Abstract
The bifunctional trypanothione synthetase-amidase catalyzes biosynthesis and hydrolysis of the glutathione-spermidine adduct trypanothione, the principal intracellular thiol-redox metabolite in parasitic trypanosomatids. These parasites are unique with regard to their reliance on trypanothione to determine intracellular thiol-redox balance in defense against oxidative and chemical stress and to regulate polyamine levels. Enzymes involved in trypanothione biosynthesis provide essential biological activities, and those absent from humans or for which orthologues are sufficiently distinct are attractive targets to underpin anti-parasitic drug discovery. The structure of Leishmania major trypanothione synthetase-amidase, determined in three crystal forms, reveals two catalytic domains. The N-terminal domain, a cysteine, histidine-dependent amidohydrolase/peptidase amidase, is a papain-like cysteine protease, and the C-terminal synthetase domain displays an ATP-grasp family fold common to C:N ligases. Modeling of substrates into each active site provides insight into the specificity and reactivity of this unusual enzyme, which is able to catalyze four reactions. The domain orientation is distinct from that observed in a related bacterial glutathionylspermidine synthetase. In trypanothione synthetase-amidase, the interactions formed by the C terminus, binding in and restricting access to the amidase active site, suggest that the balance of ligation and hydrolytic activity is directly influenced by the alignment of the domains with respect to each other and implicate conformational changes with amidase activity. The potential inhibitory role of the C terminus provides a mechanism to control relative levels of the critical metabolites, trypanothione, glutathionylspermidine, and spermidine in Leishmania.
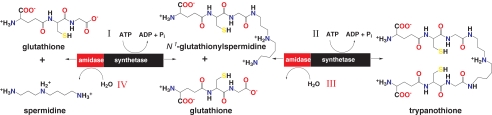
Parasitic Leishmania and Trypanosoma species cause serious tropical diseases such as kala-azar, African sleeping sickness, and Chagas diseases (1, 2). The few drugs used to combat these infections are unsatisfactory due to issues of toxicity, high cost, poor efficacy, and increasing levels of drug resistance (3, 4). The detailed genomic and proteomic data now available for these organisms advance fundamental knowledge of eukaryote biology and provide opportunities to identify and exploit novel aspects of parasite metabolism in drug discovery (5). The search for new trypanocidal agents is focused on unique metabolic pathways found in the parasites such as those involved in polyamine metabolism (6, 8) including the peptide-polyamine conjugate trypanothione (N1,N8-bis(glutathionyl)spermidine (T[SH]2),2 see Fig. 1 (9, 10)).
FIGURE 1.
Ligation and hydrolytic reactions catalyzed by trypanothione synthetase-amidase. Trypanothione is produced by stepwise ligation of glutathione with spermidine (reaction I), then glutathionylspermidine (reaction II). Hydrolysis of trypanothione (reaction III) then glutathionylspermidine (reaction IV) is performed by the N-terminal amidase domain.
Mammals use glutathione (l-γ-Glu-Cys-Gly) in conjunction with glutathione reductase and glutathione peroxidase to regulate intracellular thiol levels and redox metabolism. Trypanosomatid parasites are different and exploit the properties of T[SH]2 together with the distinct enzymes trypanothione reductase, tryparedoxin, and tryparedoxin peroxidase to form the tryparedoxin peroxidase pathway, which contributes in defense against oxidative stress (9, 10). This divergence from the host and reliance on different enzymes is of basic interest and the structure-function relationships for the tryparedoxin peroxidase pathway components have been elucidated (11-14). The host-parasite differences present opportunities to target trypanothione metabolism for drug discovery, and trypanothione reductase, for example, is a potential drug target (15). We now focus on the enzyme responsible for the synthesis and degradation of T[SH]2, trypanothione synthetase-amidase (TSA). This enzyme is uniquely positioned to regulate the supply of T[SH]2, critical for maintenance of redox metabolism and the level of polyamines that are essential for cell proliferation and differentiation (6-8).
TSA activity was first identified in the model trypanosomatid Crithidia fasciculata (16, 17). Subsequently it was observed that the active enzyme is actually a TSA glutathionylspermidine synthetase (GSPS) heterodimer (18, 19). In the human pathogens, Trypanosoma brucei, Trypanosoma cruzi, Leishmania major, and Leishmania donovani, a monomeric protein, mass ∼74 kDa, is responsible for TSA activity (20-25) and catalyzes four reactions by acting as a GSPS, a trypanothione synthetase, a trypanothione amidase, and a glutathionylspermidine amidase. Based on similarities to Escherichia coli GSPS (EcGSPS), TSA is predicted to comprise two domains with separate activities (26-28). The C-terminal domain catalyzes T[SH]2 biosynthesis by the stepwise addition of two molecules of glutathione (GSH) onto spermidine with hydrolysis of two ATP molecules (Fig. 1). The N-terminal domain is capable of hydrolyzing T[SH]2 and/or glutathionylspermidine back to spermidine and glutathione (Fig. 1) and is classified as a cysteine-histidine-dependent amidohydrolase/peptidase amidase (29, 30).
TSA is important in T. brucei as shown by RNA interference experiments, which indicate a loss of proliferation and viability with increased sensitivity to oxidative stress (21, 22). Such data are consistent with studies on T. cruzi where TSA inhibitors displayed potent trypanocidal effects (31). These observations strongly suggest that the enzyme is a potential drug target. Here, we present the structure of the recombinant L. major enzyme (LmTSA), determined in three crystal forms. We have been unable to obtain structures of TSA ligand complexes and, therefore, exploited similarities with related enzymes, in particular EcGSPS and cysteine proteases, to construct models and investigate the determinants of specificity and reactivity. These models allow us to address a key question; How can LmTSA balance or regulate conflicting biosynthetic and hydrolytic reactions that ultimately determine the critical levels of thiols and polyamines?
EXPERIMENTAL PROCEDURES
Reagents—Recombinant LmTSA was obtained by published methods (22). Selenomethionine (SeMet)-labeled protein was obtained using the methionine auxotroph strain E. coli B834- (Novagen). Cells were grown in M9 media supplemented with amino acids, where l-methionine was replaced with l-SeMet (12). Full incorporation of SeMet was confirmed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry.
Crystallization—The crystallization solution contained 5 mg ml-1 LmTSA in 40 mm HEPES, pH 7.4, 1 mm tris(2-carboxyethyl)phosphine hydrochloride, and 1 mm ethylenediamine tetraacetic acid. Protein concentration was determined using the theoretical extinction coefficient 131.8 mm-1cm-1 at 280 nm calculated using Protparam (32).
Three crystal forms were obtained at 18 °C with the hanging γ-drop vapor diffusion method and grew to full size (maximum dimension 0.2 mm) in 2 days. The drops were constructed from 1 μl of protein solution mixed with 1 μl of reservoir. Conditions for crystal form I were optimized to a reservoir of 1.4 m (NH4)2SO4, 100 mm HEPES, pH 7.0, 200 mm NaBr, crystal form II to 1.6 m (NH4)2SO4, 100 mm HEPES, pH 7.0, and 200 mm NaBr, and crystal form III to 14% polyethylene glycol 8000, 15% glycerol and 100 mm KCl. Crystal appearance and diffraction properties degraded quickly, and crystals were harvested 2 days after attaining full size. Crystal forms I and II were cryo-protected by soaking for 5 s in artificial mother liquor supplemented with 25% ethylene glycol. Form III crystals were used directly from the crystallization drops. Crystals were characterized on a Rigaku MicroMax HF-007 rotating anode generator and R-Axis IV2+ image plate detector, then stored under liquid nitrogen. Form I and II crystals required in situ annealing to optimize diffraction quality. The crystals displayed space groups P212121 (form I) and P21 (form II and III). Crystals were also obtained by co-crystallization in the presence of adenosine 5′-(β,γ-imino)triphosphate, glutathione, and spermidine, although none of these ligands was resolved in electron density maps (data not shown).
X-ray Data Collection and Processing—Data were collected at the European Synchrotron Radiation Source (Grenoble, France). Native data for form I and II crystals were collected on beam lines ID14-4 and BM14 using an ADSC Q315 CCD and a MAR MOSAIC 225 CCD detector, respectively, and for form III on ID29 with an ADSC Q315 CCD detector. Multiwavelength anomalous dispersion data were collected from a single orthorhombic form I SeMet LmTSA crystal on ID14-4. Processing and scaling were performed with XDS (33) or a MOSFLM (34) SCALA (35, 36) combination, and statistics are given in Table 1.
TABLE 1.
Crystallographic statistics
| Data set | SeMet (λ1) | SeMet (λ2) | SeMet (λ3) | Form I | Form II | Form III | |
|---|---|---|---|---|---|---|---|
| Space group | P212121 | P212121 | P212121 | P212121 | P21 | P21 | |
| Wavelength (Å) | 0.9794 | 0.97960 | 0.97780 | 0.97897 | 0.9199 | 0.9755 | |
| Unit cell dimensions a,b,c (Å) | 71.1, 85.9, 167.4 | 70.7, 85.4, 168.0 | 70.5, 167.1, 84.92 | 67.1, 127.7, 88.8 | |||
| β = 94.1° | β = 94.6° | ||||||
| Resolution range (Å) | 40-3.4 | 40-3.6 | 40-3.6 | 20-2.75 | 20-2.8 | 25-2.3 | |
| Reflections | 260,907 | 138,289 | 139,006 | 94,640 | 180,449 | 496,759 | |
| Unique reflections | 14,854 | 13,455 | 13,529 | 26,468 | 48,090 | 65,387 | |
| Completeness (%) | 95.9 (94.1)a | 83.3 (77.5) | 83.1 (76.4) | 96.5 (88.6) | 99.7 (100) | 98.7 (96.5) | |
| Rmergeb | 10.1 (23.7) | 9.0 (41.6) | 9.2 (45.6) | 9.2 (45.6) | 7.3 (41.5) | 6.6 (24.1) | |
| Redundancy | 3.4 (3.2) | 1.9 (1.9) | 2.0 (1.9) | 3.5 (3.2) | 3.8 (3.8) | 7.6 (7.6) | |
| <I/σ(I)> | 12.2 (2.9) | 8.3 (2.2) | 8.4 (2.1) | 21.7 (3.6) | 14.5 (2.5) | 17.2 (5.0) | |
| Wilson B (Å2) | 31.5 | 36.2 | 36.8 | 53.1 | 71.1 | 39.3 | |
| Rworkc/Rfreed,a | 21.0/26.1 | 19.8/24.8 | 17.7/23.3 | ||||
| Number of residues/waters/halides | 603/45/7 | 1196/161/9 | 1189/603/2 | ||||
| r.m.s.d. from ideal values for covalent bonds | |||||||
| Lengths (Å)/angles (°) | 0.014/1.46 | 0.009/1.21 | 0.013/1.37 | ||||
| Average B-factors (Å2) | 66.5 | 63.3 | 46.0 | ||||
| Ramachandran analysis (%) | |||||||
| Favored regions | 87.7 | 87.9 | 90.9 | ||||
| Allowed regions | 12.1 | 11.9 | 8.9 | ||||
| Outliers | 0.2 | 0.2 | 0.2 | ||||
Values in parentheses refer to the highest resolution bin.
Rmerge = ∑h∑i||(h,i) - <I(h)>||∑h∑i I(h,i), where I(h,i) is the intensity of the ith measurement of reflection h, and <I(h)> is the mean value of I(h,i) for all i measurements.
Rwork = ∑hkl||Fo| - |Fc||/∑|Fo|, where Fo is the observed structure factor amplitude, and the Fc is the structure-factor amplitude calculated from the model.
Rfree is the same as Rwork except calculated with a subset, 5%, of data that are excluded from refinement calculations.
Structure Determination, Refinement, and Model Analysis—Initial phases were obtained by multiwavelength anomalous dispersion phasing. Six of the 12 possible selenium positions were identified using the program SOLVE (37) giving a figure-of-merit of 0.4 for data to 4 Å resolution and a Z-score of 21.9. Density modification and phase extension was performed using the program RESOLVE (38) and increased the figure-of-merit to 0.77. Subsequent data collection (BM14) on a second orthorhombic crystal permitted phase extension to 3 Å producing an overall figure-of-merit of 0.6, with the figure-of-merit to 4 Å significantly improved to 0.87. Automated procedures (RESOLVE) produced a partial model, which formed the basis for model building in the program COOT (39). The model at this stage contained 526 residues, mainly poly-Ala. Crystal form III became available and provided data to 2.3 Å of resolution, and the form I poly-Ala derived model was used for molecular replacement with MOLREP (40). Two molecules were positioned giving an R-factor of 49.8 and a correlation coefficient of 0.40. The phases were improved by exploiting non-crystallographic symmetry averaging combined with solvent flattening (supplemental Fig. S1).
The resulting map was interpreted to produce a model with 605 residues for each molecule. Refinement was performed using REFMAC5 (41) with TLS (Translation/Libration/Screw) refinement (42), model manipulation, and inclusion of water molecules and halide ions. Once completed, this model was used to solve the other crystal forms by molecular replacement, and similar refinement protocols were implemented. MOLPROBITY (43) was used to investigate the model geometry along with the validation tools in COOT and PROCHECK (44), and figures were prepared using PyMOL (45). Least-squares superpositions and comparison of domain orientations and protein structures used LSQKAB (46), DynDom (47), and SSM (48). COOT and PyMOL were used to model substrate/product complexes. Refinement and model geometry statistics are presented in Table 1.
RESULTS AND DISCUSSION
General Comments—The structure of LmTSA was determined with phases derived from a multiwavelength anomalous dispersion experiment applied to a SeMet derivative. The model from this analysis was used to determine the structures of two additional crystal forms, one of which diffracted to 2.3 Å resolution. The statistics and model geometry (Table 1) indicate that the analyses have produced acceptable medium-resolution models. The structures derived from the three crystal forms are very similar. The root mean square deviation (r.m.s.d.) from overlap of ∼650 Cα positions of the different models ranges from 0.6 to 1.1 Å with an average of 0.8 Å. The most ordered crystal form, III, has two molecules in the asymmetric unit, labeled A and B, with an r.m.s.d. of 0.7 Å when they are superimposed. Molecule A has three disordered, presumably flexible loops, comprising residues 251-263, 551-579, 614-625. Molecule B has five loops missing, between 227 and 234, 250 and 263, 551 and 578, 617 and 625, and 632 and 637. Given the high degree of similarity between the molecules, it is only necessary to detail one, and we have arbitrarily selected molecule A.
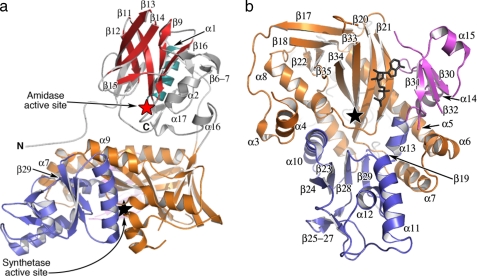
Overall Structure—The secondary and tertiary structures of LmTSA are depicted in Fig. 2 (the sequence matched to the secondary structure in supplemental Fig. S2). LmTSA comprises an N-terminal amidase domain of residues 1-215 and 634-652 and C-terminal synthetase domain of residues 216-633. The amidase domain displays a papain-like fold, (49), with two α-helices (α1 and α2) and a β-barrel created by β9to β16 (Fig. 2a).
FIGURE 2.
Secondary, tertiary, and domain structure of LmTSA. a, the fold. Red and black stars mark amidase and synthetase active sites, respectively. Selected elements of secondary structure are labeled. α1 is blue, and the β-barrel is red. b, the subdomain structure of the ATP-grasp synthetase domain viewed orthogonal to a. Subdomain A is colored orange, subdomain B is blue, and subdomain C is purple. A model of ADP (black sticks, based on structural comparisons) is included.
The synthetase domain comprises three subdomains and displays the ATP-grasp fold (Fig. 2b (50)). Subdomain A, residues 216-393 and 595-633, contains a four-stranded anti-parallel β-sheet (β20, β21, β33, β34). Helix α5 is buried under one side of this β-sheet, whereas the other side creates an open cleft, adjacent to subdomain C where ATP binds. Subdomain B, residues 394-511, consists of a four-stranded parallel β-sheet (β23, β24, β28, β29) with α10 on one side, α11, α12, and α13 on the other, and a small region of anti-parallel β-sheet (β25 to β27). A short linker region then spans to the smaller subdomain C (residues 514-595), which as mentioned, creates one side of the ATP binding cleft. This subdomain contains a three-stranded anti-parallel β-sheet (β30, β31, and β32) and with α14 and α15 forms part of the protein surface. The links between β31 and β32 and between β32 and β33 are disordered. Such flexible segments at the interface of the subdomains are a prominent feature of ATP-grasp proteins (51, 52) and are discussed below. Another linker region (residues 587-595) spans the ATP binding cleft to the C-terminal section of subdomain A, which comprises β33 to β35.
The N- and C-terminal polypeptide segments extend from the amidase and synthetase domains, respectively (Fig. 2a). The N terminus adopts an extended conformation toward α7, α9, and β29 of the synthetase domain. The C-terminal 20 residues actually contribute to the amidase domain with implications for regulation of amidase activity; this will be discussed below.
The arrangement of molecules in the three crystal forms, whether involving non-crystallographic symmetry or crystallographic symmetry, is similar, with N-terminal residues from one molecule occupying the ATP binding site of the adjacent molecule (supplemental Fig. S3). This is an extensive area of contact, important for the stability of the LmTSA crystal lattice, and has hindered efforts to obtain ligand complexes by crystal soaking or co-crystallization. To investigate the determinants of specificity and activity, we identified related structures for comparative modeling.
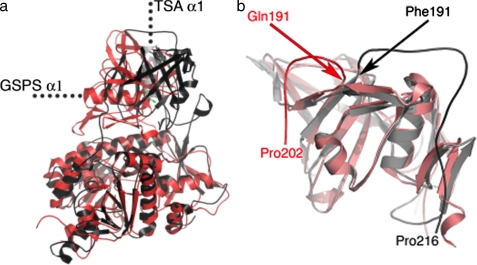
The amino acid sequences of LmTSA and EcGSPS share ∼30% identity overall, and this applies to individual domains. A least-squares superposition of the amidase domains produces an r.m.s.d. of 1.4 Å for 167 Cα atoms; the r.m.s.d. for 338 Cα positions of the synthetase domain is 1.8 Å. However, superimposition of synthetase domains reveals a striking difference in the relative orientation of the amidase domains (Fig. 3a), with a rotation of 130° center-of-mass translation of 3.5 Å. The scale and influence of this difference is illustrated by the placement of the major helix in the amidase domain (α1) in the two structures (Fig. 3a). In LmTSA α1 extends away from the synthetase domain with the amidase active site close to the interface region, whereas in EcGSPS, α1 is perpendicular to LmTSA α1. The linker regions of polypeptide that span from amidase to synthetase domains in LmTSA and EcGSPS are also significantly different (Fig. 3b). In both cases the linkers extend from the final strand of the β-barrel (β15) to the first β-strand in the synthetase domain (β16). The linker segment in LmTSA (residues 191-216) is longer compared with EcGSPS (residues 191-202). LmTSA is also extended by 18 amino acids at the C terminus, and this segment interacts with both amidase and synthetase domains.
FIGURE 3.
Comparison with EcGSPS. a, superposition of LmTSA (black ribbon, gray α1) in the same orientation as Fig. 2b and EcGSPS (red), based on the synthetase domains, to highlight the difference in the amidase domains. b, the distinct amidase-synthetase linkers based on superposition of amidase domains.
The relative orientation and nature of the interaction between amidase and synthetase domains is, therefore, distinct in the two enzymes and manifests itself ultimately in creating distinct amidase active sites. The EcGSPS amidase active site is in the middle of a large, solvent-accessible cleft on the surface of the N-terminal domain (not shown). In LmTSA the domain positions in conjunction with the structure of the C terminus renders the catalytic site inaccessible, a point discussed later.
The Synthetase Active Site—The synthetase domain displays the ATP-grasp fold, which binds nucleotide in a well defined fashion. We identified EcGSPS as the most closely related structure to LmTSA, and a comparison of the synthetase domains gives an r.m.s.d. of 1.54 Å over 343 Cα positions. Human glutathione synthetase is also structurally related, although with a low sequence identity of about 10%. Comparison of LmTSA with Human glutathione synthetase using SSM gave an r.m.s.d. of 3.1 Å over 272 Cα positions. These structural overlays indicate that the synthetase active sites of human glutathione synthetase, EcGSPS, and LmTSA are similar, and the excellent conservation of residues important for ATP and glutathione binding (supplemental Fig. S4) allowed us to model LmTSA complexes at the onset of both reactions I and II (Figs. 1 and 4). Comparison with the structure of EcGSPS in complex with an inhibitor that contained a spermidine-like moiety (28) allowed us to position the spermidine in LmTSA. This model was then extended to glutathionylspermidine by positioning the tripeptide to avoid any obvious steric clash. We made no effort to compute or dock positions of the peptide component for two reasons. There are disordered loops in this area of the synthetase domain, and this would, in conjunction with the high degree of rotational freedom of glutathionylspermidine, compromise such modeling.
FIGURE 4.
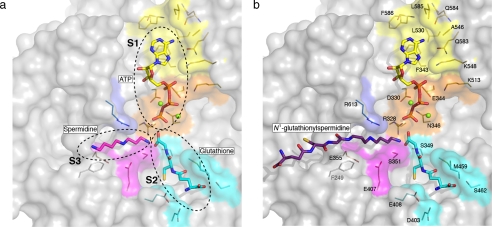
The synthetase active site. a, model of substrates at the onset of reaction I (ATP, GSH, and spermidine). The protein surface is depicted as a gray semi-transparent van der Waals surface, except for areas that interact with adenine (yellow, S1), ATP phosphates (orange, S1), GSH (cyan, S2), spermidine (purple, S3), and Arg-613 (blue). The substrates are colored according to atom type, N is blue, O is red, P is orange, ATPCis yellow, GSHCis cyan, and spermidine C is purple. Selected side chains are also shown in stick mode colored by atom type with C atoms of residues colored depending upon with which substrate component they may interact. Mg2+ ions are green spheres. b, model for the onset of reaction II (ATP, GSH, and glutathionylspermidine). The S1, S2, and S3 binding pockets are marked by dashed lines in a, and key residues labeled in b.
The synthetase active site is a triangular shaped cavity that accommodates each of the three substrates roughly at the vertices of the triangle. The catalytic center is placed at the orthocenter. The ATP binding S1 site lies between subdomains A and C, the GSH-binding cleft is formed mainly by subdomain B, and the polyamine-binding S3 site (spermidine and glutathionylspermidine) is a cleft between subdomains A and B (Fig. 4).
The S1 ATP binding site is formed by the anti-parallel β-sheet of subdomain A and the anti-parallel β-sheet of subdomain C. There are two poorly ordered segments of polypeptide around S1; these include residues that link β31 with β32 and β32 with β33. A prominent feature of the ATP-grasp family of enzymes is the presence of such flexible loops around the nucleotide binding site that clamp down on a nucleotide phosphate (52).
Adenine is predicted to bind LmTSA in a hydrophobic pocket created by Phe-343, Leu-530, Ala-546, Leu-585, Val-618, and Ile-619 (latter two not shown). Adenine N6 would donate hydrogen bonds to the carbonyl groups on the side chain of Gln-583 and main chain of Gln-584, N1 could accept a hydrogen bond from the main chain amide of Phe-586, and N7 accept a hydrogen bond from Lys-548 NZ. Six of these 10 residues, directly implicated in adenine binding, are strictly conserved in EcGSPS (supplemental Figs. S4 and S5). The remainder, Phe-343, Phe-586, Val-618, and Ile-619 of LmTSA, correspond to Tyr-329, Trp-571, Leu-603, and Val-604 in EcGSPS, respectively, and represent conservative substitutions. In addition, there are seven strictly conserved residues in LmTSA and EcGSPS important for binding and orientation of the triphosphate component of ATP during catalysis (53). In LmTSA these are Arg-328, Asp-330, Glu-344, Asn-346, Lys-513, Lys-548, and Arg-613. The basic Lys-513 and Lys-548 are placed to interact with phosphates. Asp-330, Glu-344 (strictly conserved in the ATP-grasp family), and Asn-346 coordinate Mg2+ ions, which in turn interact with the phosphate groups of ATP. These interactions orient and activate the γ-phosphate to participate in the synthetase reaction. Arg-613 helps to form the S1 and S3 pockets (see below) and may stabilize the transition state. Arg-328 is at the bottom of the catalytic site and is held in place with a hydrogen bond to the main chain carbonyl of Ser-351 (not shown). The guanidinium is directed to align the GSH carboxylate and, with Arg-613, to generate and stabilize the transition state.
EcGSPS ligand complexes with GSH and an inhibitor comprising γ-Glu-Ala-Gly linked to spermidine by a phosphinate group identify two GSH binding sites (28). The GSH-like moiety of this inhibitor occupies a cleft formed at one end by the β23-α10 and β28-α11 regions, with the other end created by the polypeptide that links β21 to α8. This is the S2 binding pocket. The GSH-like segment forms hydrogen bonds and van der Waals interactions with Ser-335, Asp-387, Glu-392, Ala-443, and Thr-446 in EcGSPS (28). The first three of these are conserved in GSPS and TSA sequences (Ser-349, Asp-403, and Glu-409 in LmTSA). Thr-446 is conserved in GSPS and either threonine or serine in TSA (Ser-462 in LmTSA). Ala-443 of EcGSPS is replaced by methionine in TSA (Met-459 in LmTSA) and either Ala or Ser in GSPS.
In the EcGSPS glutathione complex, the ligand binds in a cleft that extends away from S1 ATP and S2 GSH binding sites. This cleft is occupied by a spermidine moiety in the EcGSPS inhibitor complex and is the S3 polyamine substrate binding site. When glutathione occupies the EcGSPS S3 site, it forms a disulfide bond with Cys-338, and the glycine forms an isopeptide link with Lys-607 NZ (28). This complex is incompatible with synthetase activity. Mutagenesis of Cys-338, conserved in all GSPS sequences, to alanine results in a marked reduction in affinity for GSH and spermidine, and Pai et al. (28) suggest that this non-catalytic binding site may regulate the enzyme with a translocation mechanism required to correctly position GSH in the S2 site. Cys-338 of EcGSPS is not conserved in TSA, where it is replaced by threonine (Thr-352 in LmTSA). The presence of Cys-338 combined with the experimental conditions under which the structure was obtained may have complicated the study of EcGSPS by providing the capacity to bind glutathione in a non-productive manner. Under physiological conditions, the reducing environment in the cytosol would likely prevent such binding.
The N-terminal sections of α4, α8, and α10 create the S3 pocket of LmTSA, with contributions from β34 and the linker to β35. The polypeptide linking these two β-strands is disordered, as is the segment between α4 and β18, suggesting that conformational changes on the edge of the S3 pocket might accompany polyamine substrate binding. In EcGSPS, two acidic residues, Glu-391 and Asp-610, and the main chain carbonyl group of Lys-607 are predicted to form hydrogen bonds with the spermidine moiety of the inhibitor, and by inference these are interactions that bind the polyamine substrate in the S3 pocket. Glu-391 is conserved (Glu-407), but Lys-607 and Asp-610 are not. As mentioned, in LmTSA this section of polypeptide is disordered and likely to adjust position when substrate binds. Ser-351 OG donates a hydrogen bond to Glu-407 OD2, an arrangement providing functional groups to interact with the polyamine substrates. The Ser-351 OG in particular may align and prepare the terminal amine to participate in catalysis. Glu-355, conserved in TSA, is on α8, forming the base of the S3 cleft and placed to interact with and position polyamine components. In a structural alignment (not shown), this residue aligns with Cys-338 of EcGSPS.
The positioning of S1, S2, and S3 pockets and identification of where the substrates bind suggests how TSA catalyzes two synthetic reactions without recourse to an ill-defined translocation mechanism. In reaction I (Fig. 1), the S1, S2, and S3 pockets are occupied by ATP, GSH, and spermidine, respectively, with the γ-phosphate of ATP, the glycine carboxylate of GSH, and the terminal amine group of spermidine at the orthocenter of the synthetase active site (Fig. 4a). The GSH carboxyl group is activated by phosphorylation, facilitating the generation of an acylphosphate intermediate. The reaction is assisted and the intermediate stabilized by contributions from Arg-328 and two Mg2+ ions. The amine carries out nucleophilic attack on the anionic intermediate, which collapses to produce an amide linkage (N1-glutathionylspermidine), with release of ADP and phosphate in a manner similar to that described for glutathione synthetase (EC 6.3.2.3) and γ-glutamylcysteine synthetase (54). The reaction I products vacate the active site before reaction II (Fig. 1) can proceed. Here the S1 and S2 pockets are again occupied by ATP and GSH, respectively. Glutathionylspermidine binds in S3 (Fig. 4b) with the peptide component directed away from the active site, and the terminal amine group placed at the catalytic center. Reaction II occurs like reaction I to produce trypanothione, ADP, and phosphate.
The most noticeable differences between the synthetase domains of EcGSPS and LmTSA are localized to the extremities of the S3 pocket (not shown), where the sequence identity is poor (supplemental Fig. S5). This can be rationalized in terms of the substrate structures because in EcGSPS the S3 site does not accommodate glutathionylspermidine, only the smaller spermidine. A more open S3 pocket bounded by flexible loops allows LmTSA to bind spermidine, the larger spermine (26), and N1-glutathionylspermidine.
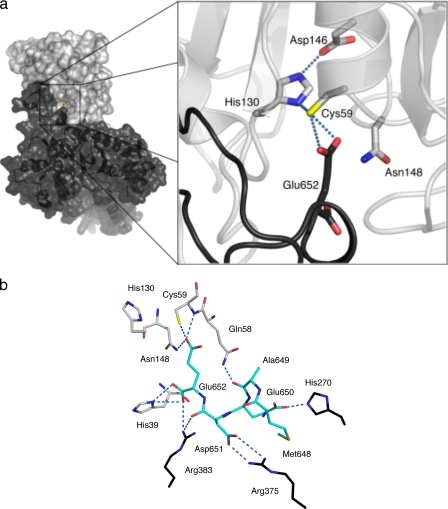
The Amidase Active Site—The amidase active site is near the N-terminal segment of α1. Site-directed mutagenesis identified Cys-59 as the catalytic cysteine (55). The catalytic triad is completed by His-130 and Asp-146 (Fig. 5).
FIGURE 5.
The amidase active site is blocked. a, a van der Waals surface representation of LmTSA is shown on the left side with the synthetase domain colored black and the amidase domain gray. The amidase active site is expanded on the right side. The secondary structure around the active site with the catalytic triad and Asn-148 side chains colored according to atom type, C is gray, N is blue, O is red, and S is yellow. The C terminus is a black coil with the final residue shown in stick representation with O positions in red. b, hydrogen bonding interactions in the amidase active site. Residues are depicted as sticks colored according to atom type, N is blue, O is red, S is yellow, and C atoms from the residues in the amidase domain are gray from the synthetase domain (black) and the C terminus (cyan). Dashed lines represent possible hydrogen bonding interactions.
The biochemistry of cysteine proteases, especially papain, is well documented (56). A superposition of the amidase domain of LmTSA with a papain structure containing peptide fragments in the active site (57) identifies important similarities (supplemental Figs. S6 and S7) and informs on the orientation of the amide bond to be cleaved with respect to the catalytic machinery. In LmTSA, the C-terminal Glu-652 carboxylate is on one side of Cys-59 SG (Fig. 5). When LmTSA is superposed on the papain-peptide complex, then the N terminus of the peptide fragment is positioned on the other side of Cys-59 SG. We take these groups to represent the approximate positions of a carboxylate and a terminal amine at the end of a reaction. The amidase mechanism involves a Cys-59—His-130 thiolate-imidazolium pair with His-130 oriented by a hydrogen bond to the Asp-146 carboxylate. Generation of a thiolate allows nucleophilic attack by Cys-59 onto the glycine-spermidine peptidic bond to form an acyl thioester. In reaction III (Fig. 1), hydrolysis of the thioester, exploiting an activated water, releases glutathione and glutathionylspermidine. In reaction IV the thioester is formed with glutathionylspermidine, and cleavage releases glutathione and spermidine.
Two related features of the LmTSA amidase active site are significant. First, Asn-148 occupies the position of Gln-19 in papain. In papain, the Gln-19 side chain, in combination with the main chain amide of the catalytic cysteine, forms an oxyanion hole to support the thiolate attack on the amide linkage by stabilizing the tetrahedral intermediate before formation of an acyl-enzyme complex (56). However, for LmTSA Asn-148 to contribute a similar function requires a different rotamer. Second, and most striking, is the observation that the side chain of the C-terminal residue (Glu-652) is directed into the amidase active site, interacting with Cys-59, forcing Asn-148 into a non-productive conformation.
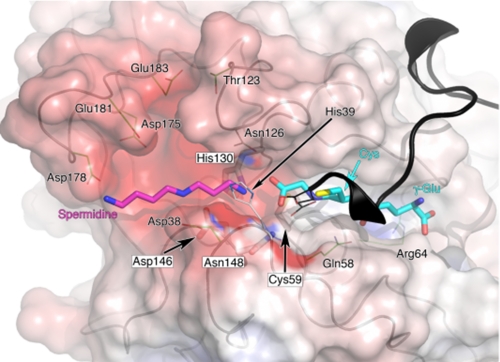
The C terminus of LmTSA breaks away from the synthetase domain at Lys-632 to interact with the amidase domain (Figs. 3a and 5). The C-terminal segments of Leishmania TSA sequences are highly conserved and acidic (supplemental Fig. S5). The last three residues in LmTSA are Glu-650—Asp-651-Glu-652. These residues participate in hydrogen bonding and salt bridge interactions with basic amino acid side chains that hold the amidase and synthetase domains in the arrangement observed and block access to the catalytic Cys-59 (Fig. 5). Glu-650 interacts with His-270, and Asp-651 associates with Arg-375 in addition to a main chain carbonyl hydrogen bonding interaction with Arg-383 NH2. The C-terminal carboxylate on Glu-652 interacts with Arg-383, and His-39 and the side chain accepts hydrogen bonds donated by Cys-59 SG and Asn-148 ND2. As with the synthetase active site, all attempts to co-crystallize with or soak ligands into the amidase active site failed. We attribute this to the presence of the C-terminal residues blocking the active site. Nevertheless, alignment of the C terminus in the active site and comparisons with the papain peptide complex (supplemental Fig. S7) indicate how substrates are aligned for catalysis. To correctly position the scissile bond with respect to Cys-59 for reaction III, then one γ-Glu-Cys-Gly component of trypanothione must lie along the groove (Fig. 6). Residues 647-652 must vacate the groove to allow access to the catalytic center. The spermidine and other γ-Glu-Cys-Gly components are directed out of the active site toward an acidic cleft. Here, Asp-38, Thr-123, Asp-178, Glu-181, and Glu-183 are placed to attract and interact with the substrate (Fig. 6). Near the catalytic site His-39 and Asn-126 may interact with the central amine of spermidine. At the onset of reaction IV (Fig. 1), glutathionylspermidine binds with the γ-Glu-Cys-Gly moiety aligned as before, the positively charged spermidine extending out toward the negatively charged surface of the cavity, allowing the second hydrolytic reaction to occur.
FIGURE 6.
A model for one of the amidase product complexes. A model for the end of reaction IV is shown. The protein surface is a semitransparent van der Waals surface, colored gray for neutral residues, red for acidic residues, and blue for basic residues. The main chain is traced in gray. The black ribbon is the C-terminal segment, which has been removed from the surface calculation to indicate the cleft shape and charge leading into Cys-59. The surface for His-30 has been omitted for the purpose of clarity. GSH and spermidine are depicted as sticks with C atoms colored cyan and purple, respectively. Residues discussed under The Amidase Active Site are shown as sticks, colored and labeled as in Fig. 4.
The Regulation of Conflicting Activities—In EcGSPS a 15-fold activation of glutathionylspermidine hydrolysis occurs when the synthetase substrates GSH and ATP are present and up to a 70-fold activation of amidase activity, dependent on substrate type, when the domain is isolated from the synthetase domain (27). This suggests communication between or interdependence of enzyme activities. The entrance to the EcGSPS amidase active site, near the synthetase domain, is accessible by a solvent-filled cavity. The enzyme is a homodimer, but the subunit interface is distant from the amidase active site (not shown), and the mechanism of activation or communication in that enzyme remains unclear. Our attempts to obtain a soluble N-terminal amidase domain of LmTSA have failed. The C-terminal segment of the protein contributes to the amidase domain structure, and its loss likely destabilizes the fold.
The bifunctional TSA must regulate or balance conflicting amidase and synthetase activities to achieve the relative concentrations of GSH, spermidine, glutathionylspermidine, and T[SH]2. In LmTSA the structure of the C terminus, which blocks the amidase active site, provides the mechanism to regulate such activity whereby a conformational change is required to vacate the active site for substrate binding. This may result from changes in domain orientation or could be driven by substrate concentrations. The amino acids responsible for placing the C terminus in the amidase active site are strictly conserved in Leishmania and C. fasciculata TSA with the exception of Glu-181 in the latter enzyme, conservatively replaced by aspartate. In Trypanosoma species the amino acid sequences at the C-terminal segment differs from Leishmania and C. fasciculata TSA sequences (supplemental Fig. S4). This implies differences at this part of the amidase active site and, therefore, a different mode of regulation. Further studies are required to investigate this issue in Trypanosoma.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility, Grenoble for synchrotron beam time and excellent staff support.
Author's Choice—Final version full access.
The atomic coordinates and structure factors (codes 2vps, 2vpm, and 2vob) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by the Biotechnology and Biological Sciences Research Council (Structural Proteomics of Rational Targets) United Kingdom and Wellcome Trust Grants 082596, 079838, and 083481. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S7.
Footnotes
The abbreviations used are: T[SH]2, N1,N8-bis(glutathionyl)spermidine; GSPS, glutathionylspermidine synthetase amidase (EC 6.3.1.8; EC 3.5.1.78); r.m.s.d., root mean square deviation; TSA, trypanothione synthetase-amidase (EC 6.3.1.9; EC 3.5.1.78); LmTSA, L. major TSA; SeMet, selenomethionine; EcGSPS, E. coli GSPS.
References
- 1.Reithinger, R., Dujardin, J. C., Louzir, H., Pirmez, C., Alexander, B., and Brooker, S. (2007) Lancet Infect. Dis. 7 581-596 [DOI] [PubMed] [Google Scholar]
- 2.Barrett, M. P., Boykin, D. W., Brun, R., and Tidwell R. R. (2007) Br. J. Pharmacol. 152 1155-1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croft, S. L., Sundar, S., and Fairlamb, A. H. (2006) Clin. Micro. Rev. 19 111-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delespaux, V., and de Koning, H. P. (2007) Drug Resist. Updat. 10 30-50 [DOI] [PubMed] [Google Scholar]
- 5.Chaudhary, K., and Roos, D. S. (2005) Nat. Biotechnol. 23 1089-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marton, L. J., and Pegg, A. E. (1995) Annu. Rev. Pharmacol. Toxicol. 35 55-91 [DOI] [PubMed] [Google Scholar]
- 7.Bacchi, C. J., and Yarlett, N. (2002) Mini Rev. Med. Chem. 2 553-563 [DOI] [PubMed] [Google Scholar]
- 8.Heby, O., Persson, L., and Rentala, M. (2007) Amino Acids 33 359-366 [DOI] [PubMed] [Google Scholar]
- 9.Fairlamb, A. H., and Cerami, A. (1992) Annu. Rev. Microbiol. 46 695-729 [DOI] [PubMed] [Google Scholar]
- 10.Krauth-Siegel, R. L., Bauer, H., and Schirmer, R. H. (2005) Angew. Chem. Int. Ed. Engl. 44 690-715 [DOI] [PubMed] [Google Scholar]
- 11.Bond, C. S., Zhang, Y., Berriman, M., Cunningham, M. L., Fairlamb, A. H., and Hunter, W. N. (1999) Structure 7 81-89 [DOI] [PubMed] [Google Scholar]
- 12.Alphey, M. S., Leonard, G. A., Gourley, D. G., Tetaud, E., Fairlamb, A. H., and Hunter, W. N. (1999) J. Biol. Chem. 274 25613-25622 [DOI] [PubMed] [Google Scholar]
- 13.Alphey, M. S., Bond, C. S., Tetaud, E., Fairlamb, A. H., and Hunter, W. N. (2000) J. Mol. Biol. 300 903-916 [DOI] [PubMed] [Google Scholar]
- 14.Alphey, M. S., Gabrielsen, M., Micossi, E., Leonard, G. A., McSweeney, S. M., Ravelli, R. B., Tetaud, E., Fairlamb, A. H., Bond, C. S., and Hunter, W. N. (2003) J. Biol. Chem. 278 25919-25925 [DOI] [PubMed] [Google Scholar]
- 15.Krieger, S., Schwarz, W., Ariyanayagam, M. R., Fairlamb, A. H., Krauth-Siegel, R. L., and Clayton, C. (2000) Mol. Microbiol, 35 542-552 [DOI] [PubMed] [Google Scholar]
- 16.Henderson, G. B., Yamaguchi, M., Novoa, L., Fairlamb, A. H., and Cerami, A. (1990) Biochemistry 29 3924-3929 [DOI] [PubMed] [Google Scholar]
- 17.Smith, K., Nadeau, K., Bradley, M., Walsh, C., and Fairlamb, A. H. (1992) Protein Sci. 1 874-883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tetaud, E., Manai, F., Barrett, M. P., Nadeau, K., Walsh, C. T., and Fairlamb, A. H. (1998) J. Biol. Chem. 273 19383-19390 [DOI] [PubMed] [Google Scholar]
- 19.Comini, M., Menge, U., Wissing, J., and Flohé, L. (2005) J. Biol. Chem. 280 6850-6860 [DOI] [PubMed] [Google Scholar]
- 20.Oza, S. L., Tetaud, E., Ariyanayagam, M. R., Warnon, S. S., and Fairlamb, A. H. (2002) J. Biol. Chem. 277 35853-35861 [DOI] [PubMed] [Google Scholar]
- 21.Comini, M., Guerrero, S. A., Haile, S., Menge, U., Lünsdorf, H., and Flohé, L. (2004) Free Radic. Biol. Med. 36 1289-1302 [DOI] [PubMed] [Google Scholar]
- 22.Oza, S. L., Shaw, M. P., Wyllie, S., and Fairlamb, A. H. (2005) Mol. Biochem. Parasitol. 139 107-116 [DOI] [PubMed] [Google Scholar]
- 23.Ariyanayagam, M. R., Oza, S. L., Guther, M. L., and Fairlamb, A. H. (2005) Biochem. J. 391 425-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oza, S. L., Wyllie, S., and Fairlamb, A. H. (2006) Mol. Biochem. Parasitol. 149 117-120 [DOI] [PubMed] [Google Scholar]
- 25.Oza, S. L., Ariyanayagam, M. R., Aitcheson, N., and Fairlamb, A. H. (2003) Mol. Biochem. Parasitol. 131 25-33 [DOI] [PubMed] [Google Scholar]
- 26.Bollinger, J. M., Jr., Kwon, D. S., Huisman, G. W., Kolter, R., and Walsh, C. T. (1995) J. Biol. Chem. 270 14031-14041 [DOI] [PubMed] [Google Scholar]
- 27.Kwon, D. S., Lin, C. H., Chen, S. J., Coward, J. K., Walsh, C. T., and Bollinger, J. M., Jr. (1997) J. Biol. Chem. 272 2429-2436 [DOI] [PubMed] [Google Scholar]
- 28.Pai, C. H., Chiang, C. C., Ko, T. P., Chou, C. C., Chong, C. M., Yen, F. J., Chen, S., Coward, J. K., Wang, A. H. J., and Lin, C. H. (2006) EMBO J. 25 5970-5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bateman, A., and Rawlings, N. D. (2003) Trends Biochem. Sci. 28 234-237 [DOI] [PubMed] [Google Scholar]
- 30.Rigden, D. J., Jedrzejas, M. J., and Galperin, M. Y. (2003) Trends Biochem. Sci. 28 230-234 [DOI] [PubMed] [Google Scholar]
- 31.Ravaschino, E. L., Docampo, R., and Rodriguez, J. B. (2006) J. Med. Chem. 12 426-435 [DOI] [PubMed] [Google Scholar]
- 32.Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., and Bairoch, A. (2005) The Proteomics Protocols Handbook (Walker, J. M., ed.) pg. 571-607, Humana Press Inc., Totowa, NJ
- 33.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795-800 [Google Scholar]
- 34.Leslie, A. G. W. (1992) Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, pp. 26-30, SERC, Daresbury Laboratory, Warrington, UK
- 35.Evans, P. (2006) Acta Crystallogr. Sect. D Biol. Crystallogr. 62 72-82 [DOI] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 37.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terwilliger, T. C. (2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56 965-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 40.Vagin, A., and Teplyakov, A. (1997) J. Appl. Crystallogr. 30 1022-1025 [Google Scholar]
- 41.Murshudov, G. N., Lebedev, A., Vagin, A. A., Wilson, K. S., and Dodson, E. J. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 247-255 [DOI] [PubMed] [Google Scholar]
- 42.Winn, M., Isupov, M., and Murshudov, G. N. (2001) Acta Crystallogr. Sect. D Biol. Crystallogr. 57 122-133 [DOI] [PubMed] [Google Scholar]
- 43.Lovell, S. C., Davies, I. W., Arendall, W. B., III, de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J., and Richardson D. C. (2003) Proteins 50 437-450 [DOI] [PubMed] [Google Scholar]
- 44.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 45.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA
- 46.Kabsch, W. (1976) Acta Crystallogr. Sect. A 32 922-923 [Google Scholar]
- 47.Qi, G., Lee, R., and Hayward, S. (2005) Bioinformatics 21 2832-2838 [DOI] [PubMed] [Google Scholar]
- 48.Krissinel, E., and Henrick, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2256-2268 [DOI] [PubMed] [Google Scholar]
- 49.Anantharaman, V., and Aravind, L. (2003) Genome Biology 4 R11.1-R11.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galperin, M. Y., and Koonin, E. V. (1997) Protein Sci. 6 2639-2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gogos, A., and Shapiro, L. (2002) Structure 10 1669-1676 [DOI] [PubMed] [Google Scholar]
- 52.Hara, T., Kato, H., Katsube, Y., and Jun'ichi, O. (1996) Biochemistry 35 11967-11974 [DOI] [PubMed] [Google Scholar]
- 53.Dinescu, A., Cundari, T. R., Bhansali, V. K., Luo, J. L., and Anderson, M. E. (2004) J. Biol. Chem. 279 22412-22421 [DOI] [PubMed] [Google Scholar]
- 54.Hiratake, J. (2005) Chem. Rec. 5 209-228 [DOI] [PubMed] [Google Scholar]
- 55.Oza, S. L., Ariyanayagam, M. R., and Fairlamb, A. H. (2002) Biochem. J. 364 679-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Storer, A. C., and Ménard, R. (1994) Methods Enzymol. 244 486-500 [DOI] [PubMed] [Google Scholar]
- 57.Alphey, M. S., and Hunter, W. N. (2006) Acta Crystallogr. F Struct. Biol. Crystalliz. Comm. 62 504-508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.