Abstract
Parental origin-specific DNA methylation regulates the monoallelic expression of the mammalian imprinted genes. The methylation marks or imprints are established in the parental germline and maintained throughout embryonic development. However, it is unclear how the methylation imprints are maintained through extensive demethylation in cleavage-stage preimplantation embryos. Previous reports suggested that DNA methyltransferase(s) other than Dnmt1 is involved in the maintenance of the imprints during cleavage. Here we demonstrate, by using conditional knockout mice, that the other known DNA methyltransferases Dnmt3a and Dnmt3b are dispensable for the maintenance of the methylation marks at most imprinted loci. We further demonstrate that a lack of both maternal and zygotic Dnmt1 results in complete demethylation of all imprinted loci examined in blastocysts. Consistent with these results we find that zygotic Dnmt1 is expressed in the preimplantation embryo. Thus, contrary to the previous reports, Dnmt1 alone is sufficient to maintain the methylation marks of the imprinted genes.
Keywords: Genomic imprinting, DNA methylation, preimplantation embryos, Dnmt1
Genomic imprinting is an epigenetic gene-marking phenomenon that causes parental origin-specific monoallelic expression of a small subset of mammalian genes. The epigenetic imprints are established in the parental germline and then maintained throughout embryonic development (Reik and Walter 2001). The imprinted genes play important roles in diverse biological phenomena such as embryonic development, placental formation, fetal and postnatal growth, and maternal behaviors (Reik and Walter 2001). In humans, disruptions of imprinting cause malformation syndromes such as Angelman syndrome (AS) and Beckwith-Wiedemann syndrome (BWS) (Robertson 2005).
DNA methylation is a major epigenetic modification involved in genomic imprinting. Many imprinted genes possess differentially methylated regions (DMRs), which are methylated differently between the parental alleles (Edwards and Ferguson-Smith 2007). Furthermore, the differential methylation marks or imprints at some of the DMRs are derived from sperm and oocytes. Indeed, at least some of these DMRs behave as imprint control regions (Edwards and Ferguson-Smith 2007). The first evidence that DNA methylation is essential for imprinting comes from a gene knockout study performed by Li et al. (1993). Mouse embryos lacking the maintenance DNA methyltransferase Dnmt1 showed genome-wide demethylation and loss of monoallelic expression of the imprinted genes. Furthermore, our previous studies demonstrated that a de novo DNA methyltransferase Dnmt3a is essential for the establishment of the differential methylation in both male and female germline (Kaneda et al. 2004; Kato et al. 2007). These results indicate that DNA methylation is a critical component of genomic imprinting.
Although the role of Dnmt1 in the maintenance of the methylation imprints in postimplantation embryos is well established (Li et al. 1993), it is still unclear how the imprints are maintained in cleavage-stage preimplantation embryos. After fertilization, extensive nuclear reprogramming including active and passive demethylation occurs (Reik et al. 2001). While the genome is globally demethylated during cleavage, the monoallelic methylation marks of the imprinted genes escape demethylation and are faithfully maintained. The imprint maintenance during preimplantation reprogramming is also critical for the production of somatic clones by nuclear transfer (Humpherys et al. 2002; Inoue et al. 2002; Tamada and Kikyo 2004). Moreover, it has been shown that expression and methylation of imprinted genes in preimplantation embryos are affected by the in vitro culture conditions (Sasaki et al. 1995; Doherty et al. 2000; Khosla et al. 2001; Fernandez-Gonzalez et al. 2004; Mann et al. 2004). Clinical studies also revealed an increased frequency of imprinting disorders such as AS and BWS in children conceived in vitro by assisted reproductive technology (Cox et al. 2002; DeBaun et al. 2003; Gicquel et al. 2003; Maher et al. 2003; Orstavik et al. 2003).
Nakamura et al. (2006) reported that a maternal factor PGC7/Stella protects DNA methylation at several imprinted DMRs from reprogramming during preimplantation development. In addition to such a factor, however, maintenance of DNA methylation through the S phase requires a methyltransferase(s). An enzyme known to be involved in the methylation imprint maintenance in preimplantation embryos is Dnmt1o, which is an oocyte-specific isoform of Dnmt1. Dnmt1o accumulates at high levels in the cytoplasm of oocytes and is present in the cytoplasm of embryos throughout the preimplantation stages (Carlson et al. 1992; Mertineit et al. 1998; Cardoso and Leonhardt 1999; Howell et al. 2001; Ratnam et al. 2002). Interestingly, it has been reported that Dnmt1o is trafficked to the nuclei only at the eight-cell stage (Carlson et al. 1992; Mertineit et al. 1998; Cardoso and Leonhardt 1999; Howell et al. 2001; Ratnam et al. 2002). Moreover, offspring of females lacking Dnmt1o exhibits an ∼50% reduction in the number of normally methylated alleles of the imprinted genes (Howell et al. 2001). These observations lead the authors to suggest that Dnmt1o maintains the methylation imprints only during one embryonic S phase at the eight-cell stage (Howell et al. 2001). Because Dnmt1o is the only Dnmt1 isoform that was detected in preimplantation embryos, it was proposed that a DNA methyltransferase(s) other than Dnmt1 might play a role in the imprint maintenance (Howell et al. 2001; Ratnam et al. 2002). Very recently, however, Kurihara et al. (2008) and Cirio et al. (2008) reported that Dnmt1s, a somatic isoform of Dnmt1, is present at very low levels in the nucleus of oocytes and preimplantation embryos.
To identify the DNA methyltransferases responsible for imprint maintenance in cleavage-stage preimplantation embryos, we used conditional mouse mutants of Dnmt1, Dnmt3a, and Dnmt3b. We demonstrate that Dnmt3a and Dnmt3b are not required for the maintenance of the imprints. Instead, our results demonstrate that Dnmt1 alone is sufficient to maintain the methylation imprints during cleavage.
Results
Expression of maternal Dnmt3a and zygotic Dnmt3b in preimplantation embryos
To assess the possibility that Dnmt3a and/or Dnmt3b is involved in the preimplantation maintenance of DNA methylation imprints, we first examined the expression and subcellular localization of these proteins in preimplantation embryos by immunostaining using monoclonal antibodies. To determine whether Dnmt3a and Dnmt3b were produced in the oocyte or were expressed in the embryo, we used embryos derived from oocytes lacking Dnmt3a and/or Dnmt3b. Such oocytes and embryos were obtained from conditional knockout female mice (Kaneda et al. 2004; Dodge et al. 2005) harboring a zona-pellucida glycoprotein 3 (Zp3)-Cre transgene, which is exclusively expressed in growing oocytes (de Vries et al. 2000). The timing and efficiency of conditional deletion of Dnmt3a and Dnmt3b by Zp3-Cre are described elsewhere (M. Kaneda, R. Hirasawa, H. Chiba, M. Okano, E. Li, and H. Sasaki, in prep.). The mutant oocytes and embryos also served as negative controls for immunostaining.
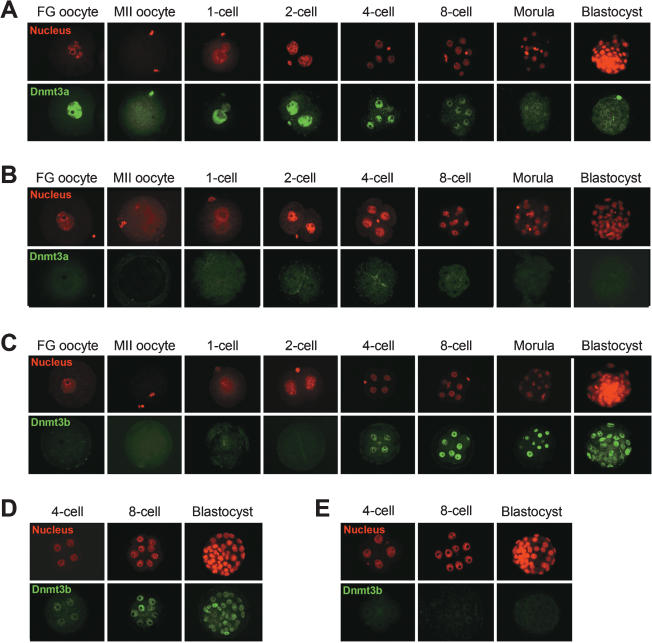
We found that Dnmt3a proteins are abundantly present in the nucleus of wild-type fully grown (FG) oocytes and diffusely present in the cytoplasm of metaphase II (MII) oocytes (or unfertilized eggs) (Fig. 1A). After fertilization, Dnmt3a relocalized to the pronuclei of one-cell embryos and remained in the nucleus up to the eight-cell stage. However, the Dnmt3a signal was significantly weaker at the eight-cell stage and became almost undetectable by the blastocyst stage (Fig. 1A). These results suggested that the nuclear localized Dnmt3a proteins present in preimplantation embryos are derived from the oocyte and thus of maternal origin. Indeed, when embryos obtained from [Dnmt3a2lox/2lox, Zp3-Cre] females crossed with wild-type males were examined, no Dnmt3a signal was detectable (Fig. 1B), confirming the maternal origin of this protein. The lack of the Dnmt3a signal in these heterozygous embryos indicated that there is little, if any, zygotic expression of this protein during the preimplantation stages. In addition, these results clearly demonstrated the specificity of the antibody.
Figure 1.
Expression and subcellular localization of Dnmt3a and Dnmt3b in mouse oocytes and preimplantation embryos. (A) Immunostaining of wild-type FG oocytes, MII oocytes, and preimplantation embryos with an anti-Dnmt3a antibody. Dnmt3a signals (green) were detectable in the nucleus of FG oocytes and embryos from the one-cell through to the eight-cell stage. Dnmt3a was diffusely present in the cytoplasm of MII oocytes. Small intense signals represent the nuclei of pole bodies. (B) Absence of detectable Dnmt3a in oocytes and preimplantation embryos from [Dnmt3a2lox/2lox, Zp3-Cre] females. This confirms the maternal origin of the protein detected in the wild-type embryos. (C) Immunostaining of wild-type oocytes and preimplantation embryos with an anti-Dnmt3b antibody. Dnmt3b (green) was not detectable in oocytes, one-cell embryos, or two-cell embryos and became detectable in the later stages. (D) Zygotic production of Dnmt3b in preimplantation embryos. Dnmt3b was detected in embryos obtained from [Dnmt3b2lox/2lox, Zp3-Cre] females crossed with wild-type males. (E) Absence of detectable Dnmt3b signals in embryos obtained from [Dnmt3b2lox/2lox, Zp3-Cre] females crossed with [Dnmt3b2lox/1lox, Tnap-Cre] males. The cell nucleus was counterstained with propidium iodide (PI) (red).
The expression pattern of Dnmt3b was significantly different from that of Dnmt3a. Dnmt3b proteins were undetectable in wild-type FG oocytes, MII oocytes, and one-cell and early two-cell embryos (Fig. 1C). It was first detected in the nucleus of late two-cell embryos (Supplemental Fig. S2) and the signal became stronger at the subsequent stages (Fig. 1C), suggesting that Dnmt3b proteins are not maternally expressed but only produced in the embryo. Consistent with this interpretation, Dnmt3b was detected in embryos obtained from the uteri of [Dnmt3b2lox/2lox, Zp3-Cre] females crossed with wild-type males (Fig. 1D). This also suggested that the paternally derived Dnmt3b allele is active and contributes to the production of the protein. Furthermore, when the tissue-nonspecific alkaline phosphatase (Tnap)-Cre gene (Lomeli et al. 2000) was used to knock out the conditional alleles in male germ cells, Dnmt3b was detected in embryos obtained from wild-type females crossed with [Dnmt3b2lox/1lox, Tnap-Cre] males (data not shown). These results strongly suggested that Dnmt3b is produced from both parental alleles of the wild-type embryo. Lastly, Dnmt3b was undetectable in homozygous Dnmt3b-null embryos obtained by crossing [Dnmt3b2lox/2lox, Zp3-Cre] females with [Dnmt3b2lox/1lox, Tnap-Cre] males (Fig. 1E), indicating that the anti-Dnmt3b antibody was highly specific.
Neither Dnmt3a nor Dnmt3b are involved in the maintenance of methylation imprints at two paternally methylated DMRs
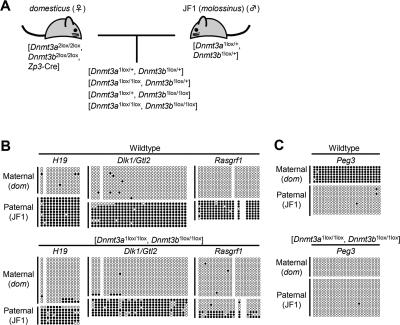
Having established the presence and nuclear localization of both Dnmt3a and Dnmt3b in preimplantation embryos, we asked whether either or both of the proteins may be involved in the maintenance of methylation imprints. To this end, we produced Dnmt3a/Dnmt3b double mutants lacking both maternally and zygotically produced enzymes. Such mutants were obtained at the expected frequency of 25% when [Dnmt3a2lox/2lox, Dnmt3b2lox/2lox, Zp3-Cre] females were crossed with [Dnmt3a1lox/+, Dnmt3b1lox/+] males (Fig. 2A). Since all embryos from this cross, including those of the desired genotype, lacked the maternal methylation imprints (see Fig. 2C) due to the loss of Dnmt3a during oocyte growth (Kaneda et al. 2004; M. Kaneda, R. Hirasawa, H. Chiba, M. Okano, E. Li, and H. Sasaki, in prep.), the imprint maintenance could not be studied at the maternally methylated DMRs. (The morphology of embryos of all genotypes is shown in Supplemental Fig. S3.) We therefore analyzed the methylation status of the paternally methylated H19, Dlk1/Gtl2, and Rasgrf1 DMRs by bisulfite sequencing at embryonic day 9.5 (E9.5). The parental origin of the DMR alleles was determined by strain-specific single nucleotide polymorphisms (SNPs).
Figure 2.
DNA methylation status of the imprinted DMRs in Dnmt3a/Dnmt3b double mutants. (A) The mouse crossing scheme for the production of embryos for bisulfite sequencing. Oocyte-specific conditional double knockout females ([Dnmt3a2lox/2lox, Dnmt3b2lox/2lox, Zp3-Cre]) were crossed with double heterozygous males ([Dnmt3a1lox/+, Dnmt3b1lox/+]), to obtain embryos with four different genotypes. The males used for this cross had a JF1-strain background. Among the embryos obtained from this cross, the [Dnmt3a1lox/1lox, Dnmt3b1lox/1lox] embryos completely lacked maternal and zygotic proteins of Dnmt3a and Dnmt3b and were used for the analysis. Strain-specific SNPs were used to determine the parental origin of the DMRs. (B) Methylation status of the paternally methylated DMRs. The paternal alleles of the H19 and Dlk1/Gtl2 DMRs were maintained methylated in [Dnmt3a1lox/1lox, Dnmt3b1lox/1lox] embryos at E9.5, whereas the paternal allele of the Rasgrf1 DMR was partially demethylated. (C) Methylation status of the maternally methylated Peg3 DMR. Maternal methylation imprint was not established, due to the lack of Dnmt3a in oocytes, confirming the effective conditional knockout. Open circles and filled circles indicate unmethylated cytosines and methylated cytosines, respectively. (JF1) JF1-derived allele; (dom) domesticus-derived allele.
The allelic methylation imprints at the H19 and Dlk1/Gtl2 DMRs were clearly maintained in the absence of either Dnmt3a or Dnmt3b (Fig. 2B). Furthermore, we observed the maternal-specific monoallelic expression of H19 (Supplemental Fig. S4). These results demonstrated that neither Dnmt3a nor Dnmt3b are essential for the imprint maintenance at these two DMRs. The only exception was the Rasgrf1 DMR, where a partial reduction in methylation was observed at the paternal allele (Fig. 2B; Supplemental Fig. S5A). Further studies with embryos of the other genotypes indicated that zygotic Dnmt3b is required for the methylation maintenance at Rasgrf1 (Supplemental Fig. S5B–D). Lastly, the maternally methylated Peg3 DMR was found to be completely unmethylated at both alleles in the mutants (Fig. 2C), confirming that the conditional deletion occurred efficiently and that the establishment of the maternal methylation imprints requires the activity of Dnmt3a during oogenesis. These results indicate that during cleavage neither Dnmt3a nor Dnmt3b are required for the maintenance of the methylation imprints at least at H19 and Dlk1/Gtl2.
Expression of Dnmt1 in oocytes and preimplantation embryos
The only other functional DNA methyltransferase known is Dnmt1, which is the well-known maintenance methyltransferase. To reinvestigate the role of Dnmt1 in the methylation imprint maintenance in preimplantation embryos, we examined its expression and subcellular localization in oocytes and preimplantation embryos using polyclonal antibodies. The antibodies were different from those used in the previous reports and detected both Dnmt1o and Dnmt1s (the somatic isoform) (ab5208-100 and the other from Takagi et al. 1995). To ascertain the origin of the detected proteins, and to obtain negative controls for immunostaining, we produced Dnmt1 conditional knockout mice (Jackson-Grusby et al. 2001) harboring Zp3-Cre (de Vries et al. 2000). This conditional knockout would disrupt both Dnmt1o and Dnmt1s isoforms. A genotype analysis of oocytes from [Dnmt12lox/2lox, Zp3-Cre] females confirmed highly efficient Cre-mediated deletion in growing oocytes by postnatal day 10 (P10) (Supplemental Fig. S6A). Immunoblotting studies further showed that Dnmt1o, the only isoform observed in wild-type oocytes, was undetectable in the mutant oocytes (Supplemental Fig. S6B).
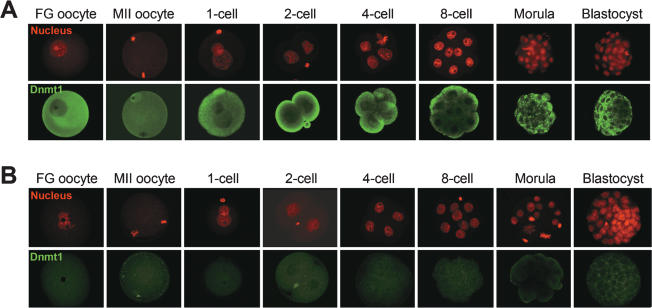
We observed strong Dnmt1 signals in the cytoplasm of wild-type oocytes and preimplantation embryos at all stages (Fig. 3A) as reported previously (Mertineit et al. 1998; Cardoso and Leonhardt 1999; Howell et al. 2001; Ratnam et al. 2002). In clear contrast with the previous reports (Mertineit et al. 1998; Cardoso and Leonhardt 1999; Howell et al. 2001; Ratnam et al. 2002), however, no clear nuclear signal was detected in eight-cell embryos with the two antibodies that we used (43 embryos stained with ab5208-100 and 15 embryos stained with the other antibody). When we examined the oocytes and preimplantation embryos obtained from [Dnmt12lox/2lox, Zp3-Cre] females crossed with wild-type males, no signal was detected either in the nucleus or in the cytoplasm (Fig. 3B), confirming the specificity of the antibodies. This also suggested that the vast majority of the Dnmt1 proteins present in preimplantation embryos are of maternal origin (but see later).
Figure 3.
Expression and subcellular localization of Dnmt1 in oocytes and preimplantation embryos. (A) Immunostaining of wild-type FG oocytes, MII oocytes, and preimplantation embryos with an anti-Dnmt1 antibody recognizing both Dnmt1o and Dnmt1s. Dnmt1 proteins (green) were mainly detected in the ooplasm and the cytoplasm of preimplantation embryos. Nuclear translocation of the proteins at the eight-cell stage was not observed. (B) Absence of detectable Dnmt1 in oocytes and preimplantation embryos from [Dnmt12lox/2lox, Zp3-Cre] females. The cell nucleus was counterstained with PI (red).
Recently, Kurihara et al. (2008) reported that another anti-Dnmt1 antibody (H-300) did not detect nuclear translocation of Dnmt1 at the eight-cell stage, consistent with our observation. This raised the possibility that the strong nuclear signal observed in the previous reports could be due to the particular antibodies that the authors used. In an effort to confirm this result, we immunostained wild-type eight-cell embryos with one of these antibodies, PATH52 (a kind gift from T.H. Bestor). Again, we observed strong Dnmt1 signals in the cytoplasm (data not shown), suggesting that the previously observed nuclear signals may be due to the particular experimental condition.
Both maternal and zygotic Dnmt1 proteins are required for the maintenance of methylation imprints in preimplantation embryos
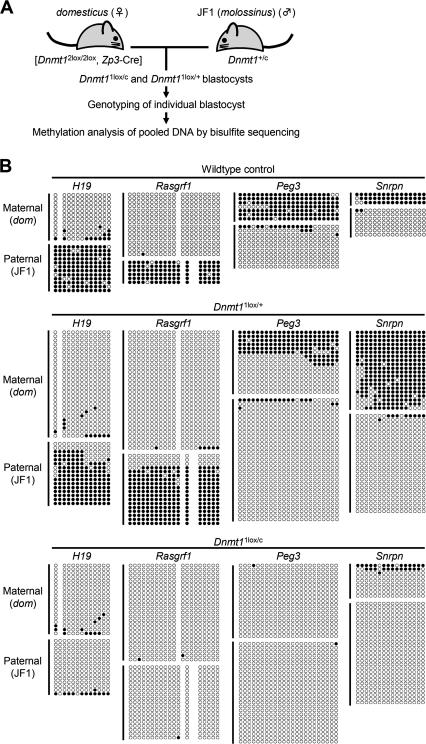
To investigate directly whether Dnmt1 is involved in the maintenance of the methylation imprints in preimplantation embryos, we studied the methylation status of the DMRs in Dnmt1 mutant blastocysts at E3.5. We first produced blastocysts lacking maternal Dnmt1, but not zygotic Dnmt1, by crossing [Dnmt12lox/2lox, Zp3-Cre] females with wild-type males (Fig. 4A). Since [Dnmt12lox/2lox, Zp3-Cre] females can establish the maternal methylation imprints, we examined both paternally methylated DMRs and maternally methylated DMRs (the Peg3 and Snrpn DMRs). The blastocysts showed a partial reduction in methylation at the paternal alleles of the H19 and Rasgrf1 DMRs and at the maternal alleles of the Peg3 and Snrpn DMRs (Fig. 4B, middle). The observed methylation defects were very similar to those described for the Dnmt1o mutants (Howell et al. 2001) except for the one at the Snrpn DMR.
Figure 4.
Methylation status of the DMRs in blastocysts lacking both maternal and zygotic Dnmt1. (A) A schematic representation of the flow of the experiment. [Dnmt12lox/2lox, Zp3-Cre] females were crossed with Dnmt1c/+ males, and then the obtained E3.5 blastocysts were genotyped by PCR with primers that specifically amplify the Dnmt1c allele. DNA from blastocysts of the same genotype (74 blastocysts of Dnmt11lox/c and 111 blastocysts of Dnmt11lox/+) was pooled and subjected to bisulfite sequencing. (B) Methylation status of the H19, Rasgrf1, Peg3, and Snrpn DMRs in wild-type (top) and mutant blastocysts (middle and bottom). (Middle) Blastocysts lacking maternal Dnmt1 (Dnmt11lox/+) showed a partial reduction in methylation at the normally methylated allele of all DMRs. (Bottom) Blastocysts lacking both maternal and zygotic Dnmt1 (Dnmt11lox/c) showed near complete loss of methylation at the normally methylated DMR alleles. (JF1) JF1-derived allele; (dom) domesticus-derived allele.
To obtain blastocysts lacking both maternal and zygotic Dnmt1 proteins, we crossed [Dnmt12lox/2lox, Zp3-Cre] females with Dnmt1c/+ males. The Dnmt1c allele is a null allele of Dnmt1 (Lei et al. 1996). Genomic DNA was extracted from individual blastocysts and the genotype was determined by PCR using half of the DNA. Seventy-four blastocysts lacked both maternal and zygotic Dnmt1 (Dnmt11lox/c) while 111 lacked maternal Dnmt1 alone (Dnmt11lox/+) (Fig. 4A). Then, pooled genomic DNA from these blastocysts was analyzed for methylation. The methylation imprints at the H19, Rasgrf1, Peg3, and Snrpn DMRs were completely lost in the Dnmt11lox/c blastocysts (Fig. 4B, bottom), indicating that not only maternal Dnmt1 but also zygotic Dnmt1 are required for the methylation imprint maintenance during preimplantation development. Combined with the results obtained with the Dnmt3a/Dnmt3b double mutants, we conclude that Dnmt1 alone maintains the methylation imprints at most of the DMRs.
Detection of zygotic Dnmt1s in preimplantation embryos
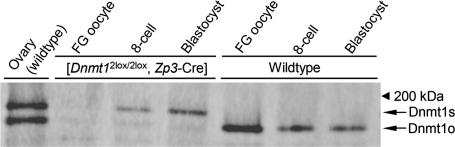
The above results argue that, although we did not observe Dnmt1 signals in the nucleus of preimplantation embryos (Fig. 3A,B), there must be a small amount of nuclear localized Dnmt1 proteins that maintain the methylation imprints. Very recently, Kurihara et al. (2008) used an antibody against the Dnmt1s-specfic N-terminal region for immunostaining and detected this isoform in MII oocytes and in the nucleus of embryos throughout preimplantation development. Cirio et al. (2008) also detected Dnmt1s in oocytes and preimplantation embryos by immunostaining and immunoblotting using another Dnmt1s-specific antibody. Both groups estimated that Dnmt1s is ∼2000-fold less abundant than Dnmt1o in MII oocytes. We attempted to detect Dnmt1s in preimplantation embryos by immunoblotting using our antibodies. Because the presence of the large amount of Dnmt1o would hamper the detection of Dnmt1s, we used embryos lacking maternal Dnmt1. As a result, while the band corresponding to Dnmt1o was completely absent, the band representing Dnmt1s was clearly detected in proteins extracted from pools of 250 eight-cell embryos and 250 blastocysts obtained from [Dnmt12lox/2lox, Zp3-Cre] females (Fig. 5). Since Dnmt1s signal was undetectable in [Dnmt12lox/2lox, Zp3-Cre] FG oocytes, it is apparent that most, if not all, of the proteins detected are produced in the embryo.
Figure 5.
Detection of zygotic Dnmt1s in preimplantation embryos by immunoblotting. Zygotic Dnmt1s was detected in eightcell embryos and blastocysts obtained from [Dnmt12lox/2lox, Zp3-Cre] females. These embryos lacked maternal Dnmt1 (mostly Dnmt1o) and thus allowed the detection of small amounts of zygotic Dnmt1s. Proteins extracted from pools of 250 oocytes or embryos from [Dnmt12lox/2lox, Zp3-Cre] females and those extracted from pools of 10 wild-type oocytes or embryos were loaded. Proteins extracted from a wild-type ovary were loaded as a control.
Discussion
The major conclusion of this study is that, in contrast to the previous suggestions, Dnmt1 alone is sufficient to maintain the methylation imprints in the preimplantation embryo at most of the DMRs. In particular, our data indicate that, in addition to maternal Dnmt1, zygotically expressed Dnmt1 is functional in the preimplantation embryo and maintains the parental imprints. It is noteworthy that the maintenance of imprints is achieved by Dnmt1 also in embryonic stem cells, which are derived from the inner cell mass of blastocysts (Okano et al. 1999). We further showed by immunoblotting that the zygotic Dnmt1 present in the preimplantation embryo have the molecular size of Dnmt1s. This is consistent with the recent reports that Dnmt1s is detectable by Dnmt1s-specific antibodies in the nucleus of preimplantation embryos at most of the stages (Cirio et al. 2008; Kurihara et al. 2008). Kurihara et al. (2008) further showed that specific inactivation of Dnmt1s in preimplantation embryos by RNAi-mediated knockdown or antibody neutralization causes a partial reduction in methylation at the H19 DMR. Because the amount of Dnmt1s per embryo increases during preimplantation development (Fig. 5), we infer that most of the proteins are translated in the embryo. Furthermore, because Dnmt1s transcripts are almost undetectable at the one-cell stage but become detectable from the two-cell stage (Ratnam et al. 2002), the RNA templates for this translation are also considered as zygotic origin.
Importantly, we did not observe previously reported nuclear translocation of Dnmt1o at the eight-cell stage using three different antibodies (including the one used to detect the nuclear translocation). Kurihara et al. (2008) recently reported that yet another antibody did not detect the eight-cell nuclear localization. These results make the maintenance role of Dnmt1o at the eight-cell stage unlikely. Considering the available evidence, a likely scenario is that maternal Dnmt1 maintains the methylation imprints during the first cell cycle and that zygotically expressed Dnmt1s maintains the imprints from the two-cell stage onward. The maternal Dnmt1 that acts at the one-cell stage can be either Dnmt1o or Dnmt1s. Kurihara et al. (2008) reported the detection of Dnmt1s in the pronuclei while Cirio et al. (2008) described the detection of Dnmt1s only in the cytoplasm of one-cell embryos. Therefore, further studies are needed to answer the question of which Dnmt1 isoform maintains the imprints at the first cell cycle.
The only DMR for which Dnmt1 was not sufficient for the methylation imprint maintenance was the Rasgrf1 DMR. This DMR required zygotic Dnmt3b for the imprint maintenance in either preimplantation embryos, postimplantation embryos, or both (Fig. 2B; Supplemental Fig. S5). Our previous study showed that, unlike the H19 and Dlk1/Gtl2 DMRs, the Rasgrf1 DMR requires not only Dnmt3a but also Dnmt3b for the establishment of the methylation imprint in male germ cells (Kato et al. 2007). It is noteworthy that this DMR is extraordinarily rich in retrotransposon sequences and flanked by a direct repeat necessary for the establishment and maintenance of the methylation imprints (Yoon et al. 2002; Holmes et al. 2006). Perhaps, these unusual structural features may be the reason for the special requirement for the methylation maintenance.
In conclusion, our results indicate that the maternal and zygotic Dnmt1 isoforms are necessary and sufficient for the maintenance of the methylation imprints during preimplantation development at all imprinted DMRs except the Rasgrf1 DMR. At present, we do not know how Dnmt1 maintains the methylation imprints against the active and passive genome-wide demethylation that occurs in preimplantation embryos. We speculate that some unknown mechanism may act and recruit Dnmt1 specifically to the DMRs. Understanding of the selective maintenance of the methylation imprints during cleavage when the genome is globally demethylated should provide a basis for improvements of reproductive engineering, animal cloning, and regenerative medicine.
Materials and methods
Mice
Production of mice with the conditional alleles, referred to as Dnmt12lox, Dnmt3a2lox, and Dnmt3b2lox, was described previously (Supplemental Fig. S1; Jackson-Grusby et al. 2001; Kaneda et al. 2004; Dodge et al. 2005). To disrupt the conditional alleles in oocytes and male germ cells, mice with the Zp3-Cre gene (de Vries et al. 2000) (Jackson Laboratory) and those with the Tnap-Cre gene (Lomeli et al. 2000), respectively, were crossed with mice with the conditional alleles. Mice possessing the Dnmt1c allele were described previously (Lei et al. 1996). All these mice had a genetic background of Mus musculus domesticus. Dnmt1c/+ mice and [Dnmt3a1lox/+, Dnmt3b1lox/+] mice were crossed with wild-type JF1 mice (of which genome is basically from Mus musculus molossinus) several times to introduce JF1-specific SNPs into the DMRs.
Preparation of oocytes and embryos
Growing oocytes and FG oocytes were obtained from the ovaries of females at P3–20 and adult females, respectively, according to the protocol described previously (Hiura et al. 2006). MII oocytes and one-cell embryos were collected from the oviducts and treated with hyaluronidase to remove the cumulus cells. Two-cell, four-cell, eight-cell and morula stage embryos were flushed from the oviducts with phosphate buffered saline (PBS). Blastocysts were flushed from the uteri with PBS. Postimplantation embryos were obtained at E9.5. All embryos were obtained by natural mating.
Antibodies
Two rabbit polyclonal antibodies were used to detect Dnmt1: One of them recognized two portions of Dnmt1 (amino acids 121–141 and 304–317) (ab5208-100, lot no. 112043, Abcam) while the other recognized the C-terminal region of the protein (amino acids 1037–1386) (Takagi et al. 1995). These antibodies detected both Dnmt1s and Dnmt1o isoforms. In most of the experiments, we used ab5208-100; the other antibody was used for the confirmation of the results obtained with ab5208-100. The anti-Dnmt3a monoclonal antibody (IMG-268, IMGENEX) recognized the C-terminal region of Dnmt3a and detected all known Dnmt3a isoforms. The anti-Dnmt3b monoclonal antibody (IMG-184, IMGENEX) recognized all known active isoforms of Dnmt3b. Alexa488-conjugated goat anti-rabbit IgG and anti-mouse IgG antibodies (Invitrogen/Molecular Probe) were used as the second antibody for immunostaining. A horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody (Jackson Immuno Research) was used as the second antibody for immunoblotting.
Immunostaining and microscopy
Embryos and oocytes were fixed and stained in microtiter plate wells and moved from one solution to another with handmade capillaries under a stereo microscope (Leica). The embryos and oocytes were fixed for 30 min in 4% paraformaldehyde in PBS on ice and washed with PBS. After incubation with a pretreatment buffer (1% bovine serum albumin [BSA] and 2% Triton-X100 in PBS) for 30 min, the embryos and oocytes were incubated overnight with the anti-Dnmt3a or Dnmt3b antibody diluted to 1:1000, or for 2 h with either of the anti-Dnmt1 antibodies diluted to 1:2000, at 4°C. All dilutions were made with antibody buffer (1% BSA and 0.1% Triton-X100 in PBS). After extensive washes, the embryos and oocytes were incubated for 30 min with an appropriate Alexa488-conjugated second antibody diluted to 1:100 at room temperature. After treatment with RNase A in PBS, DNA was counterstained with propidium iodide. Stained embryos and oocytes were mounted in Vectorshield mounting medium (Vector Laboratory) and observed with an Olympus FV500 confocal laser scanning microscope with a 40× objective lens.
Isolation of genomic DNA and genotyping
Pooled growing oocytes and FG oocytes were collected into 50 μL of PBS, and then 50 μL of 2× lysis buffer (20 mM Tris-HCl at pH 8.5, 0.2 M EDTA, 1% SDS) was added. After incubation for 1 h at 37°C, 1 μL of a Proteinase K solution (10 mg/mL) was added and the lysate was incubated for 1 h at 50°C. After phenol/chloroform extraction and ethanol precipitation, DNA was resuspended in 10 μL of distilled water. For single blastocyst genotyping, each blastocyst was boiled in 10 μL of distilled water for 5 min and the extract of 5 μL was used for genotyping. The remaining half of the extracts from blastocysts of the same genotype were combined and subjected to bisulfite sequencing. Isolation of genomic DNA from whole E9.5 embryos was prepared using a standard protocol. The sequences of the primers used for genotyping are available upon request.
DNA methylation analysis by bisulfite sequencing
Bisulfite treatment of DNA was performed with the EZ DNA Methylation Kit (Zymo Research) or BisulFAST (TOYOBO). Briefly, genomic DNA was denatured in 0.3 M NaOH for 10 min at 37°C, treated with 9 M sodium bisulfite for 1 h at 70°C, collected by using a microcolumn and desulphonated with 0.3 M NaOH. After the desulphonation, DNA was eluted with 10–20 μL of elution buffer. The DMRs of interest were amplified by PCR and subjected to sequence analysis. The primer sequences were described previously (Kato et al. 2007) except for those for the Snrpn DMR (forward, 5′-AATTTGTGTGATGTTTGTAAT TATTTGG-3′; reverse, 5′-AATAAACCCAAATCTAAAATAT TTTAATC-3′; reverse nested, 5′-ATAAAATACACTTTCACT ACTAAAATCC-3′).
Immunoblotting
FG oocytes and preimplantation embryos were treated with acidified Tyrode’s solution to remove the zona-pellucida. Oocytes, embryos, and other tissue samples were collected in a sample buffer (62.5 mM Tris-HCl at pH 6.8, 0.5× PBS, 2% SDS, 10% Glycerol, 5% 2-mercaptothanol), and then sonicated to cleave genomic DNA. Proteins were denatured by heating at 95°C for 5 min, separated by electrophoresis on SDS-5% polyacrylamide gels, and transferred onto nitrocellulose membranes (Amersham). Blots were blocked with 5% skimmed milk or ECL Advance Blocking Reagent (Amersham), incubated with a 1:10,000 dilution of the anti-Dnmt1 antibody ab5208-100. After several washes, blots were incubated with a 1:10,000 dilution of HRP-conjugated anti-rabbit IgG antibody, and detected by using ECL Advance Western Blotting Detection Kit (Amersham) and LAS1000 lumino-image analyzer (Fuji).
Isolation of RNA and allelic expression analysis
Total RNA was isolated from E9.5 embryos using ISOGEN (Nippon Gene). cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) and random primers. PCR was carried out using the following primers specific for H19 transcripts: forward, 5′-GACTCAAAGCACCCGTGAC-3′; reverse, 5′-TGATGGACCCAGGACCTCT-3′. The amplified products were digested with BglI to detect the JF1-specific SNP.
Acknowledgments
We thank Dr A. Nagy for providing Tnap-Cre mice; Dr. K. Mitsuya for information on the SNPs in H19 transcripts; Dr. T.H. Bestor for providing PATH52 antibody; Drs. M. Okano, H. Kurihara, Y. Kurihara, Y. Kato, T. Sado, and K. Hata for helpful discussion; H. Furuumi, K. Takada, C. Suda, and H. Inoue for technical assistance. This work was supported in part by Grant-in-Aid for Scientific Research on Priority Area from the Ministry of Education, Culture, Sports, Science and Technology of Japan to H.S. R.H. thanks the Japanese Society for the Promotion of Science for Young Scientists Fellowship.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1667008.
References
- Cardoso M.C., Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J. Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson L.L., Page A.W., Bestor T.H. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: Implications for genomic imprinting. Genes & Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- Cirio M.C., Ratnam S., Ding F., Reinhart B., Navara C., Chaillet J.R. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev. Biol. 2008;8:9. doi: 10.1186/1471-213X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G.F., Burger J., Lip V., Mau U.A., Sperling K., Wu B.L., Horsthemke B. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am. J. Hum. Genet. 2002;71:162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries W.N., Binns L.T., Fancher K.S., Dean J., Moore R., Kemler R., Knowles B.B. Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- DeBaun M.R., Niemitz E.L., Feinberg A.P. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am. J. Hum. Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge J.E., Okano M., Dick F., Tsujimoto N., Chen T., Wang S., Ueda Y., Dyson N., Li E. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- Doherty A.S., Mann M.R., Tremblay K.D., Bartolomei M.S., Schultz R.M. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Edwards C.A., Ferguson-Smith A.C. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., Moreira P., Bilbao A., Jimenez A., Perez-Crespo M., Ramirez M.A., De Rodriguez Fonseca F., Pintado B., Gutierrez-Adan A. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc. Natl. Acad. Sci. 2004;101:5880–5885. doi: 10.1073/pnas.0308560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicquel C., Gaston V., Mandelbaum J., Siffroi J.P., Flahault A., Le Bouc Y. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am. J. Hum. Genet. 2003;72:1338–1341. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura H., Obata Y., Komiyama J., Shirai M., Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11:353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- Holmes R., Chang Y., Soloway P.D. Timing and sequence requirements defined for embryonic maintenance of imprinted DNA methylation at Rasgrf1. Mol. Cell. Biol. 2006;26:9564–9570. doi: 10.1128/MCB.00058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell C.Y., Bestor T.H., Ding F., Latham K.E., Mertineit C., Trasler J.M., Chaillet J.R. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Humpherys D., Eggan K., Akutsu H., Friedman A., Hochedlinger K., Yanagimachi R., Lander E.S., Golub T.R., Jaenisch R. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc. Natl. Acad. Sci. 2002;99:12889–12894. doi: 10.1073/pnas.192433399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Kohda T., Lee J., Ogonuki N., Mochida K., Noguchi Y., Tanemura K., Kaneko-Ishino T., Ishino F., Ogura A. Faithful expression of imprinted genes in cloned mice. Science. 2002;295:297. doi: 10.1126/science.295.5553.297. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L., Beard C., Possemato R., Tudor M., Fambrough D., Csankovszki G., Dausman J., Lee P., Wilson C., Lander E., et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kato Y., Kaneda M., Hata K., Kumaki K., Hisano M., Kohara Y., Okano M., Li E., Nozaki M., Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Khosla S., Dean W., Brown D., Reik W., Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol. Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Kawamura Y., Uchijima Y., Amano T., Kobayashi H., Asano T., Kurihara H. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase I. Dev. Biol. 2008;313:335–346. doi: 10.1016/j.ydbio.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Lei H., Oh S.P., Okano M., Juttermann R., Goss K.A., Jaenisch R., Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Lomeli H., Ramos-Mejia V., Gertsenstein M., Lobe C.G., Nagy A. Targeted insertion of Cre recombinase into the TNAP gene: Excision in primordial germ cells. Genesis. 2000;26:116–117. [PubMed] [Google Scholar]
- Maher E.R., Brueton L.A., Bowdin S.C., Luharia A., Cooper W., Cole T.R., Macdonald F., Sampson J.R., Barratt C.L., Reik W., et al. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART) J. Med. Genet. 2003;40:62–64. doi: 10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M.R., Lee S.S., Doherty A.S., Verona R.I., Nolen L.D., Schultz R.M., Bartolomei M.S. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- Mertineit C., Yoder J.A., Taketo T., Laird D.W., Trasler J.M., Bester T.H. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2006;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Orstavik K.H., Eiklid K., van der Hagen C.B., Spetalen S., Kierulf K., Skjeldal O., Buiting K. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intarcytoplasmic semen injection. Am. J. Hum. Genet. 2003;42:218–219. doi: 10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnam S., Mertineit C., Ding F., Howell C.Y., Clarke H.J., Bestor T.H., Chaillet J.R., Trasler J.M. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev. Biol. 2002;245:304–314. doi: 10.1006/dbio.2002.0628. [DOI] [PubMed] [Google Scholar]
- Reik W., Walter J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Ferguson-Smith A.C., Shum A.S., Barton S.C., Surani M.A. Temporal and spatial regulation of H19 imprinting in normal and uniparental mouse embryos. Development. 1995;121:4195–4202. doi: 10.1242/dev.121.12.4195. [DOI] [PubMed] [Google Scholar]
- Takagi H., Tajima S., Asano A. Overexpression of DNA methyltransferase in myoblast cells accelerates myotube formation. Eur. J. Biochem. 1995;231:282–291. doi: 10.1111/j.1432-1033.1995.tb20698.x. [DOI] [PubMed] [Google Scholar]
- Tamada H., Kikyo N. Nuclear reprogramming in mammalian somatic cell nuclear cloning. Cytogenet. Genome Res. 2004;105:285–291. doi: 10.1159/000078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B.J., Herman H., Sikora A., Smith L.T., Plass C., Soloway P.D. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 2002;30:92–96. doi: 10.1038/ng795. [DOI] [PMC free article] [PubMed] [Google Scholar]