Abstract
Plants use siRNAs to target cytosine DNA methylation to both symmetrical CG and nonsymmetrical (CHG and CHH) sequence contexts. DNA methylation and siRNA clusters most frequently overlap with transposons in the Arabidopsis thaliana genome. However, a significant number of protein-coding genes also show promoter DNA methylation, and this can be used to silence their expression. Loss of the majority of non-CG DNA methylation in drm1 drm2 cmt3 triple mutants leads to developmental phenotypes. We identified the gene responsible for these phenotypes as SUPPRESSOR OF drm1 drm2 cmt3 (SDC), which encodes an F-box protein and possesses seven promoter tandem repeats. The SDC repeats show a unique silencing requirement for non-CG DNA methylation directed redundantly by histone methylation and siRNAs, and display spreading of siRNAs and methylation beyond the repeated region. In addition to revealing the complexity of DNA methylation control in A. thaliana, SDC has important implications for how plant genomes utilize gene silencing to repress endogenous genes.
Keywords: Epigenetics, DNA methylation, siRNA, gene silencing
DNA cytosine methylation is an epigenetic mark frequently associated with transcriptional repression and gene silencing (Chan et al. 2005; Goll and Bestor 2005). DNA methylation is most commonly observed in the symmetrical CG sequence context, which is maintained by enzymes orthologous to mammalian Dnmt1 and plant METHLYTRANSFERASE1 (MET1) (Chan et al. 2005; Goll and Bestor 2005). Dnmt1 recognizes hemi-methylated CG sites after DNA replication to propagate this mark through cell division (Goll and Bestor 2005). Additionally, plants contain DNA methylation in the non-CG sequence contexts, CHG and CHH (asymmetric), which are redundantly maintained by DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) and CHROMOMETHYLASE3 (CMT3) (Cao and Jacobsen 2002a; Cao et al. 2003).
Although DRM2 acts redundantly with CMT3 to maintain non-CG methylation, it functions alone during de novo DNA methylation (Cao and Jacobsen 2002b). DRM2 activity can be targeted to DNA by homologous siRNA generated by the RNAi pathway (Chan et al. 2004; Zilberman et al. 2004). Components of the plant small RNA machinery are specialized for function in chromatin silencing and RNA-directed DNA methylation (RdDM), including DICER-LIKE3 (DCL3), RNA-DEPENDENT RNA POLYMERASE2 (RDR2), and ARGONAUTE4 (AGO4) (Zilberman et al. 2003; Chan et al. 2004; Xie et al. 2004). Additionally, two DNA-dependent RNA polymerase (NRPD) complexes function in this pathway (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005; Pontier et al. 2005). The NRPD1A complex is required for accumulation of heterochromatic siRNA, while the NRPD1B complex shares this function but is also required downstream for DRM2 targeting (both complexes contain the NRPD2 subunit) (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005; Pontier et al. 2005; Mosher et al. 2008).
Genome-wide mapping of cytosine methylation in the Arabidopsis thaliana genome has shown that repeats are frequently methylated and associated with siRNA (Lippman et al. 2004; Zhang et al. 2006; Zilberman et al. 2007; Cokus et al. 2008). In addition, a large class of endogenous genes have been described with CG methylation in their ORFs, which correlates with expression level (Tran et al. 2005; Zhang et al. 2006; Zilberman et al. 2007; Cokus et al. 2008). Fewer genes have been shown to posses promoter DNA methylation, although one example is FWA (Soppe et al. 2000). FWA encodes a homeodomain transcription factor that is silenced by promoter methylation at tandem repeats, which generate siRNA (Soppe et al. 2000; Chan 2006b). The met1 mutation causes FWA demethylation and mis-expression, resulting in a dominant late-flowering phenotype (Soppe et al. 2000). Demethylated fwa epialleles are extremely stable, even after backcrossing to wild type (Soppe et al. 2000). Silencing at FWA is mainly dependent upon CG methylation, as loss of non-CG methylation at the repeats in drm1 drm2 cmt3 is not associated with mis-expression (Cao and Jacobsen 2002a).
The drm1 drm2 cmt3 triple mutant shows developmental phenotypes, including curled leaves and reduced stature, indicating that genes may also be repressed by non-CG methylation (Cao and Jacobsen 2002a; Chan et al. 2006a). The drm1 drm2 cmt3 phenotype is completely penetrant, and behaves recessively after backcrossing to wild type (Cao and Jacobsen 2002a; Chan et al. 2006a). In contrast, inbreeding of met1 leads to the stochastic appearance of several epimutations, which can frequently be inherited independently of the met1 mutation (Miura et al. 2001; Kankel et al. 2003; Saze et al. 2003; Saze and Kakutani 2007). We sought to understand the molecular basis of drm1 drm2 cmt3 phenotypes and what this might reveal about these distinct modes of epigenetic inheritance.
We genetically identified an F-box gene, whose mis-expression is responsible for drm1 drm2 cmt3 developmental phenotypes; we named this gene SUPPRESSOR OF drm1 drm2 cmt3 (SDC). The SDC promoter contains seven direct repeats that generate siRNA and recruit DNA methylation. DNA methylation at SDC in all sequence contexts is redundantly dependent upon DRM2 and CMT3, which are recruited by siRNA and histone methylation, respectively. A further unusual characteristic of the SDC repeats is that DNA methylation and siRNA spread into flanking nonrepeated sequences. During establishment of silencing the SDC gene requires DRM2 and the RdDM–RNAi pathway. However, unmethylated SDC transgenes differ from FWA in that they can efficiently recruit DNA methylation after backcrossing to wild type. SDC demonstrates that non-CG methylation can play an important role in silencing, and further shows how plants have utilized repeats to silence genes.
Results
SDC misexpression is necessary and sufficient for drm1 drm2 cmt3 developmental phenotypes
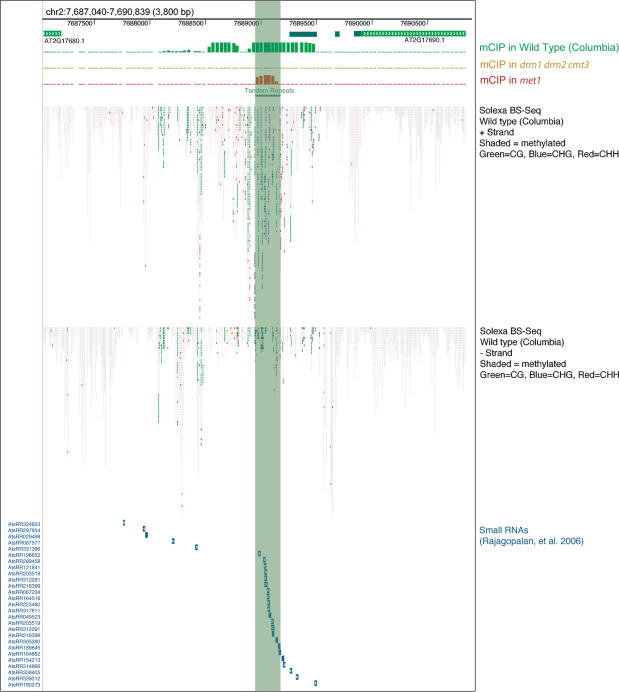
The drm1 drm2 cmt3 triple mutant has a global reduction in non-CG methylation, although the majority of CG methylation remains efficiently maintained by MET1 (Cao and Jacobsen 2002a; Chan et al. 2006a; Zhang et al. 2006; Cokus et al. 2008). Genome-wide mapping of DNA methylation combined with expression profiling had previously identified a set of promoter-methylated genes that are misexpressed in drm1 drm2 cmt3 relative to wild type (Zhang et al. 2006). These genes are strong candidates for those causing the developmental phenotypes observed in drm1 drm2 cmt3. At2g17690 had the highest fold expression change in drm1 drm2 cmt3, and was observed to posses seven promoter direct repeats (32 bp per repeat), coinciding with DNA methylation and cloned siRNAs (Zhang et al. 2006; Cokus et al. 2008) (Fig. 1). At2g17690 is predicted to encode an F-box protein of unknown function (Gagne et al. 2002).
Figure 1.
Epigenomic profile of SDC. Graphical representation of DNA methylation and siRNAs detected in the region of SDC (At2g17690). The SDC ORF is indicated as a light-green bar marked with chevrons. The SDC 5′-UTR mapped by RACE is highlighted by dark green bars. The promoter tandem repeats are marked by the light green block running down the figure. DNA methylation detected by methyl-cytosine immunoprecipitation (mCIP) and hybridization to genomic tiling arrays is highlighted beneath as colored bars in wild type (green), drm1 drm2 cmt3 (orange), and met1 (red) (Zhang et al. 2006). Methylation state of individual cytosines as measured by Solexa sequencing of bisulfite treated DNA is represented as small rectangles. Methylation is shown by shading (dark is methylated) and color-coded according to sequence context (CG, green; CHG, blue; CHH, red) (Cokus et al. 2008). Small RNAs detected by sequencing are represented by blue bars with white chevrons indicating their strandedness (Rajagopalan et al. 2006).
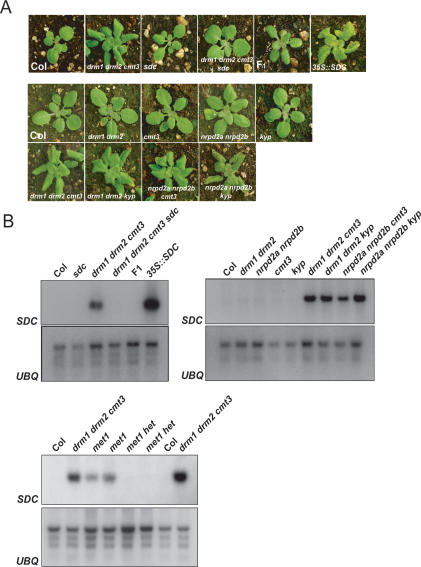
To test genetically whether misexpression of At2g17690 could account for these developmental phenotypes, we obtained a T-DNA insertion in this gene and crossed it into drm1 drm2 cmt3. The resulting quadruple mutant was identical to wild type with respect to its leaf phenotype and stature, leading us to rename At2g17690 SUPPRESSOR OF drm1 drm2 cmt3 (SDC) (Fig. 2A). Northern blotting and hybridization showed that the T-DNA effectively eliminated SDC expression in drm1 drm2 cmt3 sdc (Fig. 2B). We also generated plants overexpressing the SDC ORF from the constitutive 35S promoter, which caused phenotypes identical to those seen in drm1 drm2 cmt3 (Fig. 2A). Expression profiling in met1 indicated that SDC was also expressed in this background, although not to the level observed in drm1 drm2 cmt3 (Fig. 1; Zhang et al. 2006). We confirmed that SDC was expressed in met1 homozygous mutants, but not heterozygotes, by Northern blotting and hybridization (Fig. 2B).
Figure 2.
SDC is necessary and sufficient for drm1 drm2 cmt3 developmental phenotypes. (A) Leaf phenotypes of plants showing either wild-type (Col) flat leaves or mutant leaves that are curled downward, as in drm1 drm2 cmt3. (B) SDC expression detected by Northern blotting and hybridization. Loading was checked by hybridization with a UBIQUITIN10 (UBQ) probe.
Backcrossing drm1 drm2 cmt3 to wild type leads to reversion of developmental phenotypes in the F1 progeny (Fig. 2A; Chan et al. 2006a), suggesting that unmethylated SDC should be able to efficiently recruit the silencing machinery. Consistent with this genetic behavior we found that SDC expression was completely resilenced in the F1 after backcrossing (Fig. 2B). We also demonstrated previously that combinations of mutations between drm1 drm2 or nrpd2a nrpd2b with either cmt3 or kryptonite (kyp: a histone H3K9 methyltransferase) lead to developmental phenotypes very similar to drm1 drm2 cmt3 (Chan et al. 2006a). Northern blotting confirmed that in all of these backgrounds (drm1 drm2 cmt3, drm1 drm2 kyp, nrpd2a nrpd2b cmt3, and nrpd2a nrpd2b kyp) SDC is similarly overexpressed (Fig. 2B). Together, these results demonstrate that SDC misexpression is necessary and sufficient for the developmental phenotypes observed in drm1 drm2 cmt3 mutants.
Genetic control of DNA methylation and silencing at SDC
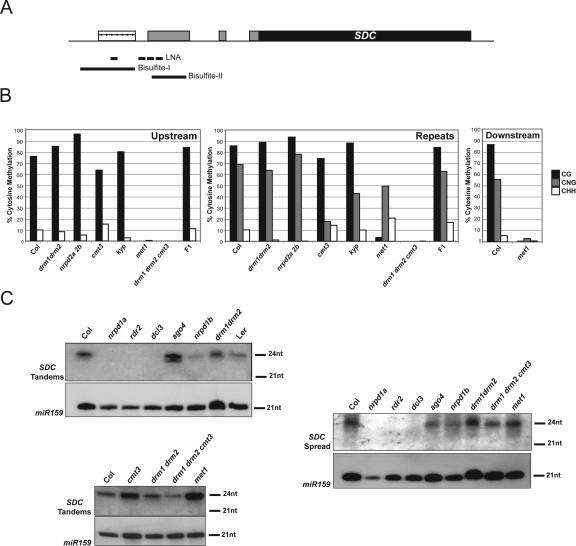
The SDC tandem repeats are located 730 bp upstream of the ATG and using 5′-RACE we mapped transcription as beginning 68 bp downstream from the repeats (in a drm1 drm2 cmt3 background) (Fig. 1). DNA methylation profiling using methyl-cytosine immunoprecipitation followed by hybridization to tiling arrays revealed a broad peak of promoter methylation coinciding with the SDC repeats and also spreading into the adjacent nonrepeated sequences (Fig. 1; Zhang et al. 2006). Whole-genome bisulfite sequencing data generated using Solexa sequencing technology allowed us to more precisely define this methylation as spreading 1218 bp upstream of and 361 bp downstream from the tandem repeats (Fig. 1; Cokus et al. 2008). To analyze the genetic requirements for SDC promoter methylation we used bisulfite sequencing of the entire tandem repeat region and also 112 bp of upstream nonrepeated sequence (Fig. 3A). Detailed analysis of bisulfite sequencing data is provided in Supplemental Figure 1 and Supplemental Table 2. In wild type, both regions are densely DNA methylated in all sequence contexts (although the region sequenced upstream contained no CHG sites) (Fig. 3B). In drm1 drm2 cmt3, we observed an elimination of all CG and non-CG methylation at SDC, although it was immediately restored to wild-type levels in the F1 after backcrossing to wild type (Columbia) (Fig. 3B). Mutation of drm1 drm2 or cmt3 alone had less dramatic effects on SDC methylation and expression (Figs. 2B, 3B). The drm1 drm2 and nrpd2a nrpd2b mutants show a similar phenotype of a reduction of CHH methylation within the repeats (Fig. 3B). In contrast, the cmt3 and kyp mutants showed a reduction in CHG methylation within the repeats, consistent with the function of these genes in maintaining this type of DNA methylation (Fig. 3B; Bartee et al. 2001; Lindroth et al. 2001; Jackson et al. 2002; Malagnac et al. 2002). This indicates that significant loss of methylation and SDC expression only occurs when both the siRNA/DRM2 and histone H3K9 methylation/CMT3 pathways are simultaneously compromised. The met1 mutant showed a strong decrease in CG methylation within the repeats, although non-CG methylation remained largely intact (Figs. 1, 3B). This indicates that DRM2 and CMT3 are still efficiently recruited to the SDC repeats in the absence of MET1 and CG methylation. Hence, SDC shows an unexpected dependence of CG methylation upon non-CG methylation.
Figure 3.
Genetic analysis of DNA methylation and siRNA accumulation at SDC. (A) Graphical representation of SDC with the tandem repeats marked as arrows within the white box. The ORF is marked by the black box and the 5′-UTR by gray boxes. Black lines show the positions of the two bisulfite sequencing products and LNA siRNA probes. (B) DNA methylation analyzed by sodium bisulfite sequencing shown as percentage CG (black), CHG (gray), and CHH (white). The Bisulfite-I region data has been separated into the tandemly repeated (Repeats) region and upstream, unique sequences (Upstream). The upstream sequence contains no CHG sites. The Bisulfite-II region data is labeled as “Downstream.” (C) Detection of SDC siRNAs by Northern blotting and hybridization. SDC LNA probes were used either for the tandem repeat siRNAs (SDC tandem) or for spreading siRNAs (SDC spread). Loading was analyzed by hybridization with a probe to microRNA159 (miR159).
A further interesting phenomenon was that met1 showed a dramatic and novel effect on spreading of methylation, which was dramatically reduced in the nonrepeated regions both upstream of and downstream from the repeats (Fig. 3B). This may mean either that MET1 itself is mediating spreading of DNA methylation, or CG methylation within the repeats is required for other cytosine methyltransferases to perform spreading. Consistent with the former model, there is accumulating evidence that MET1 can methylate non-CG sequence contexts (Aufsatz et al. 2004; Henderson et al. 2006; Cokus et al. 2008).
Genetic control of siRNA accumulation and spreading at SDC
Spreading of heterochromatic silencing is well-documented in several systems, such as fission yeast (Hall et al. 2002). However, spreading of silencing in A. thaliana has only been observed previously at the BONSAI gene in ibm1 and ddm1 mutant backgrounds (Saze and Kakutani 2007; Saze et al. 2008), or at transgenic loci (Vaistij et al. 2002). In contrast, the SDC endogene shows extensive spreading in wild type. We were interested to address whether the spreading of DNA methylation observed at the SDC promoter was also accompanied by spreading of siRNAs. To test this we queried a large database of cloned siRNAs (Rajagopalan et al. 2006). A pronounced peak of siRNAs was observed over the tandem repeats, although we also observed a broader distribution of siRNAs coincident with the observed spreading of DNA methylation (Fig. 1; Supplemental Table 4). To test the genetic requirements for siRNAs within and adjacent to the tandem repeats we designed locked nucleic acid (LNA) probes to both regions (Fig. 3A). The siRNAs homologous to the tandem repeats were 24 nt in length and undetectable in nrpd1a, rdr2, and dcl3 mutants, consistent with the proposed function of these genes upstream of siRNA accumulation (Fig. 3C). In contrast, siRNAs were detected in the ago4, nrpd1b, and drm1 drm2 mutants, which have been proposed to act largely downstream from siRNA biosynthesis (Fig. 3C). The tandem repeat siRNA were also detectable in several methyltransferase mutant backgrounds (drm1 drm2, cmt3, drm1 drm2 cmt3, and met1) (Fig. 3C). To detect the lower abundance spreading siRNAs we simultaneously hybridized three LNA probes homologous to an ∼150-bp nonrepeated region immediately downstream from the tandem repeats. In wild type we again detected 24-nt siRNAs that had the same genetic requirements for their accumulation as the tandem repeat siRNAs (Fig. 3C). Interestingly, spreading of DNA methylation was not required for siRNA spreading, as they accumulated normally in both drm1 drm2 cmt3 and met1 mutants (Fig. 3C). Equally, spreading of DNA methylation is efficiently maintained in nrpd2a nrpd2b mutants (Fig. 3B), which should lack all SDC siRNAs. Hence, the SDC repeats are unique with respect to their genetic requirements for maintenance of DNA methylation and also spreading of methylation and siRNAs.
Establishment of RNA-directed DNA methylation at SDC during transformation
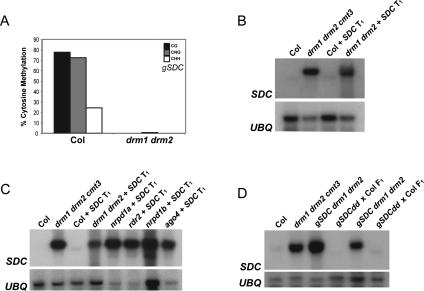
Transformation of direct or inverted repeat sequences into A. thaliana using Agrobacterium has been shown to act as an efficient trigger of RNA-directed DNA methylation (Soppe et al. 2000; Aufsatz et al. 2002; Cao and Jacobsen 2002b; Zilberman et al. 2004). To test how SDC would behave in the context of establishment of gene silencing we generated a genomic clone of the SDC region (hereafter termed gSDC) and engineered a sequence polymorphism into the repeats by mutagenesis, such that bisulfite sequencing could distinguish between endogenous and transgenic SDC. Transformation of gSDC into wild type leads to efficient silencing within the T1 transformant population and dense DNA methylation of the transgene (Fig. 4A,B). In contrast, repetition of this experiment in drm1 drm2 mutants led to high SDC expression and unmethylated transgenes (Fig. 4A,B). Although we cannot distinguish between endogenous and transgenic SDC expression by probe hybridization, we assume that the signal observed represents transgenic gSDC expression, as endogenous SDC is silent in drm1 drm2 (Fig. 2B). DRM2-dependent silencing of gSDC is likely to be siRNA-mediated, as several RNAi mutants (nrpd1a, rdr2, nrpd1b, ago4) also failed to silence gSDC expression after transformation (Fig. 4C). The genetic requirements for establishment of SDC silencing are identical to those observed for the tandem-repeat gene FWA (Cao and Jacobsen 2002b; Chan et al. 2004, 2006b). Previously, it was shown that unmethylated FWA transgenes in drm1 drm2 mutants do not become resilenced after backcrossing to wild type, suggestive of a “surveillance window” during which incoming sequences are competent to be silenced (Cao and Jacobsen 2002b; Chan et al. 2004). To test whether SDC would behave in the same way we backcrossed unmethylated gSDC transgenes from a drm1 drm2 background to wild type and analyzed expression in the F1 progeny. In contrast to FWA, we found that SDC transgenes are efficiently resilenced after backcrossing (Fig. 4D). Hence, the SDC tandem repeats act as stronger signals to recruit the de novo DNA methylation machinery and argue against the existence of a strict “surveillance window” for RNA-directed DNA methylation.
Figure 4.
DRM2 and siRNA-dependent silencing of SDC transgenes. (A) Analysis of DNA methylation using sodium bisulfite sequencing of gSDC transgenes transformed by Agrobacterium into wild-type (Columbia) or drm1 drm2 backgrounds. Percentage methylation is shown for CG (black), CHG (gray), and CHH (white) sites. (B) Analysis of SDC expression by Northern blotting and hybridization in wild-type and mutant backgrounds transformed with gSDC transgenes. Loading was analyzed by hybridization with a UBIQUITIN10 (UBQ) probe. (C) Northern blot analysis of SDC expression in wild-type and mutant plants transformed with gSDC transgenes. (D) Northern blot analysis of SDC expression after drm1 drm2 mutants with unsilenced gSDC were backcrossed to wild type.
Establishment of SDC silencing during backcrossing
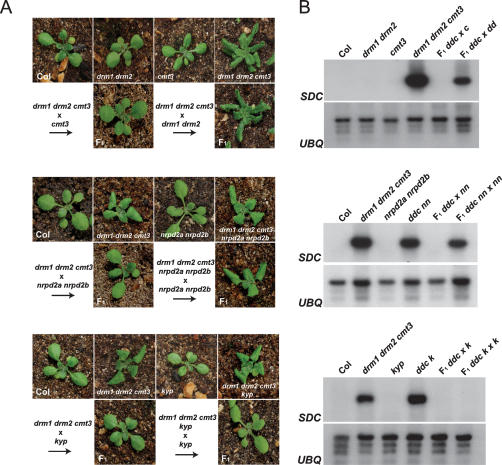
Backcrossing drm1 drm2 cmt3 to wild type provides us with an additional means to analyze establishment of silencing at SDC. We first crossed drm1 drm2 cmt3 to either drm1 drm2 or cmt3 mutants alone and analyzed rosette leaf phenotype and SDC expression in the F1 progeny. We found that crossing to cmt3 did not block resilencing, while crossing to drm1 drm2 did, consistent with the known function of DRM2 in establishment of DNA methylation (Fig. 5A,B; Cao and Jacobsen 2002b). As DRM2 can be recruited to target sequences by siRNAs we also wanted to test whether SDC promoter siRNAs would be required for establishment of silencing during backcrossing (Chan et al. 2004). We repeated the experiment by crossing drm1 drm2 cmt3 nrpd2a nrpd2b to nrpd2a nrpd2b, where both parents lack tandem repeat siRNAs (Zhang et al. 2007). As a control, we crossed drm1 drm2 cmt3 to nrpd2a nrpd2b. The absence of siRNAs was found to block establishment of SDC silencing, whereas it occurred normally in the control cross, consistent with siRNAs playing a key role in recruiting DRM2 to unmethylated SDC loci (Fig. 5A,B). As histone H3K9 methylation has been shown to be a silencing mark upstream of CMT3 activity, we predicted that loss of the histone methyltransferase KYP would not block SDC silencing (Jackson et al. 2002; Malagnac et al. 2002). Indeed, we found that backcrossing drm1 drm2 cmt3 kyp to kyp, where both parents have greatly reduced H3K9 methylation, did not block the establishment of SDC silencing (Fig. 5A,B). Hence, although the DRM2 and CMT3 pathways act redundantly to maintain silencing at SDC, during establishment DRM2 and siRNA alone are required.
Figure 5.
Genetic requirements for establishment of SDC silencing during backcrossing. (A) Leaf phenotypes and (B) SDC expression analyzed by Northern blotting and hybridization in wild-type and mutant plants used in the indicated backcrossing experiments. Loading was checked by hybridization with a UBIQUITIN10 (UBQ) probe. (dd) drm1 drm2; (c) cmt3; (nn) nrpd2a nrpd2b; (k) kyp; (ddc) drm1 drm2 cmt3; (ddcnn) drm1 drm2 cmt3 nrpd2a nrpd2b; (ddk) drm1 drm2 kyp.
Discussion
SDC demonstrates that non-CG methylation can play an important role in silencing endogenous genes in plants. The factors that determine whether a locus will be silenced by CG (e.g., FWA) or non-CG (e.g., SDC) DNA methylation are currently unclear. Comparison of cytosine context (CG, CHG, and CHH) frequencies between the SDC and FWA tandem repeats does not reveal any striking differences (data not shown). It will be important to understand the dependence on CG versus non-CG methylation as they appear to display different recruitment behavior; the non-CG DNA methylation at SDC can flexibly return once it is lost, which is in contrast to the stability of unmethylated fwa-1 epialleles (Soppe et al. 2000; Chan 2006b). The flexibility of non-CG methylation recruitment also raises the possibility that it could be used as a means of gene regulation. However, in the case of SDC, we were unable to find a developmental stage or environmental condition where SDC is expressed (data not shown), and so regulated control of SDC methylation and expression remains an open question.
The SDC tandem repeats differ from those at FWA in showing spreading of siRNAs and DNA methylation into adjacent nonrepeated sequences. Understanding the control of siRNA spreading will be important, as failure to correctly limit such phenomena can cause inappropriate silencing of endogenous genes (Saze and Kakutani 2007; Saze et al. 2008). Sequences have been defined that can act as insulators or barriers to the spread of heterochromatic silencing in a variety of systems (Gaszner and Felsenfeld 2006). As spreading of DNA methylation and siRNAs beyond repeats are not typically observed in the A. thaliana genome, this may indicate the widespread existence of similar barriers (Lippman et al. 2004; Zhang et al. 2007; Zilberman et al. 2007). Further study of the boundaries of loci such as SDC, which displays spreading in wild type, and BONSAI, which displays spreading in mutant backgrounds, may be informative in this respect (Saze and Kakutani 2007). Spreading of DNA methylation at SDC also reveals a novel and surprising role for MET1. We cannot currently determine whether MET1 is directly participating in spreading of methylation, as it is also possible that MET1 activity at the SDC repeats is required to promote spreading activity of other methyltransferases. These alternative possibilities may be resolved by analyzing the genomic localization of the methyltransferase enzymes.
Analysis of more complex, repeat-rich plant genomes is likely to uncover further examples of genes silenced by repeats and DNA methylation. For example, paramutation at the maize B’ locus has been shown to involve both an array of tandem repeats and the siRNA machinery (Stam et al. 2002; Alleman et al. 2006). The density of repeats and heterochromatin in these species means that correctly limiting silencing is likely to be of significant importance. The study of additional genomes may provide us with more examples of loci controlled by non-CG methylation, instances of heterochromatic spreading and a better understanding of the relationship between them.
Materials and methods
Plant materials
The drm1 drm2 cmt3 triple mutant generated in the Columbia background and has been previously described (Zhang et al. 2006). The SDC insertion is Salk T-DNA_017593, and was genotyped using primers JP3549, JP3550, and JP2410. All oligonucleotide sequences are listed in Supplemental Table 1. The 35S∷SDC transgene was generated by PCR amplifying the SDC ORF with primers JP3632 and JP3633, digestion with XhoI and BamHI and cloning into the pBIN-JIT binary vector, which had been digested with SalI and XhoI. The nrpd1a (Zhang et al. 2007), nrpd1b (Zhang et al. 2007), rdr2 (Chan et al. 2004), dcl3 (Chan et al. 2004), ago4 (Chan et al. 2004), nrpd2a (Zhang et al. 2007), nrpd2b (Zhang et al. 2007), met1 (Saze et al. 2003), and kyp (Chan et al. 2006a) mutations used are identical to those previously described. The SDC genomic clone was generated by PCR amplification from Columbia genomic DNA using primers JP3630 and JP3631 and cloning into pCR2.1 (Invitrogen). This clone was mutated using Quikchange (Stratagene) to change an NlaIII restriction site to BamHI using primers JP3687 and JP3688. The genomic SDC region was then cloned into a pCAMBIA1300 binary vector as a KpnI–XbaI fragment, which was introduced into Agrobacterium strain ASE and used to transform plants by floral dipping and T1 transformants selected for hygromycin resistance.
DNA methylation analysis
Genomic DNA was bisufite converted using MethylEasy (Human Genetic Signatures). The SDC upstream and tandemly repeated region (Bisulfite-I) was bisulfite sequenced using primers JP3552 for the bottom strand and JP3551 for the top strand. The region downstream from the tandem repeats (Bisulfite-II) was sequenced using primers JP5777 for the bottom strand and JP5776 for the top strand. Clone information and further bisulfite data are provided in Supplemental Tables 2 and 3 and Supplemental Figure 1.
RNA Northern blotting and hybridization
Total RNA was extracted from mature leaves using Trizol reagent (Invitogen) and analyzed by Northern blotting as described previously (Henderson et al. 2006). SDC and UBIQUITIN10 (UBQ, At4g05320) probes were amplified from Columbia cDNA using primers J3630/JP3631 and JP3483/JP3484, respectively. Small RNAs were extracted from flowers and analyzed by Northern blotting as described previously (Henderson et al. 2006). Sequences of LNA oligonucleotide probes can be found in Supplemental Table 1.
Acknowledgments
I.R.H. was supported by the Leukemia-Lymphoma Society as a Special Fellow. Thank you to X. Zhang and S. Chan for kindly providing multimutant backgrounds, W. Wong for help with growing plants, and members of the Jacobsen Laboratory for discussion and comments on the manuscript. This work was supported by NIH grant GM60398. S.E.J. is an investigator of the Howard Hughes Medical Institute. I.R.H. and S.E.J. conceived and designed experiments. I.R.H. performed experiments. I.R.H. and S.E.J. wrote the manuscript.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1667808.
References
- Alleman M., Sidorenko L., McGinnis K., Seshadri V., Dorweiler J.E., White J., Sikkink K., Chandler V.L. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- Aufsatz W., Mette M.F., van der Winden J., Matzke A.J., Matzke M. RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. 2002;99 (Suppl. 4):16499–16506. doi: 10.1073/pnas.162371499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W., Mette M.F., Matzke A.J., Matzke M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol. Biol. 2004;54:793–804. doi: 10.1007/s11103-004-0179-1. [DOI] [PubMed] [Google Scholar]
- Bartee L., Malagnac F., Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes & Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. 2002a;99:16491–16498. doi: 10.1073/pnas.162371599. (Suppl 4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002b;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Cao X., Aufsatz W., Zilberman D., Mette M.F., Huang M.S., Matzke M., Jacobsen S.E. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Zilberman D., Xie Z., Johansen L.K., Carrington J.C., Jacobsen S.E. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Henderson I.R., Jacobsen S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Henderson I.R., Zhang X., Shah G., Chien J.S., Jacobsen S.E. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet. 2006a;2:e83. doi: 10.1371/journal.p.gen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W.-L., Zhang X., Bernatavichute Y.V., Jacobsen S.E. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006b;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini S.F., Jacobsen S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Downes B.P., Shiu S.H., Durski A.M., Vierstra R.D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M., Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Kankel M.W., Ramsey D.E., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Malagnac F., Bartee L., Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A., Yonebayashi S., Watanabe K., Toyama T., Shimada H., Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Schwach F., Studholme D., Baulcombe D.C. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Nunes P.C., Pontes O., Pikaard C.S. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Kakutani T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007;26:3641–3652. doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Mittelsten Scheid O., Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Saze H., Shiraishi A., Miura A., Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- Soppe W.J., Jacobsen S.E., Alonso-Blanco C., Jackson J.P., Kakutani T., Koornneef M., Peeters A.J. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Stam M., Belele C., Ramakrishna W., Dorweiler J.E., Bennetzen J.L., Chandler V.L. The regulatory regions required for B' paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics. 2002;162:917–930. doi: 10.1093/genetics/162.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran R.K., Henikoff J.G., Zilberman D., Ditt R.F., Jacobsen S.E., Henikoff S. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr. Biol. 2005;15:154–159. doi: 10.1016/j.cub.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Vaistij F.E., Jones L., Baulcombe D.C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henderson I.R., Lu C., Green P.J., Jacobsen S.E. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Johansen L.K., Xie Z., Carrington J.C., Jacobsen S.E. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]