Abstract
Assembly and mutual proximities of α, β, and γc subunits of the interleukin 2 receptors (IL-2R) in plasma membranes of Kit 225 K6 T lymphoma cells were investigated by fluorescence resonance energy transfer (FRET) using fluorescein isothiocyanate- and Cy3-conjugated monoclonal antibodies (mAbs) that were directed against the IL-2Rα, IL-2Rβ, and γc subunits of IL-2R. The cell-surface distribution of subunits was analyzed at the nanometer scale (2–10 nm) by FRET on a cell-by-cell basis. The cells were probed in resting phase and after coculture with saturating concentrations of IL-2, IL-7, and IL-15. FRET data from donor- and acceptor-labeled IL-2Rβ-α, γ-α, and γ-β pairs demonstrated close proximity of all subunits to each other in the plasma membrane of resting T cells. These mutual proximities do not appear to represent mAb-induced microaggregation, because FRET measurements with Fab fragments of the mAbs gave similar results. The relative proximities were meaningfully modulated by binding of IL-2, IL-7, and IL-15. Based on FRET analysis the topology of the three subunits at the surface of resting cells can be best described by a “triangular model” in the absence of added interleukins. IL-2 strengthens the bridges between the subunits, making the triangle more compact. IL-7 and IL-15 act in the opposite direction by opening the triangle possibly because they associate their private specific α receptors with the β and/or γc subunits of the IL-2R complex. These data suggest that IL-2R subunits are already colocalized in resting T cells and do not require cytokine-induced redistribution. This colocalization is significantly modulated by binding of relevant interleukins in a cytokine-specific manner.
Keywords: receptor assembly, cytokine binding, cell activation
Immune responses are regulated by a series of proteins termed cytokines. Cytokines exhibit a high degree of redundancy and pleiotropy controlling a wide range of functions in various cell types. The redundancy is explained in part by the sharing of common receptor subunits among members of the cytokine receptor superfamily. Each cytokine has its own private receptor but may share public receptors with other cytokines. The multisubunit interleukin 2 receptor (IL-2R) includes three elements: the 55-kDa IL-2Rα, the 70- to 75-kDa IL-2Rβ, and the 64-kDa IL-2Rγ subunits (1–7). There are three forms of cellular receptors for IL-2 based on their affinity for ligand and receptor subunit utilization: one with a very high affinity (IL-2Rαβγ, Kd10−11 M), one with an intermediate affinity (IL-2Rβγ, 10−9 M), and one with a lower affinity (IL-2Rα, 10−8 M). IL-2Rα is the private subunit employed solely by IL-2 whereas IL-2Rβ is shared by IL-2 and IL-15, and IL-2Rγ (now termed γc) is used in common by IL-2, IL-4, IL-7, IL-9, and IL-15 (3–7). Although the physical relationship among the IL-2R subunits on the cell surface has been the subject of extended analyses, certain aspects are still being debated. It is clear that on addition of IL-2 there is heterodimerization of the IL-2Rβ and γc chain cytoplasmic domains that mediate the signal for T cell proliferation (8, 9). Although the existence of the high affinity heterotrimer in the presence of IL-2 is not in question, there are diverse views as to whether the heterotrimer can exist in the absence of IL-2 or whether it requires the presence of this cytokine for its formation. Saito, Kondo and their coworkers, on the basis of kinetic binding studies on human T cells with varying numbers of IL-2Rα chains, proposed an “affinity conversion” model for the high affinity IL-2R (10, 11). This model proposes that IL-2Rα and IL-2Rβ subunits normally exist unassociated on the cell surface in the absence of ligand. The first mandatory reaction is proposed to be between IL-2 and IL-2Rα and then between this binary complex and IL-2Rβ. Subsequent studies using anti-IL-2Rα or anti-IL-2Rβ antibodies to inhibit high affinity binding at 4°C and 37°C have been interpreted to support this affinity-conversion model (12, 13). Other studies support the contrasting view that IL-2Rα and β exist in preformed complexes. In crosslinking studies performed in the absence of IL-2 there appeared to be a close physical association of IL-2Rα and IL-2Rβ (14, 15). Furthermore, evidence for IL-2Rα–IL-2Rβ heterodimers in the absence of IL-2 was obtained by Scatchard analysis by using cell lines expressing different numbers of IL-2Rα chains (16). Finally, we used an IL-2 mutant analog, F42A, that fails to bind to the isolated IL-2Rα subunit and an antibody, HIEI, that separates the IL-2Rα and β subunits to demonstrate that IL-2Rα and β exist as preformed complexes in which the affinity of IL-2Rβ for IL-2 is altered by the proximity of IL-2Rα through mechanisms that do not require the prior binding of IL-2 to IL-2Rα (17, 18). Kuziel and coworkers made similar observations (19). Another IL-2 analog, Lys-20, that fails to bind to isolated IL-2Rβ was used to show that the proximity of IL-2Rβ affects IL-2 binding by IL-2Rα by mechanisms that do not require prior binding to IL-2Rβ (20).
The relationship of γc to the other subunits is less well defined. An interaction between IL-2 and the IL-2Rγ chain has been identified (21). Furthermore, in the presence of IL-2, immunoprecipitation using anti-IL-2Rβ monoclonal antibodies (mAbs) led to the immunoprecipitation of IL-2Rβ/γ complexes (22). However, in the absence of IL-2, antibodies to IL-2Rβ did not coprecipitate γc, raising the possibility that stable IL-2Rβ/γ complexes are not preassembled on the cell surface (22). Finally, the effect of the addition of non-IL-2 cytokines (e.g., IL-4, IL-7, IL-9, or IL-15) that bind to IL-2Rβ or γc on the topology of the IL-2R subunits has not been defined. In the present study we used fluorescence resonance energy transfer (FRET) measurements to address these issues. FRET can be successfully applied to determine intra- and intermolecular distances at the 2- to 10-nm level. When an energy donor [usually fluorescein isothiocyanate (FITC)] and an energy acceptor (e.g., Cy3) are closely associated, there is tunneling of energy from the donor to the acceptor so that the emission from the donor is reduced and from the acceptor is increased. In the present work we used FRET measurements to define the assembly and mutual proximity among the three IL-2 receptor subunits in the resting phase and after the addition of saturating concentrations of IL-2, IL-7, and IL-15 that bind to the different IL-2R subunits.
MATERIALS AND METHODS
Cell Culture and Treatment with Cytokines.
The Kit 225 K6 cell line (obtained from Takashi Uchiyama, Kyoto, Japan) that was originally derived from a human adult T cell lymphoma (23), was cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 20 units/ml of recombinant IL-2 and penicillin, and streptomycin. The IL-2 was added on a daily basis. Before the experiments the cells were washed in media without cytokine and were grown for 48–72 hr without further addition of IL-2. These cells were considered as resting control cells. The lymphokine-treated cell samples were supplemented with IL-2 (20 units/ml), IL-7 (20 pM), or IL-15 (20 pM) and cultured at 37°C for 6 hr before harvesting the cells. The cells were then washed out of media and stained at 4°C with the anti-receptor mAbs.
Monoclonal Antibodies.
The subunits of the IL-2R complex were targeted by the following antibodies: the α subunit was defined either with anti-Tac (IgG2a) or 7G7/B6 (IgG2a) mAbs. Anti-Tac competes with IL-2 for its binding site whereas 7G7/B6 does not. The Mikβ1 (IgG2a) and the Mikβ3 (IgG1κ) are specific for the β subunit, with Mikβ1 interacting with the cytokine binding site of this subunit. The γc subunit was labeled by mAb TUGh4, purchased from PharMingen. The W6/32 (IgG2a) mAb with specificity for the heavy chain of class I HLA A, B, and C molecules and L-368 (IgG1) mAb specific for β2-microglobulin were kindly provided by F. Brodsky, University of California at San Francisco.
Fab Fragment Preparation.
The Fab fragments were prepared from monoclonal antibodies by using a method described earlier by Edidin (24). Briefly, IgG mAbs were dialyzed with PBS (100 mM Na2HPO4/150 mM NaCl/1 mM EDTA, pH 8.0) and digested with activated papain at 37°C for 11 min. The enzyme activity was terminated by addition of iodoacetamide. The reaction mixture was passed through a Sephadex G-100 fine column, and the collected Fab fractions were further separated from intact Ig by using a Protein A-Sepharose column.
Conjugation of Monoclonal Antibodies with Fluorescent Dyes.
Aliquots of purified whole mAbs or Fab fragments (at least at 1 mg/ml concentration) were conjugated as described previously (25, 26) with fluorescein isothiocyanate (FITC) (Molecular Probes) or sulfoindocyanine succinimidyl bifunctional ester (Amersham). Briefly, the proteins were transferred into a carbonate–bicarbonate buffer at pH 9.4, mixed with 30- to 100-fold molar excess of freshly prepared fluorescence dyes, and incubated for 45–60 min at room temperature. For labeling with Cy3, a kit was used (Amersham). Unreacted dye molecules were removed by gel filtration through a Sephadex G-25 column. The final antibody concentration and the dye-to-protein ratio were determined spectrophotometrically. In the case of FITC this ratio varied between 1 and 4; for Cy3, it varied between 4 and 8. The fluorescently tagged antibodies retained their biological activity as assessed by competition with identical but unlabeled antibodies.
Labeling of Cells with Monoclonal Antibodies.
Freshly harvested cells were washed twice in ice-cold PBS (pH 7.4). The cell pellets were suspended in 100 μl of PBS (1 × 106 cells per ml) and labeled by incubation with approximately 10 μg of FITC- or Cy3-conjugated mAbs for 60 min on ice. The excess mAbs over the available binding sites was at least 30-fold during the incubation. To avoid possible aggregation of the antibodies they were air-fuged (at 9 × 104 rpm, for 30 min) before labeling. Staining of cells was checked by fluorescence microscopy after the labeling procedure. Special care was taken to keep the cells at an ice-cold temperature before FRET analysis to prevent unwanted aggregation of cell surface molecules and meaningful receptor internalization. The labeled cells were washed with excess cold PBS and then fixed with 1% formaldehyde. Data obtained with fixed cells did not differ significantly from those of unfixed, viable cells. The number of binding sites on the cell surface was determined from the mean values of flow-cytometric histograms of cells labeled to saturation with FITC-conjugated mAbs. The mean fluorescence intensities were converted to the number of binding sites by calibration with fluorescent microbeads (Quantum 25, Flow Cytometry Standards, San Juan, Puerto Rico). The diameter of the Kit 225 K6 cells was estimated to be 10–13 μm.
Flow Cytometric Energy Transfer Measurements.
The donor (FITC) and acceptor (Cy3) labels were targeted to cell surface molecules by the relevant mAbs. The measurements were carried out in a Becton Dickinson FACStar Plus flow cytometer as described before (27–29), with minor variations in the excitation and emission parameters. Briefly, the cells were excited at 488 and 565 (occasionally 514) nm sequentially, and the respective emission data were collected at 540 and >590 nm. The fluorescence emissions were gated on the forward-angle light scatter signal to avoid artifacts from cell debris. Corrections were made for the direct excitability of the acceptor at the donor’s excitation wavelength and the spectral spillover of the donor’s and acceptor’s fluorescence by using cell samples labeled with only FITC- or, alternatively, Cy3-conjugated mAbs. The necessary signals were collected in list mode and analyzed as described before (27–29). The efficiency of energy transfer (E) is expressed as percentage of the donor’s (FITC) excitation energy tunneled to the acceptor (Cy3) molecules. E has an inverse sixth-power dependence on the donor–acceptor separation distance and thus an extremely high sensitivity to changes in distances in the range of 2–10 nm (30), if the orientation factor, accounting for the relative position of the donor’s emission and the acceptor’s absorption dipoles, can be considered as a statistical average of isotropic random orientations (i.e., 2/3) (31). The energy transfer frequency histograms were calculated from the equations described previously (28). The data analysis was performed by a software program termed flowin (version β2). The program was written and kindly provided by L. Balkay and M. Emri (Positron Emission Tomography Center, University Medical School of Debrecen, Hungary). The mean values of the calculated energy transfer distribution curves were utilized and tabulated as characteristic energy transfer efficiencies between two epitopes.
Because data have been obtained on a cell-by-cell basis, the means of energy transfer histograms represent averages over the cell population, whereas each data point is an average for a single cell. A computerized normalization procedure was used to correct for the differences in the actual receptor densities and for the inequalities in the individual dye-to-protein labeling ratios of mAbs. Therefore, a comparison of the energy transfer efficiency data allows a two-dimensional mapping, i.e., a possible projection of mutual proximities onto the plane of the plasma membrane (32, 33).
RESULTS
Distribution of IL-2 Receptor Subunits in the Plasma Membrane of Resting T Cells.
A considerable disproportionality was found between the relative cell surface densities of the three IL-2R subunits on Kit 225 K6 T cells as was observed with other activated T cells and T cell lines (1, 2). The α subunit had a 10-fold higher expression level than the β and γ subunits (Table 1). Based on the copy numbers observed and the surface area of these cells, if a random Poissonian distribution of the subunits were present, the occurrence frequency of hetero- and homoassociations of any size was predicted to be negligible. Thus, if there were a random distribution, a lack of FRET would be expected between any pairs of labeled subunits. To determine whether there was nonrandom distribution with colocalization, the mutual proximities of α, β, and γc subunits of the IL-2 receptor complex were characterized by the measure of FRET (mean efficiency: E%) obtained from pair-by-pair measurements between donor- and acceptor-labeled subunits. Because of the larger number of α subunits, the energy transfer was measured for both technical and theoretical reasons (34) from the β or γ subunits toward the α, in a pairwise fashion.
Table 1.
Relative surface expression of IL-2Rα, β and γ subunits on Kit 225 K6 T cells
| Subunit | Number of copies (×103)* | |
|---|---|---|
| IL-2Rα | (Tac, p55) | 96 ± 12† |
| IL-2Rβ | (p70–75) | 11 ± 5 |
| IL-2Rγ | (p64) | 8 ± 4 |
The number of IL-2R subunits per cell was determined from the mean fluorescence intensities of flow cytometric histograms of cells labeled to saturation with fluorescently conjugated mAbs as described in Materials and Methods.
Data represent mean ± SEM of at least three independent measurements.
The Kit 225 K6 T cells were cultured in the presence of 20 units per ml of IL-2 administered on a daily basis. Although mAbs 7G7/B6 and Mikβ3 can bind to the α and β subunits, respectively, in the presence of IL-2, other antibodies compete with IL-2 for its binding site (e.g., anti-Tac and Mikβ1). Therefore, before FRET analysis, the added IL-2 was either removed from the cells by acid washing (in 10 mM sodium citrate, 0.14 M NaCl, pH 4.0) or the cells were grown for 48–72 hr without IL-2 addition resulting in internalization and catabolism of the available IL-2 by the cells. These latter cells were considered as “resting” T cells.
The FRET data concurrently obtained for the subunit pairs of β-α, γ-α, and β-γ (Fig. 1 B–D) by using mAbs with different epitope specificities indicated that the these subunits must be in close proximity to each other in the plasma membrane of resting T cells (FRET β-α, 25.4 ± 5.9%; γ-α, 20.6 ± 5.1%; and γ-β, 12.4 ± 5%). The FRET efficiencies obtained for these labeled subunit pairs was comparable to the intramolecular FRET efficiency (ca. 15%) measured between the labeled β2-microglobulin (by L-368 mAb) and heavy chain epitopes (by W6/32 mAb) of class I HLA molecules on Kit 225 K6 and on a number of other cells (33). Because the latter FRET efficiency coincides well with the high-resolution x-ray structure of class I HLA molecule (35), it can also serve as a reference value for any FRET analysis.
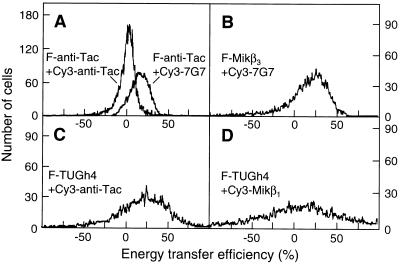
Figure 1.
Representative flow cytometric histograms of energy transfer efficiencies measured between FITC (F)- and Cy3-conjugated mAbs bound to IL-2R α, β, and γ subunits on Kit 225 K6 T cells. Averages and error estimates (E ± ΔE) of mean values of flow cytometric energy transfer histograms for the indicated donor–acceptor pairs were calculated from data of three to five independent measurements: (A) 1.5 ± 0.1% for F-anti-Tac + Cy3-anti-Tac, 18.2 ± 3.5% for F-anti-Tac + Cy3–7G7; (B) 25.4 ± 5.9% for F-Mikβ3 + Cy3–7G7; (C) 20.6 ± 5.1% for F-TUGh4 + Cy3-anti-Tac; and (D) 12.4 ± 5.0% for F-TUGh4 + Cy3-Mikβ1. Displacement from a mean value of 0 indicates energy transfer and nonrandom proximity of the epitopes.
A microaggregation of subunits by bivalent whole IgG antibodies can be excluded as a possible reason for the observed FRET, because application of fluorescent Fab fragments of the mAbs to target the subunits resulted in comparable FRET efficiencies (ranging between 14 and 20%) (data not shown). Because the subunits were labeled and kept on ice before FRET analysis, the possible effects of lateral mobility were also minimized.
There are two plausible interpretations for the high energy transfer efficiencies obtained for the β-α and γ-α subunit pairs. First, the energy transfer observed might reflect the possibility that these subunit pairs are in close proximity to each other within the effective Förster-distance range (ca. 10 nm, in this case). Alternatively, it might mean that both labeled β and γ subunits are surrounded by a large excess of acceptor-labeled α subunits, which would increase the acceptor:donor ratio, favoring a more efficient FRET (34). However, in the latter case, significant homo-transfer of excitation energy would be expected between α subunits labeled with donor- and acceptor-conjugated mAbs directed against the same epitope. This was not observed (Fig. 1A). Furthermore, a significant homo-association of α subunits supporting the second explanation is less likely in the light of recent findings showing a random distribution of gold-labeled IL-2Rα polypeptides on mouse T lymphocytes (36) and the lack of a tendency to form aggregates in the case of soluble IL-2Rα (37). Therefore, the high FRET efficiencies appear to reflect a close mutual proximity of all the three subunits rather than a high acceptor density. A similarly high FRET efficiency (12%) was found between the β and γ subunits labeled by Cy-Mikβ3 and FITC-TUGh4 mAbs, respectively (Fig. 1D), also supporting the view that the three subunits are in relatively close proximity at the surface of resting T cells, in the absence of IL-2.
IL-2Rα Conformation Probed by Intramolecular FRET.
The α subunit of IL-2 receptor complex on Kit 225 K6 resting T cells was specifically labeled at two distinct epitopes through the use of two noncompeting mAbs, FITC-anti-Tac and Cy3–7G7/B6, respectively. This analysis yielded results that were similar to those that used intramolecular FRET labels on class I major histocompatibility complex (MHC) molecules and may serve as a “conformational marker” (38) for the α subunit. A mean FRET efficiency of 18% was obtained for this anti-IL-2Rα antibody-pair (Fig. 1A), indicating that the two epitopes located on IL-2Rα are well within the critical Förster distance; thus, changes in the E% with the two anti-IL-2Rα mAbs would be expected to sensitively monitor conformational changes of the IL-2Rα molecule.
Modulation of IL-2R Subunit Organization on the T Cell Surface by IL-2, IL-7, and IL-15.
The effect of cytokines IL-2, IL-7, and IL-15 on the lateral subunit organization of the IL-2R complex was studied by means of three distinct approaches. The Kit 225 K6 T cells deprived of IL-2 for 72 hr were cultured with these cytokines (for details, see Materials and Methods) for 6 hr before harvesting. After staining the cells with mAbs, we examined the effect of the addition of these cytokines on: (i) the accessibility of subunit epitopes to the mAbs applied in FRET analysis, (ii) the intramolecular FRET within IL-2Rα (a marker of IL-2Rα conformation), and (iii) the FRET efficiencies measured between β-α, γ-α, and β-γ subunit pairs (a measure of subunit proximity).
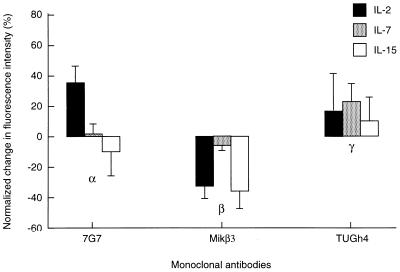
As shown in Fig. 2, IL-2 addition led to a modulation of the mAb binding (epitope accessibility) to all the three subunits. Here, the mean fluorescence intensities of cells labeled to saturation by mAbs (in the presence of cytokines) were compared with the respective intensities of resting cells without cytokine addition. With all subunits, part of the modulation is probably due to changes in the subunit conformation. The modulation was a bit smaller for the γ chain. IL-7 left the mAb binding to α and β chains unchanged while meaningfully modifying mAb binding to the γc chain to which it binds. IL-15 is a recently recognized cytokine that shares the β and γc subunits with the IL-2R (39). This is consistent with the large mAb-binding modulation observed for the β and a smaller one for the γc chain observed with IL-15 (Fig. 2). The small change in the mAb binding to IL-2Rα is possibly due to an indirect cross-modulation of its conformation by IL-15 binding to the IL-2Rβ and γc chains that normally associate with it.
Figure 2.
Effect of IL-2, IL-7, and IL-15 on the accessibility of epitopes to FITC-conjugated mAbs 7G7/B6 (against IL-2Rα), Mikβ3 (against IL-2Rβ), and TUGh4 (against IL-2Rγ) added as single agents on Kit 225 K6 wild-type T cells. Interleukins were added to cell cultures 6 hr before harvesting at the following concentrations: IL-2 (solid bars), 20 units/ml; IL-7 (shaded bars) and IL-15 (open bars), both 1 ng/ml (20 pM). The mean values of fluorescence histograms collected on cells treated with the interleukins were expressed as percent change from control values. Bars represent mean ± SEM of four to six independent measurements.
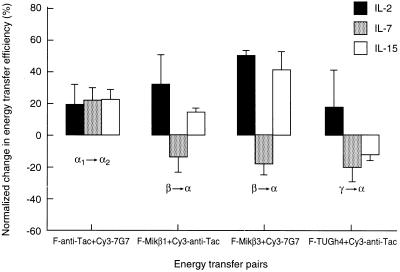
The conformation of IL-2Rα was modulated by all three of the cytokines, as reflected by the changes in intramolecular FRET (Fig. 3). The increased FRET efficiencies indicate a closer proximity of the Tac and 7G7/B6 epitopes, i.e., a more compact conformation of α chains upon addition of IL-2, IL-7, or IL-15 to the T cells.
Figure 3.
Effect of IL-2, IL-7, and IL-15 on energy transfer efficiencies measured between FITC (F)- and Cy3-conjugated mAbs specific to the IL-2R α, β, and γ subunits on Kit 225 K6 T cells. The cells were treated with interleukins as described in the legend to Fig. 2 by IL-2 (solid bars), IL-7 (shaded bars), and IL-15 (open bars). The mean values of the fluorescence energy transfer histograms collected from cytokine-treated cells were expressed as percent change from the control values. Bars represent mean ± SEM of four to six independent measurements. Greek letters with arrows indicate the subunits labeled with the mAbs and the direction of energy transfer. The subscripts 1 and 2 on the letter α designate the two epitopes recognized by mAbs anti-Tac and 7G7, respectively, on the same IL-2Rα subunit.
The effects of these cytokines on the mutual proximities of the three IL-2R subunits were tested by FRET measurements on cytokine-pretreated cells (Fig. 3). It should be noted that receptor binding of cytokines also changed the donor:acceptor ratio (influencing FRET efficiency) by modulation of mAb binding in some cases with the exception of the FRET between β and α subunits. In all cases the direction of the changes would not significantly influence the interpretation of FRET data. These FRET data show that IL-2 binding promotes a closer proximity for both β-α and γ-α subunit pairs (the FRET efficiencies increased). This might be a result of conformational changes induced by IL-2 binding that leads to stronger bridges among the subunits in the IL-2R complex or, alternatively, the movement together of IL-2R subunits in the plasma membrane. In contrast, the addition of IL-7 that only binds to γc loosened the interaction of the α subunit with both β and γ chains, as reflected by the reduced FRET efficiencies (Fig. 3). IL-15 increased the proximity between the α and β chains while weakening the interaction between the α and γ chains. This suggests that the binding of IL-15 breaks up (somewhat linearizes) the triangle geometry of the high affinity IL-2R complex, possibly to allow its private specific receptor, IL-15Rα, to complex with the IL-2Rβ/γ subunits (39).
DISCUSSION
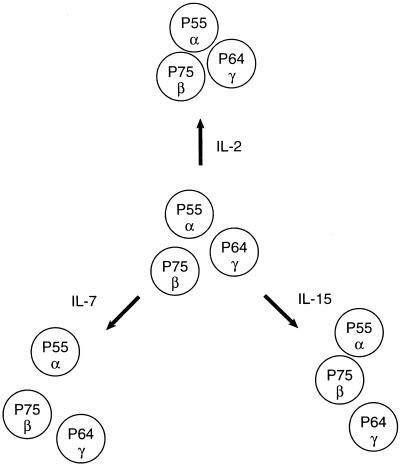
In the present study we used fluorescence energy transfer measurements to demonstrate the close proximity of IL-2 receptor subunits in the surface membrane of resting Kit 225 K6 T cells. In contrast to the predictions of the “affinity conversion model,” it appears that preassembled subunit pairs (β/γ) or heterotrimers (α/β/γ) form intermediate or high affinity IL-2Rs, respectively, in the absence of added cytokine. These observations are in accord with the IL-2 binding model that postulates the existence of preformed α/β complexes in the plasma membrane (14–20). In the case of the high affinity receptor composed of all of the three IL-2R subunits, a topological two-dimensional projection of subunit organization could be modeled as a triangle (Fig. 4) that is consistent with both the FRET data and the size of the subunits. This triangular model of subunit clustering is very likely in a dynamic equilibrium and is in a form that can be modulated by the binding of the ligands IL-2, IL-7, and IL-15 albeit in diverse directions and extents. The addition of the cognate cytokine, IL-2, strengthens the bridges between the IL-2 subunits, making the triangle more compact (Fig. 4). These data indicating a closer association of IL-2Rβ and γc on IL-2 addition are in accord with previous reports that IL-2R activation leads to heterodimerization of IL-2Rβ and γ chains, leading to signaling (8, 9). It also is in accord with parallel work that used coprecipitation analysis of IL-2Rγ and β to support this tighter assembly of cytokine receptor subunits on IL-2 addition (40, 41). In contrast, the addition of IL-7 and IL-15 acted in the opposite direction by opening the triangle, loosening the contact between the IL-2Rα and γc subunits. This may reflect the movement within the cell membrane of the γc subunit so that it becomes associated with the private specific α receptors for IL-7 and IL-15. This suggests that IL-7 and IL-15 and, although not examined in the present study, IL-4 might inhibit the action of IL-2 by diverting γc from the high affinity IL-2R complex. In support of this view, IL-4 has been shown to inhibit the expression of high affinity IL-2Rs on monoclonal human B cells (42). Furthermore, IL-4 manifested an inhibitory action on IL-2-supported human B cell proliferation and differentiation (43).
Figure 4.
Schematic representation of lateral organization of the subunits of the IL-2 receptor complex on resting Kit 225 K6 T cells and the complex’s modulation by addition of IL-2, IL-7, and IL-15. FRET data suggested that IL-2R α, β, and γ subunits are preassembled, forming heterotrimers on the surface of resting cells (Middle). The proximity between the β and γ subunits was not altered significantly with any of the interleukins. Whereas IL-2 promoted a stronger contact of the α subunit with the β and γ chains (Top), IL-7 loosened it (Bottom Left). IL-15 induced a closer proximity of β and α subunits, whereas the contact between the γ and α subunits became weaker, thereby leading to a somewhat linearized configuration of the IL-2R complex (Bottom Right).
It is of interest that the addition of IL-7 and IL-15 altered the intramolecular distances of IL-2Rα; that is, the FRET between the 7G7/B6 and Tac epitopes was increased (Fig. 3). Because neither IL-7 nor IL-15 bind to IL-2Rα, this suggests that the movement of γc away from IL-2Rα mediated by IL-7 and IL-15 addition leads to a change in the conformation of IL-2Rα. A similar change in the conformations of IL-2Rα and IL-2Rβ by their proximity was noted previously (17–20).
The present approach, FRET analysis, has been used to demonstrate that intercellular adhesion molecule-1 (ICAM-1) and class I MHC are nonrandomly associated with the IL-2 receptor (32). Such cell surface receptor patterns have also been described for other kinds of receptors. For example, significant subunit clustering in the case of the IL-1 receptor was emphasized by Guo et al. (44). Furthermore, two-dimensional models of receptor distribution have been derived from fluorescence energy transfer data for MHC class I and class II DR and DQ molecules, ICAM-1, the CD20 B cell marker, as well as CD53, CD81, and CD82 “tetraspan” molecules (32, 45–48). In general, these may be critical events involved in effective signal transduction and the cellular adhesion process (49). These studies demonstrate that FRET analysis together with scanning force microscopy and video-tracking techniques may provide new insights into the molecular organization of multisubunit receptor complexes and the rearrangement of this organization upon ligation.
Acknowledgments
S.D. was a Fulbright Scholar working on the Metabolism Branch, National Cancer Institute with T.A.W. during this work. The work was also supported by Orszagos Tudomanyos Kutatasi Alap (National Scientific Research Fund) of Hungary (T023873 and T17592) and a special award (AKP-96–316/54).
ABBREVIATIONS
- Cy3
sulfoindocyanine succinimidyl bifunctional ester
- FITC
fluorescein isothiocyanate
- FRET
fluorescent resonance energy transfer
- IL-2R
interleukin 2 receptor
- mAbs
monoclonal antibodies
References
- 1.Waldmann T A. Annu Rev Biochem. 1989;58:875–911. doi: 10.1146/annurev.bi.58.070189.004303. [DOI] [PubMed] [Google Scholar]
- 2.Taniguchi T, Minami Y. Cell. 1993;73:5–8. doi: 10.1016/0092-8674(93)90152-g. [DOI] [PubMed] [Google Scholar]
- 3.Leonard W J, Depper J M, Uchiyama T, Smith K A, Waldmann T A, Greene W C. Nature (London) 1982;300:267–269. doi: 10.1038/300267a0. [DOI] [PubMed] [Google Scholar]
- 4.Tsudo M, Kozak R W, Goldman C K, Waldmann T A. Proc Natl Acad Sci USA. 1986;83:9694–9698. doi: 10.1073/pnas.83.24.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharon M, Klausner R D, Cullen B R, Chizzonite R, Leonard W J. Science. 1986;234:859–863. doi: 10.1126/science.3095922. [DOI] [PubMed] [Google Scholar]
- 6.Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K-I, Sugamura K. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 7.Noguchi M, Nakamura Y, Russell S M, Ziegler S F, Tsang M, Cao X, Leonard W J. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura Y, Russell S M, Mess S A, Friedmann M, Erdos M, Francois C, Jacques Y, Adelstein S, Leonard W J. Nature (London) 1994;369:330–333. doi: 10.1038/369330a0. [DOI] [PubMed] [Google Scholar]
- 9.Nelson B H, Lord J D, Greenberg P D. Nature (London) 1994;369:333–336. doi: 10.1038/369333a0. [DOI] [PubMed] [Google Scholar]
- 10.Saito Y, Ogura T, Kamio M, Sabe H, Uchiyama T, Honjo T. Int Immunol. 1990;12:1167–1177. doi: 10.1093/intimm/2.12.1167. [DOI] [PubMed] [Google Scholar]
- 11.Kondo S, Shimizu A, Saito Y, Kinoshita M, Honjo T. Proc Natl Acad Sci USA. 1986;83:9026–9029. doi: 10.1073/pnas.83.23.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Audrain M, Boeffard F, Soulillou J-P, Jacques Y. J Immunol. 1991;146:884–892. [PubMed] [Google Scholar]
- 13.Kamio M, Uchiyama T, Arima N, Itoh K, Ishikawa T, Hori T, Uchino H. Int Immunol. 1990;2:521–530. doi: 10.1093/intimm/2.6.521. [DOI] [PubMed] [Google Scholar]
- 14.Saragovi H, Malek T R. J Immunol. 1988;141:476–482. [PubMed] [Google Scholar]
- 15.Yamaguchi A, Ide T, Hatakeyama M, Doi T, Kono T, Uchiyama T, Kikuchi K, Taniguchi T, Uede T. Int Immunol. 1989;1:160–168. doi: 10.1093/intimm/1.2.160. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein B, Jones D, Kevekidis I G, Perelson A S. Int Immunol. 1992;4:23–32. doi: 10.1093/intimm/4.1.23. [DOI] [PubMed] [Google Scholar]
- 17.Grant A, Roessler E, Ju G, Tsudo M, Sugamura K, Waldmann T A. Proc Natl Acad Sci USA. 1991;89:2165–2169. doi: 10.1073/pnas.89.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roessler E, Grant A, Ju G, Tsudo M, Sugamura K, Waldmann T A. Proc Natl Acad Sci USA. 1994;91:3344–3347. doi: 10.1073/pnas.91.8.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuziel W A, Ju G, Grdina T A, Greene W C. J Immunol. 1993;150:3357–3365. [PubMed] [Google Scholar]
- 20.Arima N, Kamio M, Okuma M, Ju G, Uchiyama T. J Immunol. 1991;147:3396–3401. [PubMed] [Google Scholar]
- 21.Boss S, Leary T, Sondel P, Robb R J. Proc Natl Acad Sci USA. 1993;90:2428–2432. doi: 10.1073/pnas.90.6.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeshita T, Ohtani K, Asao H, Kumaki S, Nakamura M, Sugamura K. J Immunol. 1992;148:2154–2158. [PubMed] [Google Scholar]
- 23.Hori T, Uchiyama T, Tsudo M, Umadome H, Ohno N, Fukuhara S, Kita K, Uchino H. Blood. 1987;70:1069–1072. [PubMed] [Google Scholar]
- 24.Edidin M, Wei T. J Cell Biol. 1982;95:458–462. doi: 10.1083/jcb.95.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szöllösi J, Damjanovich S, Balázs M, Nagy P, Trón L, Fulwyler M J, Brodsky F M. J Immunol. 1989;143:208–213. [PubMed] [Google Scholar]
- 26.De Petris S. In: Methods in Membrane Biology. Korn E D, editor. Vol. 9. New York: Plenum; 1978. pp. 1–201. [Google Scholar]
- 27.Szöllösi J, Trón L, Damjanovich S, Helliwell S H, Arndt-Jovin D J, Jovin T M. Cytometry. 1984;5:210–216. doi: 10.1002/cyto.990050216. [DOI] [PubMed] [Google Scholar]
- 28.Trón L, Szöllösi J, Damjanovich S, Helliwell S H, Arndt-Jovin D J, Jovin T M. Biophys J. 1984;45:939–946. doi: 10.1016/S0006-3495(84)84240-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matkó J, Szöllösi J, Trón L, Damjanovich S. Q Rev Biophys. 1988;21:479–544. doi: 10.1017/s0033583500004637. [DOI] [PubMed] [Google Scholar]
- 30.Stryer L. Annu Rev Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 31.Dale R E, Eisinger J, Blumberg W E. Biophys J. 1979;26:161–194. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damjanovich S, Gáspár R, Pieri C. Q Rev Biophys. 1997;30:67–106. doi: 10.1017/s0033583596003307. [DOI] [PubMed] [Google Scholar]
- 33.Bene L, Balázs M, Matkó J, Möst J, Dierich M, Szöllösi J, Damjanovich S. Eur J Immunol. 1994;24:2115–2123. doi: 10.1002/eji.1830240928. [DOI] [PubMed] [Google Scholar]
- 34.Fung B K, Stryer L. Biochemistry. 1978;17:5241–5248. doi: 10.1021/bi00617a025. [DOI] [PubMed] [Google Scholar]
- 35.Silver M L, Guo H-C, Strominger J L, Wiley D C. Nature (London) 1992;360:367–369. doi: 10.1038/360367a0. [DOI] [PubMed] [Google Scholar]
- 36.Breitfeld O, Kuhlcke K, Lother H, Hohenberg H, Mannweiler K, Rutter G. J Histochem Cytochem. 1996;44:605–613. doi: 10.1177/44.6.8666745. [DOI] [PubMed] [Google Scholar]
- 37.Junghans R P, Stone A L, Lewis M S. J Biol Chem. 1996;271:10453–10460. doi: 10.1074/jbc.271.18.10453. [DOI] [PubMed] [Google Scholar]
- 38.Bene L, Szöllösi J, Balázs M, Mátyus L, Gáspár R, Ameloot M, Dale R E, Damjanovich S. Cytometry. 1997;27:353–357. [PubMed] [Google Scholar]
- 39.Tagaya Y, Bamford R N, DeFilippis A P, Waldmann T A. Immunity. 1996;4:329–336. doi: 10.1016/s1074-7613(00)80246-0. [DOI] [PubMed] [Google Scholar]
- 40.Voss S D, Sondel P M, Robb R J. J Exp Med. 1992;176:531–541. doi: 10.1084/jem.176.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeshita T K, Ohtani H, Asao S, Kumaki M, Nakamura M, Sugamura K. J Immunol. 1992;148:2154–2158. [PubMed] [Google Scholar]
- 42.Karray S, Dautry-Varsat A, Tsudo M, Merle-Beral H, Debre P, Galanaud P. Am Assoc Immunol. 1990;145:1152–1158. [PubMed] [Google Scholar]
- 43.Jelinek D F, Lipsky P E. J Immunol. 1988;141:164–173. [PubMed] [Google Scholar]
- 44.Guo C, Dowers K S, Holowka D, Baird B. J Biol Chem. 1995;270:27562–27568. doi: 10.1074/jbc.270.46.27562. [DOI] [PubMed] [Google Scholar]
- 45.Szöllösi J, Damjanovich S, Goldman C K, Fulwyler M J, Aszalós A A, Goldstein G, Rao P, Talle M A, Waldmann T A. Proc Natl Acad Sci USA. 1987;84:7246–7250. doi: 10.1073/pnas.84.20.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szöllösi J, Horejsi V, Bene L, Angelisova P, Damjanovich S. J Immunol. 1996;157:2939–2946. [PubMed] [Google Scholar]
- 47.Damjanovich S, Vereb G, Shaper A, Jenei A, Matko J, Starink J P P, Fox G, Arndt-Jovin D J, Jovin T M. Proc Natl Acad Sci USA. 1995;92:1122–1126. doi: 10.1073/pnas.92.4.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Damjanovich S, Szöllösi J, Trón L. Immunol Today. 1992;13:A12–A15. doi: 10.1016/0167-5699(92)90058-F. [DOI] [PubMed] [Google Scholar]
- 49.Sheets E D, Simson R, Jacobson K. Curr Opinion Cell Biol. 1995;7:707–714. doi: 10.1016/0955-0674(95)80113-8. [DOI] [PubMed] [Google Scholar]