Abstract
The positive transcription elongation factor b (P-TEFb) is a heterodimeric complex composed of cyclin-dependent kinase 9 and its regulator cyclin T1/2. It stimulates transcription elongation by phosphorylation of serine 2 residues in the carboxy-terminal domain of polymerase II. 7SK RNA and HEXIM proteins can antagonize transcriptional stimulation by sequestering P-TEFb in a catalytically inactive ribonucleoprotein (RNP). Here, we show that the previously uncharacterized La-related protein 7 (LARP7) has a role in 7SK-mediated regulation of transcription. LARP7 binds to the highly conserved 3′-terminal U-rich stretch of 7SK RNA and is an integral part of the 7SK RNP. On stimulation, LARP7 remains associated with 7SK RNA, whereas P-TEFb is released. Interestingly, reduction of LARP7 by RNA interference enhances transcription from cellular polymerase II promoters, as well as a TAT-dependent HIV-1 promoter. Thus, LARP7 is a negative transcriptional regulator of polymerase II genes, acting by means of the 7SK RNP system.
Keywords: 7SK RNA, LARP7, P-TEFb
Introduction
A network of positive and negative regulators controls the transcription elongation phase of polymerase II (POLII). An essential component of this regulatory circuit is the positive transcription elongation factor b (P-TEFb), which is composed of cyclin-dependent kinase 9 (CDK9) and its regulatory subunit cyclin T1 (CYCT1; Peng et al, 1998; Price, 2000). It stimulates transcription elongation by phosphorylating specific serine residues in the carboxy-terminal domain of POLII, and the negative elongation factors NELF and DSIF (DRB sensitivity inducing factor), allowing for efficient messenger RNA production (Wada et al, 1998; Yamaguchi et al, 1999; Zhou & Yik, 2006).
In growing cells, a fraction of P-TEFb exists as an active CDK9–CYCT1 dimer, which associates with other factors allowing its recruitment to sites of transcription. Two established examples for these factors are bromodomain containing 4 (BRD4) and the TAT protein of HIV-1 (Jones, 1997; Zhu et al, 1997). BRD4 binds to acetylated chromatin and targets P-TEFb to sites of transcription. Transcription of HIV-1, by contrast, is activated by the TAT protein, which simultaneously binds to the transactivation response element of nascent transcripts and active P-TEFb (Karn, 1999).
P-TEFb activity can be downregulated by sequestration into a catalytically inactive ribonucleoprotein (RNP), the known components of which are the HEXIM (hexamethylene bis-acetamide inducible 1) protein 1 or 2, and the 7SK RNA (Nguyen et al, 2001; Yang et al, 2001; Yik et al, 2003). A stem–loop structure located in the 5′ part of 7SK RNA provides the binding site for homo- or heterodimeric HEXIM proteins, which recruits P-TEFb, through interaction with CYCT1, to the 7SK RNP (Egloff et al, 2006). The association of P-TEFb with the 7SK RNP is responsive to changes in growth conditions. 7SK RNA association is favoured when high-level transcription is not required, for example, under starvation conditions. By contrast, P-TEFb becomes activated in proliferating cells or when transcription elongation is compromised (Chao & Price, 2001; Chen et al, 2004). Recently, a remodelling of the 7SK RNP has been observed as a result of P-TEFb activation, which results in the replacement of HEXIM and P-TEFb with heterogeneous nuclear ribonucleoproteins (hnRNP) and RNA helicase A (Van Herreweghe et al, 2007); the trigger for this process remains unknown.
Initially, we aimed to identify proteins interacting with 5′-terminal oligopyrimidine (TOP) motifs, U-rich sequences present in many mRNAs encoding translation factors. These elements are required for translational repression of TOP mRNAs under growth arrest (Meyuhas, 2000). Here, we identify the La-related protein 7 (LARP7) as an oligopyrimidine-binding protein. Surprisingly, although LARP7 binds to TOP motifs in vitro, its primary cellular target is the 7SK RNA. LARP7 provides a bridge between 7SK RNA and components of the 7SK RNP—CDK9 and HEXIM1—and acts as a crucial factor in the regulation of P-TEFb through 7SK RNP.
Results
Identification of LARP7 as an oligo-U-binding protein
To identify proteins interacting with TOP motifs, we performed ultraviolet-crosslinking studies in HeLa extracts with 32P-labelled oligonucleotides corresponding to either the TOP motif of ribosomal protein S16 mRNA (oligo-U) or a mutated version thereof. A 75 kDa protein showed crosslinking to the oligo-U sequence but not to the control (Fig 1A, lanes 1,2). Two RNA transcripts were generated that consisted of either the oligo-U target or the control sequence, followed by a tobramycin aptamer. These transcripts were immobilized on a tobramycin matrix and used to purify the crosslinked protein. One protein that eluted from the oligo-U column in the expected size range was identified by mass spectrometry as LARP7 (accession number EU667388; Fig 1B, compare lane 1 with lane 2). LARP7 was strongly enriched in the eluate of the oligo-U column and crosslinking was severely impaired in immunodepleted extracts. Hence, LARP7 is the protein that crosslinks to the oligo-U target (Fig 1B, lanes 3,4; Fig 1C; Supplementary Fig 1 online). LARP7 belongs to a group of factors that share an amino-terminal La-like domain followed by one or two RNA-recognition motifs (Fig 1D; Wolin & Cedervall, 2002).
Figure 1.
LARP7 binds to oligo-U in vitro. (A) Ultraviolet crosslinking of 32P-labelled oligo-U (lane 1; for sequence details see Materials and Methods in the Supplementary information online) and a control oligoribonucleotide (Ctrl; lane 2) in a HeLa extract. A candidate 75 kDa protein is indicated by an arrowhead. (B) SDS–polyacrylamide gel electrophoresis of proteins isolated by affinity chromatography with tobramycin-tagged oligo-U (lane 1) or the control sequence (lane 2). Proteins of the 75 kDa range were analysed by mass spectrometry and western blot (WB) using LARP7 antibodies (lanes 3,4). (C) Characterization of a LARP7 antibody by immunoprecipitation (IP; lane 2) and western blot of HeLa extracts (lane 1). The abbreviation ‘hc' indicates heavy chains of the antibody, and ‘lc' indicates light chains of the antibody. (D) Domain structure of LARP7. LARP7, La-related protein 7; RRM1 and RRM3, RNA-recognition motif 1 and 3.
7SK RNA association of LARP7
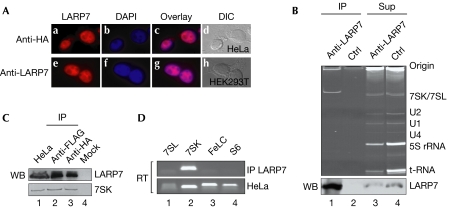
Immunofluorescence studies of endogenous and transfected haemagglutinin (HA)-tagged LARP7 showed a predominant nucleoplasmic localization, suggesting a role of LARP7 in this compartment (Fig 2A; see also Supplementary Fig 2 online). Hence, nuclear extracts were used to identify targets of LARP7 by co-immunoprecipitation. As shown in Fig 2B, LARP7 antibodies specifically immunoprecipitated LARP7 along with a single RNA of about 330 nucleotides (lanes 1,2). Identical results were obtained when transfected Flag-HA-tagged LARP7 was immunoprecipitated with antibodies against either tag (Fig 2C, lanes 2,3). The size of the RNA suggested that it might be either 7SK or 7SL RNA. To distinguish between the two, the immunoprecipitated RNA was amplified by reverse transcription–PCR (RT–PCR) with primer sets specific for 7SK, 7SL, a control mRNA (FeLC) and a U-rich mRNA (rpS6). All primer pairs amplified their respective RNA from total cellular RNA (Fig 2D, lower panel), but only the 7SK RNA-specific primer pair amplified the immunoprecipitated RNA (upper panel). These results were confirmed by sequencing (data not shown) and identified LARP7 as a 7SK RNA-associated factor.
Figure 2.
LARP7 associates with 7SK RNA. (A) HeLa cells transfected with Flag-HA-LARP7 were analysed by immunofluorescence with HA antibodies (a). Panel e shows non-transfected human embryonic kidney (HEK) 293T cells stained with LARP7 antibodies; panels b and f show 4,6-diamidino-2-phenylindole (DAPI) staining; panels c and g show merged images; and panels d and h show differential interference contrast (DIC) images. (B) Immunoprecipitates (IP) from extracts with LARP7 and control (Ctrl) antibodies (lanes 1,2) and the corresponding supernatants (Sup; lanes 3,4). RNAs of the immunoprecipitates and the corresponding supernatants were visualized by ethidium bromide staining. Specific RNAs and the origin of the gel are indicated on the right. The lower panel shows a western blot (WB) of the immunoprecipitates and supernatants probed with LARP7 antibodies. (C) Immunoprecipitation of Flag-HA-tagged LARP7 with HA or Flag antibodies (lanes 2,3). The upper panel shows a western blot of the immunoprecipitated LARP7 and the lower panel the co-precipitated RNA. The control (lane 4) was performed in the absence of primary antibodies; lane 1 represents 10% of the extract used for immunoprecipitates. (D) Identification of LARP7-associated RNA. The RNA identified in lane 1 of (B) was amplified with primer pairs specific for the indicated RNAs (upper panel). As a control, total cellular extract was used (lower panel). HA, haemagglutinin; LARP7, La-related protein 7; RT, reverse transcription.
LARP7 is a stable component of the 7SK RNP
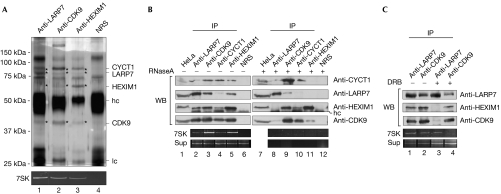
To investigate whether LARP7 is part of the known 7SK RNP implicated in the regulation of transcription, co-immunoprecipitation studies were performed using the nuclear extracts of HeLa cells (Fig 3A, lanes 1–4). Antibodies against LARP7, CDK9 and HEXIM1 efficiently precipitated 7SK RNA, whereas no RNA was found in the control (Fig 3A, lower panel, lanes 1–4). Interestingly, HEXIM1, CDK9, CYCT1 and LARP7 were present in all immunopurifications, as determined by mass spectrometry of silver-stained gels (Fig 3A; data not shown) and western blotting (Fig 3B, lanes 2,3,5; Fig 3C, lanes 1,2), but not in a purification with control antibodies. In addition, we detected substoichiometric amounts of the recently identified 7SK methylphosphate capping enzyme MePCE, as well as hnRNPA1 in the LARP7 purification (Fig 3A, lane 1; see also Supplementary Fig 3 online; Jeronimo et al, 2007; Van Herreweghe et al, 2007).
Figure 3.
LARP7 is a component of the 7SK ribonucleoprotein. (A) Immunoprecipitates from a HeLa extract with LARP7 (lane 1), CDK9 (lane 2), HEXIM1 (lane 3) antibodies and control serum (NRS; lane 4). Proteins were separated by SDS–polyacrylamide gel electrophoresis and bands marked with an asterisk were identified by mass spectrometry. The lower panel shows an ethidium bromide staining of the co-purified 7SK RNA. The abbreviation ‘hc' indicates heavy chains of the antibody, and ‘lc' indicates light chains of the antibody. (B) Mock-treated (lanes 1–7) or RNase A-treated (lanes 8–12) extracts were immunoprecipitated (IP) with the indicated antibodies. Immunoblots were probed with the indicated antibodies. 7SK RNA from the immunoprecipitates and the supernatants (Sup) are presented in the lower two rows. (C) Extracts derived from mock- or DRB-treated HeLa cells were immunoprecipitated with LARP7 and CDK9 antibodies. The indicated proteins were detected by western blot and 7SK RNA was visualized by ethidium bromide staining. CDK9, cyclin-dependent kinase 9; CYCT1, cyclin T1; DRB, 5,6-dichloro-1-β-D-ribofuranosyl benzimidazole; HEXIM1, hexamethylene bis-acetamide inducible 1; LARP7, La-related protein 7; WB, western blot.
Next, we determined whether the observed association relied on protein–protein or protein–RNA interactions. To investigate whether these factors are kept together by 7SK RNA, co-immunoprecipitation studies were carried out using either untreated or RNase-treated extracts. As shown in Fig 3B, LARP7, HEXIM1, CDK9, CYCT1 and 7SK RNA could be co-precipitated with antibodies recognizing any of these components (lanes 2–6; note that the CYCT1 antibody precipitated LARP7 and HEXIM1 only poorly). However, this pattern changed significantly when extracts were pretreated with RNase (lanes 7–12). Although LARP7 could still be precipitated efficiently, neither CDK9 nor HEXIM1 proteins co-purified. Similarly, no significant co-immunoprecipitation of LARP7 was observed after RNase treatment with antibodies against CDK9, CYCT1 or HEXIM1 (lanes 8–11). In this assay, association of a fraction of HEXIM1 with P-TEFb was still observed, although 7SK RNA was entirely degraded (Fig 3B, lanes 9–11, see the lower panel for 7SK RNA). We conclude that the interaction of LARP7 with the 7SK RNP is largely RNA mediated.
As P-TEFb dissociates from 7SK RNP after treatment with the POLII inhibitor 5,6-dichloro-1-β-D-ribofuranosyl benzimidazole (DRB), we wanted to find out how this treatment affects the association of LARP7 with this RNP (Nguyen et al, 2001). Immunoprecipitations from control or DRB-treated extracts were performed using LARP7 and CDK9 antibodies. Both co-immunoprecipitated LARP7, HEXIM1, CDK9 and 7SK RNA from the untreated extract (Fig 3C, lanes 1,2); however, after DRB treatment, LARP7 remained associated with 7SK RNA, whereas HEXIM1 and CDK9 dissociated (lane 3). Accordingly, an anti-CDK9 immunoprecipitation showed a strongly reduced association of CDK9 with 7SK RNA and LARP7 (lane 4). These data show that, in contrast to P-TEFb and HEXIM1, LARP7 is a stable component of the 7SK RNP complex.
Multiple interactions between LARP7 and the 7SK RNP
Next, we investigated how LARP7 associates with the 7SK RNP. Initially, we wanted to find out which sequence element of 7SK RNA comes in contact with LARP7. Owing to the U-rich 3′-terminal stretch, we suggested that it might be involved in binding. To test this, recombinant LARP7 was incubated with either 32P-labelled 7SK RNA or a mutant lacking the 3′-terminal 18 nucleotides and then separated on a native RNA gel. LARP7 retarded full-length but not mutant 7SK RNA (Fig 4A, lanes 3,4). Furthermore, the addition of a tenfold molar excess of unlabelled RNA encompassing the terminal 18 nucleotides strongly reduced binding, whereas the addition of LARP7 antibodies caused a supershift of the 7SK–LARP7 complex but not of 7SK RNA alone (Fig 4A, lanes 5,6,9). Thus, LARP7 makes direct contact with 7SK RNA by binding to the U-rich 3′ terminus. Remarkably, the N terminus of LARP7 containing the La motif bound to 7SK RNA with the same efficiency as the full-length protein (Fig 4A, compare lane 3 with 7). We conclude that the 3′ terminus of 7SK RNA binds to the N-terminal domain of LARP7.
Figure 4.
Interactions of LARP7 with components of the 7SK ribonucleoprotein. (A) Direct binding of LARP7 to the 3′ terminus of 7SK RNA. 32P-labelled 7SK RNA (lanes 1,3,5,6) or a mutant lacking the 3′-terminal 18 nucleotides (7SKmut; lanes 2,4,8) was incubated with either LARP7 (lanes 3–6) or an amino-terminal fragment thereof (lanes 7,8). In lane 5, a tenfold molar excess of an oligonucleotide corresponding to the terminal 18 nucleotides was added to the binding reaction. In lanes 6 and 9, a LARP7 antibody was added to the reaction mixtures. Samples were separated by native gel electrophoresis and visualized by autoradiography. (B) Immobilized GST-CYCT1 binds to 35S-labelled CDK9 (lane 2) but not to LARP7 (lane 5). Lanes 1 and 4 show 20% of translates used for binding assays, and lanes 3 and 6 show binding to the GST control. (C) Immobilized GST-HEXIM1 binds weakly to 35S-labelled CDK9 (lane 2) and strongly to LARP7 (lane 5). Lanes 1 and 4 show 20% of translates used, and lanes 3 and 6 show GST controls. (D) Binding of 35S-labelled CDK9 to immobilized GST-LARP7 and deletion mutants thereof (lanes 2–6). Lane 1 shows the input of translated protein and lane 7 shows the GST control. GST proteins are shown in the upper gel and the in vitro translates are shown in the lower gel of all binding experiments. (E) A model illustrating interactions in the 7SK RNP. CDK9, cyclin-dependent kinase 9; CYCT, cyclin T1; GST, glutathione-S-transferase; HEXIM1, hexamethylene bis-acetamide inducible 1; LARP7, La-related protein 7; RNP, ribonucleoprotein; RRM, RNA-recognition motif.
Previous studies have shown that HEXIM1 binds to the 5′ stem–loop of 7SK and provides, along with the 3′ stem–loop of 7SK, the docking site for the P-TEFb component CYCT1 (Egloff et al, 2006). To test whether LARP7 also comes in contact with the proteins of this RNP, in vitro binding studies were performed (Fig 4B–D). CYCT1 bound to CKD9 but not to LARP7 (Fig 4B, lanes 2,5, upper and lower panels). Interactions of HEXIM1 with CDK9 and LARP7 were also observed in these assays (Fig 4C, lanes 2,5, upper and lower panels). Binding of CDK9 to LARP7 occurred through its C-terminal part encompassing amino acids 375–589 (Fig 4D, lanes 2–7). Thus, although binding of LARP7 to 7SK RNP is predominantly RNA-mediated (Fig 3B), additional protein–protein interactions are likely to contribute to RNP stability. A model summarizing the observed interactions is shown in Fig 4F.
A role for LARP7 in transcription of POLII genes
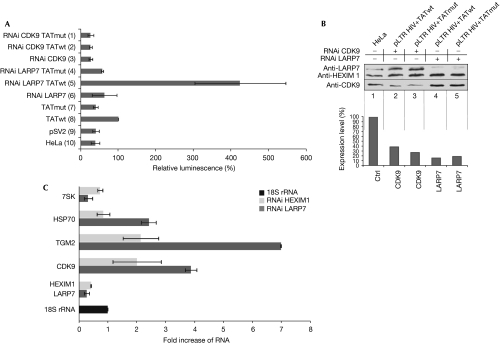
Transcription of the proviral HIV-1 genome is controlled mainly at the level of elongation and is dependent on the recruitment of P-TEFb to the active site by the transcriptional transactivator TAT (Price, 2000). To address the role of LARP7 in this process, TAT-dependent transcription from the HIV-1 long terminal repeat (LTR) promoter was analysed. TZMbl cells expressing stably integrated firefly luciferase under the control of the HIV LTR promoter were transfected with either LARP7 or CDK9 short interfering RNAs (siRNAs). After 24 h, a plasmid encoding wild-type or mutant TAT was co-transfected together with a second boost of siRNAs. Gene expression rates (Fig 5A) and knockdown efficiencies (Fig 5B) were assessed after 24 h. In the absence of TAT, or when mutant TAT was expressed, transcription was inefficient (Fig 5A, bars 7,9,10). However, transcription was stimulated when functional TAT was expressed (bar 8). Markedly, knockdown of LARP7 by approximately 75% (Fig 5B, lanes 4,5) enhanced TAT-dependent transcription three- to fourfold (Fig 5A, compare bar 5 with bar 8). By contrast, knockdown of CDK9 severely reduced expression of the reporter (Fig 5A, compare bar 2 with bar 8). The transcription rate was not influenced by these knockdowns when TAT was absent (see bars 1,3,4,6). Thus, LARP7 is a new component of 7SK RNP that represses TAT-dependent transcription in vivo. These data are further supported by experiments shown in Supplementary Fig 4 online.
Figure 5.
Knockdown of LARP7 enhances polymerase II transcription. (A) TAT-dependent transcription of a firefly luciferase reporter under the control of the HIV-1 long terminal repeat promoter in HeLa cells. The reporter is activated on transfection of a plasmid encoding the HIV-1 TAT protein (TATwt; bar 8), whereas mutant TAT (TATmut; bar 7) fails to activate (bars 9,10). The effect on reporter expression as a result of transfection of siRNAs against LARP7 (RNAi LARP7) is shown in bars 4–6 and that against CDK9 (RNAi CDK9) in bars 1–3. (B) Western blot of cellular extract treated with CDK9 (lanes 2,3) or LARP7 siRNA (lanes 4,5) and quantification of the signals normalized to HEXIM1, CDK9 and LARP7 (lane 1; lower panel). (C) Quantitative PCR levels of the indicated RNAs after RNAi treatment of LARP7 (dark bars) or HEXIM1 (light bars), normalized to 18S ribosomal RNA (18S). CDK9, cyclin-dependent kinase 9; HEXIM1, hexamethylene bis-acetamide inducible 1; LARP7, La-related protein 7; RNAi, RNA interference; siRNA, short interfering RNA.
Next, we tested whether LARP7 also has a role in the transcription of cellular POLII genes. For this, HeLa cells were transfected with siRNAs against either LARP7 or HEXIM1, and transcription from various genes was analysed by real-time PCR compared with 18S ribosomal RNA levels as control. When LARP7 expression was reduced by RNA interference treatment by about 73%, transcription of three POLII genes, transglutaminase 2 (TGM2), heat-shock protein 70 (HSP70) and CDK9, was upregulated three- to sevenfold. A similar, albeit less severe, effect was observed after knockdown of HEXIM1. Interestingly, in contrast to the stimulatory effect on POLII transcripts, 7SK RNA was significantly destabilized. In conclusion, these experiments show that LARP7 negatively regulates not only viral but also cellular POLII class genes through the 7SK P-TEFb system.
Discussion
The activity of transcription elongation factor P-TEFb is under tight control, thus ensuring adjustment of the cell to altered growth conditions. A crucial event in the regulation of this factor is its reversible association with the 7SK RNP, which determines whether P-TEFb is inactive (RNP bound) or active (dissociated from RNP; Yik et al, 2003). We have identified LARP7 as a new P-TEFb-interacting factor with a role in 7SK RNP-mediated transcriptional control. Several lines of evidence support this view: first, immunoprecipitation of LARP7 co-precipitates 7SK RNA. The N-terminal part of LARP7, composed of a La domain and an RNA recognition motif, mediates binding to the U-rich 3′ terminus of 7SK RNA. Second, LARP7 makes contact with CDK9 and HEXIM1—that is, two proteins of the 7SK RNP. Finally, on treatment of cells with DRB, LARP7 remains stably associated with 7SK RNA, although HEXIM1 and CDK9 dissociate under these conditions. We have also detected low amounts of hnRNPA1 in purifications of LARP7; hence, LARP7 might be part of not only the inhibitory 7SK RNP but also the recently discovered second 7SK RNP that forms when P-TEFb is released from 7SK RNA (Hogg & Collins, 2007; Van Herreweghe et al, 2007). Our findings are consistent with recent publications reporting a role of LARP7 in P-TEFb-mediated transcriptional regulation (Jeronimo et al, 2007; He et al, 2008; Krueger et al, 2008).
RNAi-mediated silencing shows that POLII transcription from cellular as well as viral (HIV-1) genes was stimulated after LARP7 ablation. One possible mechanism of LARP7 function is the regulated release of P-TEFb from the 7SK RNP. This process is known to depend on CDK9 phosphorylation (Chen et al, 2004), which might be facilitated directly or indirectly by LARP7. The close proximity of the 7SK RNA-binding sites for LARP7 and CYCT1, as well as the direct interaction of LARP7 with CDK9 and HEXIM1, is consistent with this view. Alternatively, and not mutually exclusive with the first model, LARP7 might exert its effect on transcription by affecting the stability of 7SK RNA. In accordance, we observed a significant destabilization of 7SK RNA after RNAi-induced LARP7 reduction. LARP7 has also been found to associate with the TOP motif in vitro. Whether it also interacts with TOP mRNAs in vivo and has a function in translational regulation remains to be investigated.
The La protein acts as a termination and maturation factor of all POLIII-transcribed RNAs, but the functions of the other LARPs are not yet clear. La binds directly through its La domain to 3′-terminal U stretches, which is a common characteristic of POLIII-transcribed RNAs including 7SK (Wolin & Cedervall, 2002). As we failed to detect La in our anti-LARP7 immunoprecipitations, it is possible that both proteins bind to 7SK in a mutually exclusive manner. Hence, La might associate with 7SK RNA only at an early stage in its maturation pathway and be replaced by LARP7 at later stages to allow the formation of inhibitory 7SK RNP. We speculate that the La protein is replaced from other POLIII-transcribed RNAs in an analogous manner by members of the LARP family, thereby directing the RNA to specific pathways and/or functions.
Methods
In vitro transcription and RNA-binding assays. In vitro-transcribed 32P-labelled wild-type or mutant 7SK RNAs were incubated with 250 ng of either recombinant full-length LARP7 or a fragment thereof (amino acids 1–197) for 20 min at 20°C in binding buffer (10 mM HEPES, pH 7.5, 10 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol, 5% glycerol and 0.1 μg transfer RNA). Before loading onto a native 6% PAA gel, 2 μg of heparin was added and incubated for 10 min. For supershifts, 1 μg of LARP7 antibody was added.
Immunoprecipitation, RT–PCR and western blotting. Polyclonal antibodies against LARP7 were raised by immunizing rabbits with bacterially expressed full-length 6 × His-LARP7 and affinity purified. Cells were incubated with or without 100 μM DRB for 1 h and immunoprecipitations and western blotting were carried out as described previously (Otter et al, 2007). RNA was phenol extracted and amplified by RT–PCR with superscript II reverse transcriptase (Invitrogen, Karlsrune, Germany).
In vitro interaction assays. 35S-labelled proteins were produced as reported previously (Otter et al, 2007). CYCT1, CDK9, LARP7 or truncated versions thereof were expressed as glutathione S-transferase (GST) fusions in Escherichia coli as described. For binding studies, in vitro-translated proteins were added to 2 μg of purified GST fusion protein, immobilized on glutathione-Sepharose (GE Healthcare, Munchen, Germany) and incubated at 4°C for 90 min. After washing, bound proteins were resolved on gels and visualized by Coomassie blue staining and autoradiography.
Luciferase assays. TZMbl cells grown in DMEM supplemented with 10% FCS and 1% Pen/Strep were transfected with siRNA using Oligofectamine (Invitrogen). After 24 h, the indicated TAT-expressing plasmids were transfected using Nanofectin (PAA). Cell extracts were prepared 24 h later and protein concentrations were normalized. Luciferase activity was determined by using standard chemiluminescence detection procedures.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
This work was supported by grants from the German Israel Foundation (no. I-819-66.13/2004) and the Rudolf-Virchow Centre (RVZ FZT-82).
Footnotes
The authors declare that they have no conflict of interest.
References
- Chao SH, Price DH (2001) Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276: 31793–31799 [DOI] [PubMed] [Google Scholar]
- Chen R, Yang Z, Zhou Q (2004) Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem 279: 4153–4160 [DOI] [PubMed] [Google Scholar]
- Egloff S, Van Herreweghe E, Kiss T (2006) Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol 26: 630–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q (2008) A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 29: 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Collins K (2007) RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 13: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C et al. (2007) Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 27: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA (1997) Taking a new TAK on tat transactivation. Genes Dev 11: 2593–2599 [DOI] [PubMed] [Google Scholar]
- Karn J (1999) Tackling Tat. J Mol Biol 293: 235–254 [DOI] [PubMed] [Google Scholar]
- Krueger BJ et al. (2008) LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res 36: 2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O (2000) Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem 267: 6321–6330 [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O (2001) 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414: 322–325 [DOI] [PubMed] [Google Scholar]
- Otter S, Grimmler M, Neuenkirchen N, Chari A, Sichmann A, Fischer U (2007) A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J Biol Chem 282: 5825–5833 [DOI] [PubMed] [Google Scholar]
- Peng J, Zhu Y, Milton JT, Price DH (1998) Identification of multiple cyclin subunits of human P-TEFb. Genes Dev 12: 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol 20: 2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T (2007) Dynamic remodelling of human 7SK RNP controls the nuclear level of active P-TEFb. EMBO J 26: 3570–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H (1998) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J 17: 7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T (2002) The La protein. Annu Rev Biochem 71: 375–403 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51 [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q (2001) The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414: 317–322 [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q (2003) Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell 12: 971–982 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Yik JH (2006) The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev 70: 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH (1997) Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev 11: 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information