wild-type activity (Fig. 2) despite completely lacking a terminal stem domain. As the apical stem in isolation is a weak activator of PKR and the two conserved tetranucleotide sequences in the central domain immediately adjacent to the Δ21 deletion are critical for efficient inhibition, the TSΔ21 RNA created here is the shortest possible VA RNA inhibitor of PKR (mini-VA RNAI).
Structural Impact of Terminal Stem Deletions—UV melting experiments were used to assess the global impact on the RNA structure of each terminal stem deletion. In line with previous observations (25), deletions in the terminal stem affected only the lower temperature apparent transition that corresponds to the coupled unfolding of the terminal stem and central domain. Even the smallest deletion, TSΔ5, uncouples the unfolding of these domains such that two apparent transitions are observed, one at lower apparent Tm and the other at the original Tm. Similarly shaped profiles are observed for TSΔ10 and TSΔ15 RNAs but with variation in both the hypochromicity and Tm of both apparent transitions (Fig. 3B).
The changes observed in apparent Tm might reflect a reorganization of the base pairing within the terminal stem, e.g. to incorporate nucleotides in the 24–30 loop as the lower stem is shortened (and presumably destabilized). Terminal stem structures corresponding to the wild-type and each deletion mutant sequence, but with a short stable stem-loop above base pair 39–120 in place of the central domain and apical stem, were assessed using mFold (39). Two identical low energy structures were identified for each RNA, with the secondary structure of Fig. 1 corresponding to a marginally less stable structure (data not shown). It is therefore plausible that the changes observed in the melting profiles correspond to a switch in the pairing scheme to the alternative structure identified as the stem is shortened. Regardless of the precise pairing, however, such alterations clearly do not have a major impact on the remaining central domain structure; for deletions up to TSΔ15, the melting profiles indicate a folded structure and the steady decrease in PKR inhibition activity correlates with the size of the deletion.
The melting profile for the remaining central domain structure of TSΔ21 RNA is, however, dramatically different with two very broad peaks of low hypochromicity (Fig. 3B). We initially took this result to indicate that the central domain of VA RNAI was largely unfolded in the absence of an intact terminal stem, supporting the idea of its role as a structural clamp to secure the functional central domain structure (19). Although this may be true, from the high activity of the TSΔ21 RNA the terminal stem is clearly not essential for PKR inhibition. It is also interesting to note that in the ITC analysis of the TSΔ21 RNA-PKR interaction, there is a large change in the enthalpic and entropic contributions to binding despite an overall identical ΔG and binding affinity as for wild-type-VA RNAI. Although we are cautious in attempting to interpret the physical meaning of these changes, both the UV melting profiles and other spectroscopic analyses we have conducted with TSΔ21 RNA5 point to a flexible, “loosely” folded central domain structure. Earlier structure probing experiments on the full-length RNA have shown that both PKR and Mg2+ can induce similar conformational changes upon binding (15). These observations suggest that VA RNAI has a dynamic flexible structure, most pronounced in the absence of a terminal stem in TSΔ21 RNA, that is folded or stabilized into the correct functional tertiary structure upon interaction with PKR.
The VA RNAI Central Domain Contains a pH-dependent Tertiary Structure—Removal of the terminal stem allowed us to assess in more detail other factors that might influence the VA RNAI central domain tertiary folding. As for wild-type VA RNAI (25), no significant specific stabilization of any apparent unfolding transition was observed in melting experiments containing various concentrations of Mg2+ or different monovalent ions (data not shown). One component of the central domain was, however, specifically stabilized by low pH with a striking difference in apparent Tm of ∼11 °C over the pH range 5.5–7.5. This result provides clear evidence for the involvement of a protonated base in creating the VA RNAI central domain structure, such as a protonated cytosine in forming base triple interactions as observed in DNA (40, 41) and RNA triplexes (42, 43), and other RNA tertiary structures (44–46). The most obvious candidates for the site(s) of protonation within the central domain are cytosines 104–105, 107, or 116 in an interaction with base pairs in the lower stem of the RNA (nts 120–130). Interestingly, C116 is immediately adjacent to the conserved pair of tetranucleotides also proposed to be involved in the RNA tertiary structure (20). However, point mutations at each of these nucleotides (to either G or A) did not dramatically reduce the inhibitory function of the resulting RNA (24). Therefore, confirmation of the site of protonation and whether this is critical for central domain tertiary structure folding and/or PKR inhibition will require further detailed investigation.
Implications for Analysis of PKR-RNA Interaction—The terminal stem is an imperfectly paired helix that could potentially provide a secondary binding site for the dsRNA binding domain of PKR. Our gel filtration data shows differences in complex formation with excess PKR between wild-type and TSΔ21 RNAs. For both RNAs similar large (nonspecific) complexes are ultimately formed, but unlike the wild-type RNA, TSΔ21 RNA initially maintains a predominantly 1:1 complex stoichiometry suggesting that it does lack some lower affinity binding site for PKR. ITC data for VA RNAI and other inhibitor and activator RNAs are best fit using a model for two sites, where the lower affinity site is ascribed to nonspecific binding and otherwise ignored (33, 34, 38). Our ITC analysis of TSΔ21 RNA, which lacks the terminal stem, shows two distinct binding events. The Kd and ΔG values for the high affinity site are in excellent agreement with previous values for VA RNAI (Table 1) although, as noted above, with large compensating changes in ΔH and ΔS. More curiously, even though the second “weak affinity” binding site is well resolved in the titration and the data apparently well fit by a two-site model, the thermodynamic parameters obtained for this site are highly unrealistic. The shape of the titration suggests a binding site of moderate affinity, but a Kd of ∼0.5 nm is obtained. We hypothesize that this “binding site” in fact arises from conformational heterogeneity in the VA RNAI apical stem (20). Rather than a single RNA molecule with two binding sites, the data reflect a mixed population of RNA structures with the fully extended apical stem predominant and producing the high affinity site. We predict that RNAs with modified apical stems that remove this heterogeneity will produce a PKR binding titration curve fit optimally with a single high affinity site, and experiments to test this hypothesis are currently underway.
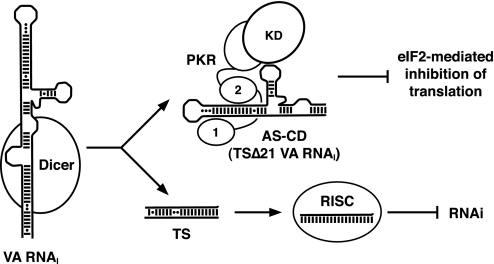
VA RNAI Function in Vivo—The VA RNAI terminal stem has been suggested to play a role in stabilizing or protecting the critical central domain against degradation in vivo (21). Although our results do agree that the terminal stem provides a large degree of structural stabilization, it is clearly not necessary for PKR inhibition. Other recent findings therefore offer another intriguing possibility. The active mini-VA RNAI we have produced in vitro is strikingly similar to the product of Dicer processing of the full-length RNA in vivo (26). Thus, adenovirus may both inhibit PKR and saturate Dicer using a single gene product where each RNA transcript contributes to both mechanisms (Fig. 6). VA RNAI may therefore be a truly bifunctional RNA and exemplify a remarkably parsimonious use of short RNA transcripts to counteract host cell defenses against viral infection.
FIGURE 6.
Dual functions of VA RNAIin vivo. VA RNAI is processed by Dicer to produce short single-stranded RNAs from the TS that are incorporated into RISC, and an apical stem-central domain (AS-CD) RNA, equivalent to TSΔ21 RNA, that remains fully active against PKR. Thus, each transcript from the VA RNAI gene is able to both saturate cellular RNA interference mechanisms and block the PKR-mediated down-regulation of general translation of cellular and viral proteins via eIF2α phosphorylation. Abbreviations used for proteins are as follows: PKR, dsRNA binding motifs (1 and 2); KD, kinase domain.
Acknowledgments
PKR expression plasmid pPET-PKR/PPase was the generous gift of Dr. James Cole (University of Connecticut). The program OD Deriv was written and provided by Prof. David E. Draper (The Johns Hopkins University). We thank Dr. Christine Dunham for discussion and comments during the preparation of the manuscript.
This work was supported by Research Career Development Fellowship 061444 from The Wellcome Trust (to G. L. C.) and studentship support from the Egyptian Ministry for Higher Education (to A. M. W.) and Medical Research Council, UK (to V. K. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: dsRNA, double-stranded RNA; PKR, double-stranded RNA-activated protein kinase; Ad2, adenovirus type 2; VA RNAI, virus-associated RNAI; TS, (VA RNAI) terminal stem; T7 RNAP, T7 RNA polymerase; ITC, isothermal titration calorimetry; nt, nucleotide; MOPS, 4-morpholinepropanesulfonic acid; MES, 4-morpholineethanesulfonic acid.
A. M. Wahid, G. L. Conn, A. J. Hobro, and Ewan W. Blanch, unpublished data.
References
- 1.Clemens, M. J., and Elia, A. (1997) J. Interferon Cytokine Res. 17 503-524 [DOI] [PubMed] [Google Scholar]
- 2.Meurs, E., Chong, K., Galabru, J., Thomas, N. S. B., Kerr, I. M., Williams, B. R. G., and Hovanessian, A. G. (1990) Cell 62 379-390 [DOI] [PubMed] [Google Scholar]
- 3.Pathak, V. K., Schindler, D., and Hershey, J. W. B. (1988) Mol. Cell. Biol. 8 993-995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowlands, A. G., Panniers, R., and Henshaw, E. C. (1988) J. Biol. Chem. 263 5526-5533 [PubMed] [Google Scholar]
- 5.Krishnamoorthy, T., Pavitt, G. D., Zhang, F., Dever, T. E., and Hinnebusch, A. G. (2001) Mol. Cell. Biol. 21 5018-5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langland, J. O., Cameron, J. M., Heck, M. C., Jancovich, J. K., and Jacobs, B. L. (2006) Virus Res. 119 100-110 [DOI] [PubMed] [Google Scholar]
- 7.Clarke, P. A., Schwemmle, M., Schickinger, J., Hilse, K., and Clemens, M. J. (1991) Nucleic Acids Res. 19 243-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp, T. V., Schwemmle, M., Jeffrey, I., Laing, K., Mellor, H., Proud, C. G., Hilse, K., and Clemens, M. J. (1993) Nucleic Acids Res. 21 4483-4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soderlund, H., Pettersson, U., Vennstrom, B., Philipson, L., and Mathews, M. B. (1976) Cell 7 585-593 [DOI] [PubMed] [Google Scholar]
- 10.Kitajewski, J., Schneider, R. J., Safer, B., Munemitsu, S. M., Samuel, C. E., Thimmappaya, B., and Shenk, T. (1986) Cell 45 195-200 [DOI] [PubMed] [Google Scholar]
- 11.Mathews, M. B., and Shenk, T. (1991) J. Virol. 65 5657-5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahr, J. A., and Gooding, L. R. (1999) Immunol. Rev. 168 121-130 [DOI] [PubMed] [Google Scholar]
- 13.Furtado, M. R., Subramanian, S., Bhat, R. A., Fowlkes, D. M., Safer, B., and Thimmappaya, B. (1989) J. Virol. 63 3423-3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellits, K. H., and Mathews, M. B. (1988) EMBO J. 7 2849-2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke, P. A., and Mathews, M. B. (1995) RNA (N. Y.) 1 7-20 [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke, P. A., Pe'ery, T., Ma, Y. L., and Mathews, M. B. (1994) Nucleic Acids Res. 22 4364-4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghadge, G. D., Malhotra, P., Furtado, M. R., Dhar, R., and Thimmapaya, B. (1994) J. Virol. 68 4137-4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghadge, G. D., Swaminathan, S., Katze, M. G., and Thimmapaya, B. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 7140-7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, Y. L., and Mathews, M. B. (1993) J. Virol. 67 6605-6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, Y. L., and Mathews, M. B. (1996) RNA (N. Y.) 2 937-951 [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, Y. L., and Mathews, M. B. (1996) J. Virol. 70 5083-5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellits, K. H., Kostura, M., and Mathews, M. B. (1990) Cell 61 843-852 [DOI] [PubMed] [Google Scholar]
- 23.Mellits, K. H., Pe'ery, T., and Mathews, M. B. (1992) J. Virol. 66 2369-2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman, A., Malhotra, P., Dhar, R., Kewalramani, T., and Thimmapaya, B. (1995) J. Virol. 69 4299-4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coventry, V. K., and Conn, G. L. (2008) Nucleic Acids Res. 36 1645-1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson, M. G., Haasnoot, P. C. J., Xu, N., Berenjian, S., Berkhout, B., and Akusjarvi, G. (2005) J. Virol. 79 9556-9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker, S. C., Avis, J. M., and Conn, G. L. (2003) Nucleic Acids Res. Methods 31 e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pe'ery, T., and Mathews, M. B. (1997) Methods (San Diego) 11 371-381 [DOI] [PubMed] [Google Scholar]
- 29.Ichetovkin, I. E., Abramochkin, G., and Shrader, T. E. (1997) J. Biol. Chem. 272 33009-33014 [DOI] [PubMed] [Google Scholar]
- 30.Conn, G. L. (2003) BioTechniques 35 682-683 [DOI] [PubMed] [Google Scholar]
- 31.Lemaire, P. A., Lary, J., and Cole, J. L. (2005) J. Mol. Biol. 345 81-90 [DOI] [PubMed] [Google Scholar]
- 32.Wagner, A. F., Bugianes, R. L., and Shen, T. Y. (1971) Biochem. Biophys. Res. Commun. 45 184-189 [DOI] [PubMed] [Google Scholar]
- 33.McKenna, S. A., Lindhout, D. A., Shimoike, T., and Puglisi, J. D. (2007) Methods Enzymol. 430 373-396 [DOI] [PubMed] [Google Scholar]
- 34.McKenna, S. A., Kim, I., Liu, C. W., and Puglisi, J. D. (2006) J. Mol. Biol. 358 1270-1285 [DOI] [PubMed] [Google Scholar]
- 35.Manche, L., Green, S. R., Schmedt, C., and Mathews, M. B. (1992) Mol. Cell. Biol. 12 5238-5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pe'ery, T., Mellits, K. H., and Mathews, M. B. (1993) J. Virol. 67 3534-3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laing, L. G., and Draper, D. E. (1994) J. Mol. Biol. 237 560-576 [DOI] [PubMed] [Google Scholar]
- 38.McKenna, S. A., Lindhout, D. A., Shimoike, T., Aitken, C. E., and Puglisi, J. D. (2007) J. Mol. Biol. 372 103-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuker, M. (2003) Nucleic Acids Res. 31 3406-3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asensio, J. L., Lane, A. N., Dhesi, J., Bergqvist, S., and Brown, T. (1998) J. Mol. Biol. 275 811-822 [DOI] [PubMed] [Google Scholar]
- 41.Roberts, R. W., and Crothers, D. M. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 4320-4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holland, J. A., and Hoffman, D. W. (1996) Nucleic Acids Res. 24 2841-2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klinck, R., Liquier, J., Taillandier, E., Gouyette, C., Huynhdinh, T., and Guittet, E. (1995) Eur. J. Biochem. 233 544-553 [DOI] [PubMed] [Google Scholar]
- 44.Conn, G. L., Gutell, R. R., and Draper, D. E. (1998) Biochemistry 37 11980-11988 [DOI] [PubMed] [Google Scholar]
- 45.Ferre-D'Amare, A. R., and Doudna, J. A. (2000) J. Mol. Biol. 295 541-556 [DOI] [PubMed] [Google Scholar]
- 46.Nixon, P. L., and Giedroc, D. P. (2000) J. Mol. Biol. 296 659-671 [DOI] [PubMed] [Google Scholar]