Abstract
Specification of germ cell fate is fundamental in development. With a highly representative single-cell microarray and rigorous quantitative PCR analysis, we defined the genome-wide transcription dynamics that create primordial germ cells (PGCs) from the epiblast, a process that exclusively segregates them from their somatic neighbors. We also analyzed the effect of the loss of Blimp1, a key transcriptional regulator, on these dynamics. Our analysis revealed that PGC specification involves complex, yet highly ordered regulation of a large number of genes, proceeding under the strong influence of mesoderm induction but specifically avoiding developmental programs such as the epithelial–mesenchymal transition, Hox cluster activation, cell cycle progression, and DNA methyltransferase machinery. Remarkably, Blimp1 is essential for repressing nearly all the genes normally down-regulated in PGCs relative to their somatic neighbors. In contrast, it is dispensable for the activation of approximately half of the genes up-regulated in PGCs, uncovering the Blimp1-independent events for PGC specification. Notably, however, highly PGC-specific genes exhibited distinct correlations to Blimp1 in wild-type embryos, and these correlations faithfully predicted their expression impairments in Blimp1 mutants. Moreover, their expression overlaps within single cells were severely damaged without Blimp1, demonstrating that Blimp1 exerts positive influence on their concerted activation. Thus, Blimp1 is not a single initiator but a dominant coordinator of the transcriptional program for the establishment of the germ cell fate in mice.
Keywords: Primordial germ cells, Blimp1/Prdm1, cell fate specification, single-cell analysis, genome-wide gene expression profiling
In the development and heredity of multicellular organisms, segregation of the germline from somatic lineages is a fundamental process, ensuring the perpetuation of the genetic and epigenetic information of a given species from generation to generation. The Metazoans have evolved two key strategies for the specification of the germ cell lineage in development. One operates through the inheritance of maternally deposited determinants, generally known as the germ plasm, into specific blastomeres at the outset of development (“preformation”); the other works through induction by specific signals of a pluripotent cell population during the course of development (“epigenesis”) (Extavour and Akam 2003; Seydoux and Braun 2006; Hayashi et al. 2007). Although the pre-formation mode is seen in many model organisms of modern biology, including Caenorhabditis elegans and Drosophila melanogaster, it has been suggested that the epigenesis mode, which is seen in mice and probably all other mammals, is more prevalent across and ancestral to the Metazoa (Extavour and Akam 2003).
Classic embryological studies have shown that germ cell fate in mice is induced in some of the proximal epiblast cells around embryonic day 6.5 (E6.5) (Lawson and Hage 1994). The primordial germ cells (PGCs), the first population of the germ cell lineage that give rise to either oocytes or spermatazoa, can first be identified as a small cluster of ∼40 alkaline phosphatase (AP)-positive cells just posterior to the primitive streak in the extraembryonic mesoderm at E7.25 (Chiquoine 1954; Ginsburg et al. 1990). Gene knockout studies have demonstrated that Bmp4 and Bmp8b signaling from extraembryonic ectoderm and Bmp2 signaling from visceral endoderm, as well as their signal transducers (the Smads), are critical for the successful induction of AP-positive PGC fate from the epiblast (Lawson et al. 1999; Ying et al. 2000; Chang and Matzuk 2001; Tremblay et al. 2001; Ying and Zhao 2001; Hayashi et al. 2002; Chu et al. 2004). However, until recently very little was known regarding the specific genes that determine PGC fate and the intrinsic properties of PGCs. In part, this was due to the difficulty of analyzing the founder population of PGCs, which are few in number and embedded deeply in somatic neighbors.
Recently, single-cell gene expression profiling of founder PGCs and their somatic neighbors has revealed that PGCs are characterized by high levels of an interferon-inducible transmembrane protein, fragilis/mil-1/ifitm3 (Saitou et al. 2002; Tanaka and Matsui 2002; Tanaka et al. 2005), and exclusive expression of a small nuclear cytoplasmic shuttling protein, stella/Pgc7/Dppa3 (Saitou et al. 2002; Sato et al. 2002; Bortvin et al. 2003). Additionally, PGCs specifically repress the Hox genes, including Hoxb1 and Hoxa1, which are highly up-regulated in somatic mesodermal neighbors (Saitou et al. 2002). Accordingly, it has been proposed that the repression of a “somatic program” represented by the Hox genes reveals one of the mechanisms by which the PGCs escape from a somatic fate and retain their pluripotency. Subsequently, Blimp1 (B-lymphocyte-induced maturation protein-1, also known as Prdm1), a potent transcriptional regulator with a PR (PRDI-BF1 and RIZ)-domain and five zinc fingers (Turner et al. 1994), has been identified as a key factor for the specification of this lineage (Ohinata et al. 2005; Vincent et al. 2005; Robertson et al. 2007). Blimp1 expression is first induced in a few of the posterior-proximal epiblast cells at E6.25 just prior to the onset of gastrulation. Blimp1-positive cells increase in number and form a cluster of ∼20 cells at E6.75 (mid-streak [MS] stage) and 40 cells with strong AP activity at E7.25 (early-bud [EB] stage). A genetic lineage-tracing experiment demonstrated that Blimp1-positive cells at these early stages contribute almost invariably to stella-positive PGCs, defining Blimp1-positive cells induced in the proximal epiblast from E6.25 onward as the lineage-restricted PGC precursors. In Blimp1 mutants, PGC specification seems to be blocked at a very early stage, with only about 20 AP-positive PGC-like cells formed. These Blimp1-deficient PGC-like cells exhibited inconsistent repression of the Hox genes and activation of some of the PGC-specific genes such as stella, indicating that transcriptional control medicated by Blimp1 plays a critical role in PGC specification (Ohinata et al. 2005).
More recently, quantitative single-cell gene expression profiling of several selected genes in Blimp1-positive cells during the 36 h of germ cell development (from E6.75 to E8.25) in the CD1 genetic background has identified a developmental heterogeneity in Blimp1-expressing cells: At least a certain proportion of them express Hoxb1 but not Sox2, a key gene for pluripotency (Avilion et al. 2003; Takahashi and Yamanaka 2006), especially at earlier stages (Yabuta et al. 2006). Since Blimp1- and stella-positive PGCs at later stages show consistent repression of the Hox genes and expression of Sox2, it has been proposed that Blimp1-positive cells may initially have properties more similar to somatic mesodermal neighbors, but specifically turn off the “somatic program” and concomitantly reacquire potential pluripotency, as reflected by Sox2 reactivation, to be established as PGCs (Yabuta et al. 2006).
These advances in turn open up further critical questions regarding the mechanism of germ cell specification. For example, what precisely is the “somatic program” that is repressed in PGCs? How does Blimp1 deficiency impair the germ cell specification program? Is the proposed sequence of events really associated with PGC specification? Are there any germline-specific genes that function upstream of Blimp1? In an attempt to understand the process of germ cell specification in a more comprehensive manner, we decided to embark on the identification of genome-wide expression dynamics associated with the entire germ cell specification process, using a single-cell cDNA microarray technology that we developed recently (Kurimoto et al. 2006, 2007). With this methodology, we measured the global expression profiles of the initial 48 h of germ cell development (from E6.25 to E8.25 with 12-h interval) in embryos from an inbred C57BL/6 background, as well as those for Blimp1-deficient PGC-like cells and somatic mesodermal neighbors. Here, we report a detailed analysis of the results that provides a comprehensive view on the process of germ cell specification in mice at a high resolution. Our study provides a novel foundation on which to define the gene regulatory network for the specification of mammalian germ cell fate.
Results
Quantitative single-cell analysis of germ cell specification in wild-type and Blimp1-deficient embryos
In order to achieve a more comprehensive understanding of the molecular program underlying germ cell specification, we set out to analyze the genome-wide gene expression changes encompassing the entire process of germ cell specification. For this purpose, we prepared single-cell cDNAs from proximal epiblast (PrE) cells of the prestreak (PS, E6.25) stage embryos, from mesodermal cells of the early/mid-streak (E/MS, E6.5–E6.75) and late-streak/no-bud (LS/0B, E6.75–E7.0) stage embryos, from the base of the allantois of the early/mid-bud (E/MB, E7.25–E7.5) and early head fold (EHF, E7.75) stage embryos, and from developing hindgut endoderm of early somite (E8.25) stage embryos. All embryos were derived from mice of an inbred C57BL/6 background (Fig. 1A). The single-cell cDNAs were screened for the expression of Blimp1, Oct4, and T (Brachyury) for the PS stage to identify precursors of the germ cell lineage, and for the expression of Blimp1 and Oct4 for all the remaining stages to identify developing germline cells. In total, we obtained 154 such single-cell cDNAs (Supplemental Fig. S1). Single cells negative for Blimp1 and positive for Oct4 at the E/MS (31 cells), LS/0B (32 cells), and E/MB (32 cells) stages were chosen randomly as somatic neighbors and used for comparison. In order to determine the effect of Blimp1 deficiency, we also generated single-cell cDNAs from homozygously mutant Blimp1−/− embryos from the same C57BL/6 background at the LS/0B and E/MB stages. These cells, which express mutant Blimp1 transcripts (Blimp1Δ) that generate a nonfunctional truncated protein (Ohinata et al. 2005), were selected in the same manner as in the wild-type analysis. We consider these cells to be those that are initially destined for a germ cell fate but ultimately fail to do so because of the absence of functional Blimp1. In total, we obtained 25 and 24 Blimp1Δ-positive, Oct4-positive cells from the LS/0B and E/MB stages, respectively (Supplemental Fig. S1). Since Blimp1-deficient cells were highly abnormal by the E/MB stages, analyses later than these stages would not provide primary information as to the function of Blimp1.
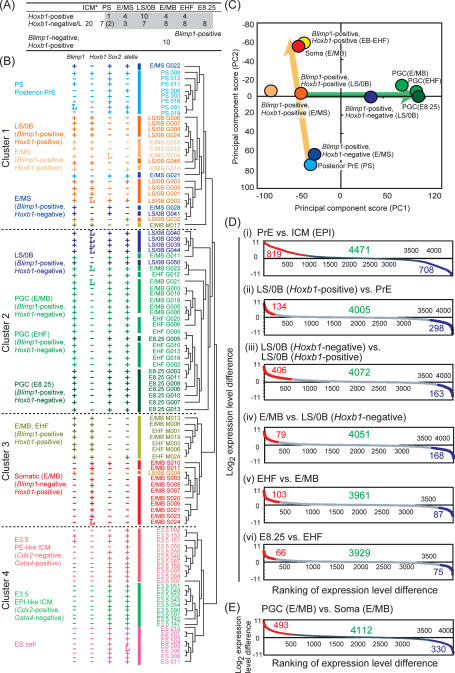
Figure 1.
Single-cell analysis of the expression of key genes during germ cell specification, both in the wild-type and Blimp1 KO embryos. (A) Schematic representation of embryos used for analysis. Dotted squares indicate the regions from which single-cell cDNAs were prepared. Arrows indicate embryonic stages covered for the analysis. (ICM) Inner cell mass; (PS) prestreak; (E/MS) early-/mid-streak; (LS/0B) late-streak/no-bud; (E/MB) early-/mid-bud; (EHF) early head fold. (B) Transition of the ratio of Hoxb1-positive (black), Sox2-positive (red), and stella-positive (blue) cells within wild-type Blimp1 or T (PS stage)-positive, Oct4-positive cells (left), Blimp1 KO Blimp1Δ-positive, Oct4-positive cells (middle), and wild-type Blimp1-negative, Oct4-positive somatic neighbors (right). The thin lines in the middle panel represent data from the wild-type embryos shown on the left. (C,D) Immunofluorescence analysis of Oct4 (panels i,ii) and Sox2 (panels iii,iv) in Blimp1-mVenus embryos at the LS (C) and MB (D) stages. (Panels ii,iv) Merged images with DAPI are shown. Blimp1-positive cells initially repressed Sox2 protein (C, panel iii) but subsequently specifically regained it (D, panel iii). This is consistent with the result obtained by the single-cell cDNA analysis (shown in B), despite the different technical sensitivities between the two methods or the differences between mRNA and protein detection. Bar, 100 μm. (E) Immunofluorescence analysis of Sox2 in Blimp1-mVenus wild-type (panels i,ii) and Blimp1 KO (panels iii,iv) embryos at the MB stage. (Panel ii,iv) Merged images with DAPI are shown. Bar, 100 μm.
We first determined the quantitative expression of key genes (Blimp1, stella, Oct4, Sox2, Hoxb1, and Gapdh as a housekeeping gene) associated with germ cell specification in all the selected cells, including those from the Blimp1 mutants (Fig. 1B; Supplemental Fig. S1). This extends observations we made previously using a limited number of wild-type samples (Yabuta et al. 2006). In wild-type embryos, the expression level of Oct4 was highest in the PrE cells at the PS stage and showed gradual reduction in both the Blimp1-positive and Blimp1-negative populations at least until the E/MB stage (Supplemental Fig. S1). The average Blimp1 expression level increased as the developmental stage progressed, consistent with observations made by in situ hybridization and transgenic reporter analyses (Ohinata et al. 2005). We identified significant variation in Blimp1 expression levels among single cells from the same developmental stages (F-test, P < 0.001) (see Supplemental Fig. S1 and below). The proportion of stella-positive cells and the stella expression level among this population increased progressively from the E/MS stage onward, demonstrating that the Blimp1-positive cells we analyzed fairly represent the developing germ cell population (Fig. 1B; Supplemental Fig. S1). Remarkably, we found that a large proportion of the Blimp1-positive cells exhibited Hoxb1 expression throughout the stages we analyzed. The proportion of Hoxb1-positive cells among Blimp1-positive cells went up to as high as 86% at the LS/0B stage and then went down thereafter. However, even at the EHF stage, 61% of the Blimp1-positive cells showed Hoxb1 expression. In contrast, a key pluripotency gene, Sox2, exhibited almost the opposite expression dynamics: The proportion of Sox2-positive cells among the Blimp1-positive population went down as low as 43% at the LS/0B stage and then rose to ∼65% after the E/MB stages. In contrast, in the somatic neighbors, all the cells were Hoxb1-positive, with expression levels increasing monotonically along with developmental progression, while the number of Sox2-positive cells decreased monotonically (Fig. 1B). Given that Blimp1-expressing cells, especially at earlier stages (until around LS/0B), are the lineage-restricted PGC precursors (Ohinata et al. 2005), these findings substantially extend the previous analysis by demonstrating that essentially all the Blimp1-positive cells destined for a germ cell fate transiently show Hoxb1 expression and lose Sox2 expression just as in somatic neighbors, but subsequently specifically revert to the Hoxb1-negative and Sox2-positive state.
In Blimp1 mutant embryos, at the LS/0B stage, the proportions of Blimp1Δ-positive cells expressing Sox2 or Hoxb1 were similar to those of wild-type cells (Fig. 1B). However, the proportion of those cells expressing stella was only 4%, significantly lower than in their wild-type counterparts (39%). Subsequently, at the E/MB stage, the proportion of Blimp1Δ-positive cells expressing Sox2 went down as low as 8%, and the proportion of Blimp1Δ-positive cells expressing Hoxb1 went up as high as 96%. These dynamics were somewhat similar to those of the wild-type somatic neighbors. However, the proportion of Blimp1Δ-positive cells expressing stella went up to 21%, which was significantly higher than that of the wild-type somatic neighbors (3%) but lower than that of the wild-type Blimp1-positive cells (77%). These findings indicate that in the absence of Blimp1, although the cells destined toward the germ cell fate may acquire germ cells properties to a certain extent, they fail to revert to the Hoxb1-negative and Sox2-positive state.
To validate these observations, we examined Oct4 and Sox2 expression in wild-type and Blimp1 mutant embryos bearing the Blimp1-mVenus reporter, by immunofluorescence analysis (Fig. 1C). In wild-type embryos, Oct4 was detected at a similar intensity both in Blimp1-positive cells and neighboring embryonic cells at both LS and MB stages. In contrast, we found that Sox2 protein was greatly reduced or undetectable in Blimp1-positive cells and posterior mesodermal cells, compared to anterior epiblast cells (or the entire epiblast at the PS stage) (data not shown) and extraembryonic ectoderm at the LS stage. However, at the MB stage, Blimp1-positive cells regained exclusive Sox2 expression, demonstrating that cells destined toward the germ cell fate indeed initially turn Sox2 off but dynamically reacquire it soon thereafter. In the Blimp1 mutants at the MB stage, however, Blimp1-mVenus-positive cells were few in number and very weak or negative for Sox2. Based on these data, we conclude that Blimp1 plays a critical role in the specific reversion of the cells destined toward the germ cell fate to the Hoxb1-negative and Sox2-positive state.
Global outlook for the germ cell specification pathway in wild-type embryos
In order to elucidate the gene expression dynamics associated with germ cell specification and its impairment by the loss of Blimp1 at the genome-wide level, we next subjected the single-cell cDNAs from all the time points we analyzed both in the wild-type (Fig. 2A) and Blimp1-deficient conditions (Fig. 4A, below) to microarray analysis using Affymetrics GeneChip Mouse Genome 430 2.0 Array. We included data on 20 cDNAs from E3.5 single inner cell mass (ICM) cells and eight embryonic stem (ES) cell cDNAs (Kurimoto et al. 2006) for comparison. In total, data sets from 134 single-cell cDNAs were processed, using analysis of variance (ANOVA) to detect statistically significant changes. In all, 5694 differentially expressed genes were selected for the subsequent analyses.
Figure 2.
Global profile of the germ cell specification pathway. (A) Number of cells subjected to the microarray analysis. Developmental stages and marker gene expressions are indicated. (*) Data from the previous study (Kurimoto et al. 2006) are used for the E3.5 ICM cells. (L) <10 copies per cell. (B) UHC analysis of the global expression profiles of the wild-type Blimp1-positive single cells. Gene expression levels of Blimp1, Hoxb1, Sox2, and stella determined by Q-PCR are indicated with symbols: (+) >10 copies per cell; (L) <10 copies per cell; (−) not detected. (C) PCA. Each circle represents the averaged expression profile of each indicated cell group. (D,E) Gene expression differences between the indicated cell groups are shown by scatter plots. Among the 5694 genes selected by ANOVA, those expressed in either of the indicated cell groups (the number indicated by green) are ranked in the order of their expression level differences between the two groups. Genes and their numbers significantly up-regulated (red) or down-regulated (blue) in the left-hand cells are indicated.
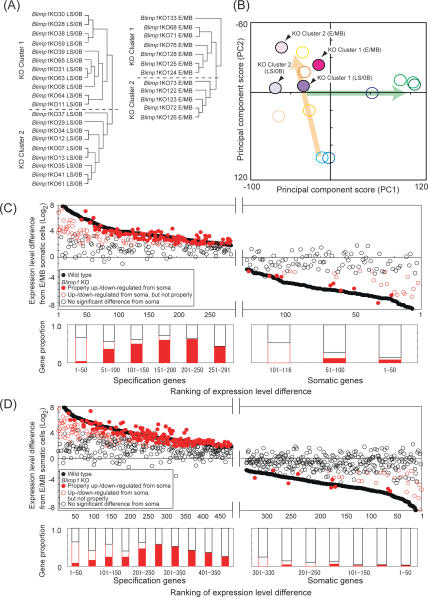
Figure 4.
Single-cell microarray analysis of the Blimp1 KO PGC-like cells. (A) UHC analysis of the Blimp1 KO cells at the LS/0B (left) and E/MB (right) stages, using the 493 specification genes. (B) PCA. The wild-type cells are represented with open circles, with the color code as in Figure 2C. Blimp1 KO cells are represented with filled circles, with indications for the KO clusters and developmental stages. (C,D, top) Scatter plots of the specification gene activation (left) and somatic gene repression (right) in Blimp1 mutants at the LS/0B (C) and E/MB (D) stages. To estimate the impact of Blimp1 deficiency at the LS/0B stage, wild-type LS/0B Blimp1-positive, Hoxb1-negative/low cells were used for the comparison to the LS/0B Blimp1 KO cells; in wild-type LS/0B Blimp1-positive, Hoxb1-negative cells, 291 of 493 specification and 116 of 330 somatic genes were up-regulated and down-regulated in comparison to the E/MB somatic cells, respectively. The vertical axis indicates log2 expression level difference between the Blimp1- or Blimp1Δ-positive cells and the E/MB stage somatic neighbors. The horizontal axis represents the ranking of the expression level difference between the wild-type Blimp1-positive, Hoxb1-negative cells and the E/MB stage somatic neighbors. Black filled circles indicate genes in the wild-type Blimp1-positive, Hoxb1-negative/low cells. Red filled circles indicate specification and somatic genes in the Blimp1 KO Cluster 1 cells that are properly up- and down-regulated (expression level difference from the somatic neighbors was significant and expression level difference from the wild-type cells was within twofold), respectively. Red open circles indicate specification and somatic genes in the Blimp1 KO Cluster 1 cells that are up- and down-regulated in comparison to the somatic neighbors (expression level difference from somatic neighbors was significant), respectively, but not properly compared to the wild-type cells (expression level difference from the wild-type cells was more than twofold). Black open circles indicate genes in the Blimp1 KO Cluster 1 cells that are indistinguishable from the somatic neighbors (expression level difference was not significant). For the definition of a significant expression level difference from the somatic neighbors, see Materials and Methods. (Bottom) Histogram of the Blimp1 KO Cluster 1 genes with the color code as in the top panel. The proportion of the specification genes properly up-regulated in the Blimp1 KO embryos (red-filled bars) is negatively correlated with the ranking of expression-level differences between the wild-type cells and somatic neighbors (i.e., magnitude of specificity); this notion is supported by the χ2 test with the null hypothesis that the proportion of the properly up-regulated genes is the same irrespective of the magnitude of specificity (P < 0.05).
We first analyzed the germ cell specification program in wild-type embryos. The selected wild-type single cells were subject to the unsupervised hierarchical clustering (UHC) (Eisen et al. 1998), which classified them into four major clusters (Fig. 2B). The PS-stage PrE cells (all Sox2-positive and mostly Hoxb1-negative) were tightly grouped in Cluster 1, irrespective of their Blimp1 expression, suggesting that Blimp1 expression is one of the earliest signs of germ cell specification, at which stage there might not yet be any major transcriptome difference in posterior-proximal epiblast cells. The E/MS-stage Blimp1-positive cells with or without Hoxb1 expression and the LS/0B-stage Blimp1-positive, Hoxb1-positive cells, three of which expressed stella, were intermingled with each other and were clustered next to the PS-stage PrE cells in Cluster 1. In contrast, the LS/0B-stage Blimp1-positive, Hoxb1-negative cells with Sox2 and stella expression were essentially grouped in Cluster 2, which includes all the Blimp1-positive, Hoxb1-negative cells with Sox2 and stella of E/MB, EHF, and E8.25 stages, which are considered to be established PGCs. Therefore, the UHC of Blimp1-expressing cells based on their genome-wide expression profiles is consistent with the notion that cells destined for a PGC fate originate from a Sox2-positive and Hoxb1-negative state, going through a Sox2-negative and Hoxb1-positive state, and eventually reverting to a Sox2-positive and Hoxb1-negative state with lineage-specific overall properties. On the other hand, the E/MB-stage Blimp1-negative, Hoxb1-positive somatic mesodermal cells were grouped in Cluster 3. Notably, in contrast to a majority of LS/0B-stage Blimp1-positive, Hoxb1-positive cells, the E/MB-, EHF-stage Blimp1-positive, Hoxb1-positive cells were grouped next to the somatic mesoderm in Cluster 3. This suggests that although these cells express Blimp1, they may no longer become PGCs (see below and Discussion for the potential fate of Blimp1-positive and Hoxb1-positive cells after the E/MB stages). The E3.5 ICM cells and ES cells were grouped distantly in Cluster 4. It is of note that ES cells (undifferentiated E14tg2a) were closer to ICM cells than to PGCs in all stages analyzed.
Principal component analysis (PCA) (Yeung and Ruzzo 2001) with cell populations after the PS stage yielded a consistent outcome (Fig. 2C). The PS-stage PrE cells; E/MS-stage Blimp1-positive, Hoxb1-negative cells; LS/0B-stage Blimp1-positive, Hoxb1-positive cells; E/MB-stage somatic neighbors; and EB-EHF Blimp1-positive, Hoxb1-positive cells exhibited very similar PC1 scores and were plotted along the PC2 axis in a developmental stage-dependent manner (Fig. 2C, orange arrow). In contrast, the LS/0B-stage Blimp1-positive, Hoxb1-negative cells and the established PGCs of E/MB, EHF, and E8.25 stages were plotted closely and aligned along the PC1 axis, also in a developmental stage-dependent manner (Fig. 2C, green arrow). It therefore appears that PC2 reflects the progress of the somatic mesodermal program and PC1 represents a progression toward PGC properties. Euclidean distances of the expression profiles of the developing Blimp1-positive cells along the defined pathway exhibited consistent increments from the PS stage PrE cells (Supplemental Fig. S2). Collectively, these analyses suggest that a major transition of genome-wide expression profile during germ cell specification takes place mainly at the LS/0B stage, the point at which the Blimp1-positive, Hoxb1-positive cells transform into Blimp1-positive, Hoxb1-negative cells with distinct PGC properties.
In good agreement, pairwise comparison of Blimp1-positive cells at different developmental stages along with the progression of PGC specification showed that it is between Blimp1-positive, Hoxb1-positive, and Hoxb1-negative cells at the LS stage, where the largest numbers of the genes show statistically significant expression level changes (Fig. 2D). Consequently, in comparison to somatic mesodermal neighbors at the E/MB stage, the established PGCs at the E/MB stage up-regulate 493 genes and down-regulate 330 genes (Fig. 2E; Supplemental Table S1), which, hereafter, we refer to as “specification genes” and “somatic genes,” respectively.
Unraveling the specification and somatic genes
Expression dynamics
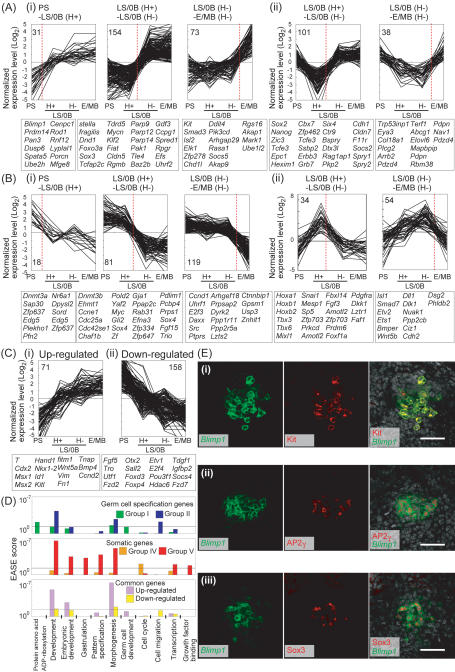
We then went on to scrutinize the specification and somatic genes (Fig. 3). We first classified them based on their expression dynamics along the defined germ cell specification pathway. The specification genes were classified largely into three groups: The Group I genes (258, ∼52%) (Fig. 3A, panel i) show consistent up-regulation from the epiblast, whereas the Group II genes (139, ∼28%) (Fig. 3A, panel ii) show transient down-regulation from the epiblast, followed by substantial up-regulation. The Group III genes (96, ∼20%) (Supplemental Fig. S3A) do not show significant changes from the epiblast but do show specific down-regulation in the somatic neighbors. Similarly, the somatic genes were classified into three large groups, with the Group IV genes (218, ∼66%) (Fig. 3B, panel i) showing consistent down-regulation from the epiblast, the Group V genes (88, ∼27%) (Fig. 3B, panel ii) showing transient up-regulation followed by substantial down-regulation, and the Group VI genes (24, ∼6%) (Supplemental Fig. S3B) showing specific up-regulation only in the somatic neighbors. Genes in each group were subclassified based on the timing of their most prominent up-/down-regulation, which may reflect the potential order of gene activation and repression during germ cell specification.
Figure 3.
Complex expression dynamics of the specification and somatic genes. (A–C) Expression dynamics of the specification genes (A), somatic genes (B), and genes commonly regulated in the E/MB stage PGCs and their somatic neighbors (C). The specification and somatic genes are classified based on their dynamics: group I (A, panel i), group II (A, panel ii), group IV (B, panel i), and group V (B, panel ii) (see text for details). Each group is subclassified based on the developmental intervals where the genes’ greatest expression level changes are observed (red dashed line): Intervals between the PS-stage and LS/0B-stage Hoxb1-negative cells, between the LS/0B-stage Hoxb1-positive and Hoxb1-negative cells, and between the LS/0B-stage Hoxb1-positive and E/MB-stage PGCs. Normalized log2 expression levels are plotted. Numbers of the genes that belong to each subclass and representative genes are shown. (D) Functional annotation of the genes over-represented in the indicated groups. EASE scores (see the Materials and Methods) are represented with bars. (E) Immunofluorescence analysis of Kit (panel i), AP2γ (panel ii), and Sox3 (panel iii) in Blimp1-mVenus wild-type embryos at the MB stage. Merged images with DAPI are shown in the right panel. Bar, 50 μm.
Two notable findings emerged from this analysis. First, both for the specification and somatic genes, the major transcriptional dynamics are the consistent, specific up-/down-regulation from the epiblast (Groups I and IV), rather than those reflecting a reversion to the epiblast state (Groups II and V). Specifically, genes that are cosegregated with Sox2 and Hoxb1 represent only 20% and 10% of the specification and somatic genes, respectively. Second, a majority (∼52%) of the specification genes are acquired between LS/0B Hoxb1-positive and LS/0B Hoxb1-negative stages, whereas repression of the somatic genes occurs gradually, with the greatest numbers of the somatic genes (∼52%) repressed between LS/0B Hoxb1-negative and E/MB Hoxb1-negative stages.
Functional classification
We next went on to identify functional properties of the specification and somatic genes (Fig. 3D). Based on the Gene Ontology (GO) functional annotation, the Group I specification genes were apparently not particularly enriched in specific functional categories. However, this group included genes known to be associated with germ cell development (Blimp1, fragilis, stella, Dnd1, Kit), and the timing of their greatest up-regulation was highly consistent with previous findings made by other methodologies, reinforcing the possibility that the identified factors may indeed function in germ cell specification pathway in an ordered manner. Accordingly, this group includes transcriptional regulators (e.g., Prdm14, Sox3, Sox17, Tcfap2c, Elf3, Elk1, Isl2, Mycn, Klf2, Fiat, Sp8, Smad3), membrane-associated receptors (e.g., Celsr1, Cdon), and intracellular signal transducers (e.g., Socs5, Pak1, Spred1, Plxdc2, Mark1, Akap9). Notably, genes for “protein ADP ribosylation” (Parp9, Parp12, Parp14) were specifically enriched in this group. Specific expression of Kit, AP2γ (encoded by Tcfap2c), and Sox3 in the Blimp1-expressing cells was validated by immunostaining (Fig. 3E).
In contrast, the Group II genes were enriched with genes functioning in “development,” particularly “morphogenesis.” Such genes include transcription factors (e.g., Sox2, Nanog, Zic3, Tcfe3, Epc1, Six4, Eya3), a Polycomb group gene (Epc1), membrane-associated receptors (e.g., Erbb3), and intracellular signal transducers (e.g., Grb7, Socs2, Spry1, Spry2, Phf17, Rag1ap1, Bspry).
The Group IV genes were again poorly enriched with specific GO functional categories. However, genes for “cell cycle” were specifically enriched in this group. These included key genes promoting the S phase such as Ccne1 (CyclinE1), Ccnd1 (CyclinD1), Cdc25a, Cdc6, Pold2 (DNA polymerase delta2 regulatory subunit), E2F3, and Myc. Additionally, Ccne2 (CyclinE2) and Cdk2 were significantly down-regulated in PGCs compared to the epiblast. Furthermore, this group includes DNA methyltransferases Dnmt3a and 3b and histone methyltransferase Glp (Ehmt1). Remarkably, although Dnmt1 showed consistently high expression during the course of germ cell development, Np95 (Uhrf1), which was shown recently to be essential for recruitment of Dnmt1 to replication foci (Bostick et al. 2007; Sharif et al. 2007), was identified as a member of this group and exhibited specific down-regulation after the E/MB stage. Therefore, essentially all the known enzymes required for DNA methyltransferase activity were revealed to be inactive in PGCs after the E/MB stage.
The Group V genes, which include Hoxb1, the first gene designated to represent a “somatic mesodermal program” (Saitou et al. 2002), showed prominent enrichment of genes for “embryonic development,” “gastrulation,” “pattern specification,” and “morphogenesis.” This group includes Hox cluster genes (Hoxa1, Hoxb1, Hoxb2), transcription factors associated with mesoderm induction and organ development (e.g., Snail, Tbx3, Tbx6, Mesp1, Sp5, Mixl1, Sall3, Cdx1, Isl1, Ets1, Etv2, Nfkbia, Foxf1, Plxna2, Smad7), signaling molecules (e.g., Wnt5b, Fgf3, Dkk1, Bmper, Cfc1, Dll1, Nmb), membrane-associated receptors (e.g., Pdgfra, Il11ra1), and intracellular signal regulators (e.g., Ppp2cb, Prkcd, Faf1, Nkd1, Nuak1, Pdlim7). Furthermore, regarding the other Hox cluster genes, we found that Hoxd9, which belongs to Group VI, was repressed in PGCs; Hoxa2, Hoxa3, and Hoxd1 (average expression level less than five copies per cell) were also exclusively detected in somatic neighbors at the E/MB stage.
Notably, in addition to these findings, our analysis revealed that PGC specification avoids the archetypal epithelial mesenchymal transition (EMT) (Lee et al. 2006; Peinado et al. 2007) (Supplemental Fig. S4): Fgfr1, which is involved in the initiation of EMT (Ciruna and Rossant 2001), showed down-regulation in the E/MB-stage PGCs, Spry1 and 2, which are negative regulators of Fgfr signaling (Hacohen et al. 1998), belonged to the Group II genes and showed up-regulation in PGCs; and Snai1, a key transcription factor for EMT, belonged to the Group V genes and was repressed in PGCs. Consistently, the epithelial cell adhesion molecule E-cadherin and tight junction molecules Cln (Claudin) 5, 7, 12, ZO1, and ZO2 were up-regulated in PGCs, whereas mesenchymal markers including N-cadherin, desmoplakin, and vimentin were down-regulated. Collectively, these analyses revealed, for the first time and with unprecedented clarity, the genes, functional properties, and activation/repression dynamics that distinguish the nascent germ cell lineage from its somatic neighbors at the genome-wide scale.
Common programs for PGCs and their somatic neighbors and complex dynamics of the specification and somatic genes
Our analysis also identified a significant number of genes commonly up-regulated (71) or down-regulated (158) in E/MB-stage PGCs and their somatic neighbors when compared to PS-stage PrE cells (Fig. 3C,D). Specifically, the commonly up-regulated genes showed significant enrichment of genes for “development,” particularly for “embryonic development” and “morphogenesis,” which include transcription factors (e.g., T, Cdx2, Msx1, Msx2, Nkx1.2, Hand1, Id1) and signaling molecules (e.g., Bmp4, Wnt5a, Kitl). This may reflect a developmental similarity of these neighboring cell populations.
We also noted that a relatively small but significant proportion of the specification (78, 16%) and somatic (107, 32%) genes exhibit down- and up-regulation, respectively, in the E8.25 migrating PGCs, although they did not show notable enrichment for specific GO functional categories (Supplemental Fig. S3C,D). It is therefore likely that PGCs continue to develop by altering some of their expression profiles, although their key properties remain unchanged.
The global impact of Blimp1 deficiency on the germ cell specification pathway
We next determined the global impact of Blimp1 deficiency on the germ cell specification pathway by analyzing the microarray data of 20 and 12 Blimp1Δ-positive cells from LS/0B and E/MB mutant embryos, respectively (Fig. 4A). As demonstrated above, in the wild type, PGC specification is actively progressing in these stages; accordingly, Blimp1-positive cells from these stages showed considerable developmental heterogeneity, as reflected by their Blimp1 expression levels and Hoxb1 and Sox2 expression states. Therefore, the mutant cells may also bear developmental heterogeneity. In such a case, for the precise evaluation of the impact of Blimp1 deficiency, it would be critical to compare the wild-type and mutant cells in a similar developmental state. However, since all the Blimp1Δ-positive cells essentially failed to revert to the Hoxb1-negative and Sox2-positive state (Fig. 1), it was impossible to use these markers for the evaluation of their potential developmental states. We therefore decided to perform UHC of these cells with the expression of all 493 specification genes. Consequently, Blimp1Δ-positive cells from the mutants at both stages were roughly classified into two major clusters (Fig. 4A), with the KO Cluster 1 (11 of 20 at the LS/0B stage and seven of 12 at the E/MB stage) exhibiting generally higher expression levels of the specification genes (Supplemental Fig. S5A). The UHC analysis using all the cells subjected to the microarray analyses exhibited similar results (Supplemental Fig. S5B).
We then performed PCA on Blimp1 mutant cells, comparing their genome-wide expression profiles with those of wild-type cells (Fig. 4B). This analysis revealed that, consistent with their expression of specification genes, the KO Cluster 1 cells at both stages had characteristics more similar to those of PGCs than did the KO Cluster 2 cells. It would therefore be logical to consider that KO Cluster 1 cells are those that were developmentally more advanced and went on to acquire PGC properties to the highest degree in the absence of Blimp1. However, even the KO Cluster 1 cells had very low PC1 scores compared with those of the established PGCs. Both KO Cluster 1 and 2 cells at the E/MB stage showed higher PC2 scores than the cells at the LS/0B stage, indicating that in the absence of Blimp1, cells otherwise destined for a germ cell fate were propelled instead toward a somatic mesodermal fate as the mouse embryo developed.
Differential impact of Blimp1 on the activation of specification genes and repression of somatic genes
We then compared the expression states of individual specification and somatic genes between wild-type Blimp1-positive, Hoxb1-negative cells and KO Cluster 1 cells both at the LS/0B and E/MB stages. As shown in Figure 4, C and D, we found that both at the LS/0B and E/MB stages, in KO Cluster 1 cells, approximately half of the specification genes were up-regulated significantly from the somatic cell levels (61% and 48% for LS/0B and E/MB stages, respectively) (Supplemental Table S2). Specifically, more than one-third of the specification genes showed indistinguishable expression levels from those in the wild-type PGCs (43% and 33% for LS/0B and E/MB stages, respectively). This finding indicates the presence of Blimp1-independent mechanisms that confer some PGC properties to epiblast cells. However, we found that the properly up-regulated specification genes were restricted mostly to those that showed relatively low specificity (Fig. 4C,D, bottom panels). It was only 4% and 8% of the top 50 specification genes at the LS/0B and E/MB stages, respectively, that showed proper up-regulation in the absence of Blimp1. Indeed, there existed a negative relationship between the magnitude of the specificity and the proportion of the genes properly up-regulated in the absence of Blimp1 (P < 0.05). Additionally, we noted that the specification genes that showed dynamic expression-level changes from the epiblast (Group I and II) were more predisposed to the impairment than those with no prominent expression-level changes (Group III) (Supplemental Fig. S5C).
In striking contrast, it became evident that a great majority of the somatic genes failed to be repressed properly in the Blimp1-deficient condition (91% and 97% at the LS/0B and E/MB stages, respectively) and many of these exhibited expression levels indistinguishable from those in the somatic neighbors (74% and 89% at the LS/0B and E/MB stages, respectively) (Fig. 4C,D; Supplemental Table S2). Indeed, we found almost complete failure of repression in all the expression dynamics groups (Groups IV, V, and VI) (Fig. 3; Supplemental Fig. S3) of somatic genes. Taken together, these findings demonstrate that Blimp1 plays a key role in the lineage-specific regulation of specification genes and is the primary factor for the repression of almost all somatic genes. It also became evident that a Blimp1-independent mechanism is responsible mainly for the activation of specification genes with relatively low specificity.
Quantitative correlations of highly specific specification genes with Blimp1
We next went on to obtain a more precise insight into the role of Blimp1 in the regulation of specification genes with high specificity. Considering the difficulty of biochemical analysis of Blimp1 function in the context of germ cell specification, we set out to explore the correlations between the levels of Blimp1 expression and the levels and frequencies of a selective set of germ cell specification genes in all the relevant single cells we analyzed. This analysis is based on two ideas: (1) If Blimp1 is required for the efficient induction of a gene, the probability (frequency) of its expression should depend on the presence and expression level of Blimp1 (level-to-frequency correlation between Blimp1 and a gene); and (2) if Blimp1 is required for the up-regulation of a gene’s expression level, it should depend on the presence and expression level of Blimp1 (level-to-level correlation between Blimp1 and a gene).
We therefore measured the expression levels of 22 selected specification genes (the Group I genes Prdm14, stella, fragilis, Dnd1, Tdrd5, Tcfap2c, Klf2, Parp9, Sp8, Fiat, Kit, Ddit4; the Group II genes Sox2, Nanog, Elf3, Zic3, Zfp462, E-cadherin; four genes found to be significantly up-regulated in E/MB stage PGCs by Q-PCR; Klf5, Gadd45a, Nanos3, Tnap) in Blimp1-positive wild-type cells (E/MS-E/MB stages), Blimp1Δ-positive mutant cells (LS/0B and E/MB stages), and somatic neighbors (E/MS-E/MB stages) (Supplemental Fig. S6). We also measured the expression levels of some other genes for comparison (the Group IV genes Hoxa1 and Hoxb1; the Group V genes Dnmt3b and Gja1; the commonly expressed genes Oct4 and T) in the same cell population.
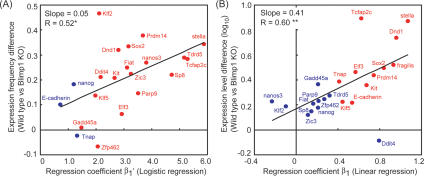
We then calculated logistic and linear regression coefficients to evaluate the level-to-frequency and level-to-level correlations, respectively, in the wild-type cells (Supplemental Figs. S7–S9). While Oct4 and T, which were equally expressed in the Blimp1-positive and Blimp1-negative cells, did not exhibit correlations with Blimp1, of the 22 specification genes, 18 (82%) and 11 (50%) showed expression frequencies and levels, respectively, positively correlated with the expression level of Blimp1. This analysis thus classified the specification genes into four categories in terms of their correlations with Blimp1: The first category of genes (Prdm14, Sox2, stella, Dnd1, Kit, Tcfap2c, Klf5, and Elf3) showed correlated expression frequencies and levels; the second (Tdrd5, Nanos3, Klf2, Fiat, Parp9, Zic3, Sp8, Zfp462, Ddit4, Gadd45a) showed only correlated frequencies; the third (fragilis, Tnap, E-cadherin) showed only correlated levels; and the fourth (Nanog) showed no correlations. Among the somatic genes examined, Dnmt3b and Hoxb1 showed both negative level-to-frequency and level-to-level correlations with Blimp1 expression levels, whereas Gja1 and Hoxa1 exhibited only negative level-to-frequency correlations.
We then compared the differences in expression frequencies and levels of the specification genes between wild-type and mutant cells (Fig. 5A,B). Consistent with the microarray analysis, all of these genes showed impaired expression levels and/or frequencies. We next asked whether there would be any correlation between the extent of expression impairments and the logistic and linear regression coefficients in the wild-type cells. Remarkably, we found a good correlation between the logistic regression coefficients of specification genes and their expression frequency differences between wild-type and mutant cells (Fig. 5A). Moreover, there existed a prominent correlation between the linear regression coefficients of the specification genes and their expression level differences between wild-type and mutant cells (Fig. 5B). These findings indicate that Blimp1 positively influences both the induction and expression levels of those specification genes that exhibit both significant logistic and linear regression coefficients (Prdm14, Sox2, stella, Dnd1, Kit, Tcfap2c, Klf5, Elf3), whereas it has positive influence only on the induction of those genes that exhibit significant logistic regression coefficients only (Tdrd5, Nanos3, Klf2, Fiat, Parp9, Zic3, Sp8, Gadd45a, Ddit4, Zfp462). Thus, the distinct level-to-frequency and level-to-level correlations of the representative specification genes with Blimp1 predict their impairment in expression in Blimp1 mutant cells.
Figure 5.
Correlations between the selected specification genes and Blimp1. (A) Expression frequency differences between the wild-type Blimp1-positive cells and Blimp1 KO Blimp1Δ-positive cells, plotted against the logistic regression coefficient between the Blimp1 expression level and the expression frequency of the selected genes in the wild-type Blimp1-positive cells. Red and blue dots represent genes that show statistically significant logistic regression with the Blimp1 expression level (P < 0.01) (see also Supplemental Figs. S7, S9) and that do not, respectively. (*) Statistically significant correlation (P < 0.05). (B) Expression level differences between the wild-type Blimp1-positive cells and Blimp1 KO Blimp1Δ-positive cells, plotted against the linear regression coefficient between the Blimp1 expression level and the expression levels of the selected genes in the wild-type Blimp1-positive cells. Averaged expression level differences are calculated using cells expressing individual genes. Red and blue dots represent genes that show statistically significant correlation coefficients (P < 0.01) (see also Supplemental Figs. S8, S9) and that do not, respectively. (**) Statistically significant correlation coefficient (P < 0.01).
Coordinated activation of specification genes in the Blimp1-expressing cells
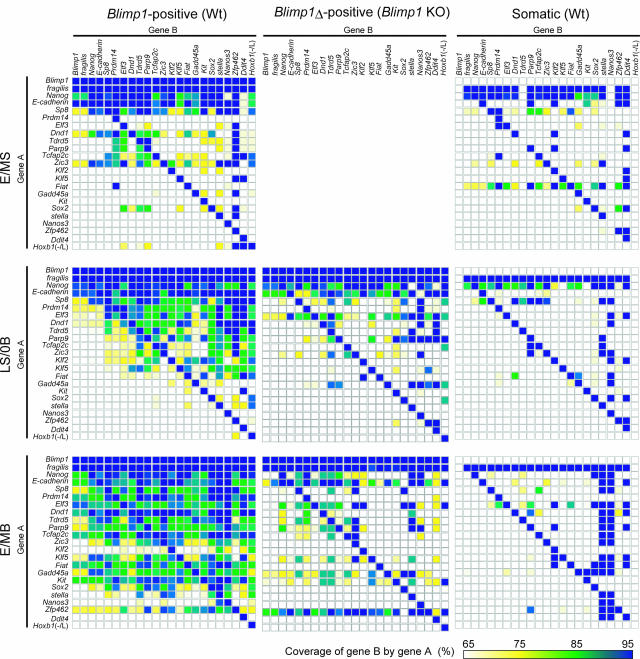
With the extensive Q-PCR analysis described above, we noted that even highly specific specification genes that exhibited both level-to-frequency and level-to-level correlations with Blimp1 showed expression in somatic neighbors and Blimp1 mutant cells at certain levels and frequencies (Supplemental Fig. S6). We therefore wanted to determine whether there might be any factors other than Blimp1 that can coordinate the expression of many specification genes. For this purpose, we calculated the expression coverage of gene B by gene A, defined as [(the number of single cells expressing both genes)/(the number of single cells expressing gene A)] in all possible pairs of the selected specification genes in the Blimp1-positive wild-type cells (E/MS–E/MB stages), Blimp1Δ-positive mutant cells (LS/0B and E/MB stages), and somatic neighbors (E/MS–E/MB stages). This analysis is based on the idea that if such factors exist, some of the specification genes should be coordinately activated in the overlapping Blimp1 mutant single cells.
As shown in Figure 6, at the E/MS stage, the expression of fragilis, Nanog, E-cadherin, and Sp8 all exhibited high coverage of other genes in the wild-type Blimp1-positive cells. This reflects the fact that these four genes were expressed in a majority of the Blimp1-positive cells at this stage. Although the other genes were expressed at various frequencies, cells expressing these genes did not significantly overlap in either the Blimp1-positive or Blimp1-negative cells, indicating most likely that the expression of specification genes was just starting to be initiated around this stage. At the LS/0B stage, however, in addition to the above four genes, Prdm14, Elf3, Dnd1, and Tdrd5 exhibited good coverage ratios over many specification genes in the Blimp1-positive cells. On the other hand, genes such as Sox2, stella, Zfp462, and Nanos3 showed lower expression frequencies, and their expression did not considerably overlap. Genes such as Parp9, Tcfap2c, Zic3, Klf2, and Klf5 exhibited moderate coverage ratios: Cells expressing these genes were covered by the cells expressing fragilis, Nanog, E-cadherin, Sp8, Prdm14, Elf3, Dnd1, and Tdrd5, and covered the cells expressing Sox2, stella, Zfp462, and Nanos3. At the E/MB stage, the expression of many specification genes overlapped considerably. However, the cells expressing stella and Nanos3, and those that were negative or low for Hoxb1 [Hoxb1 (−/low)], still comprised a relatively minor portion of the cells expressing the specification genes. These findings suggest that a hierarchically overlapping, ordered activation of the specification genes may be involved in germ cell specification; as in the activation of stella and Nanos3, repression of the Hox genes denotes one of the final events for germ cell specification.
Figure 6.
Expression coverage analysis of the selected genes. The expression coverages of genes shown as Gene B by genes shown as Gene A in the wild-type Blimp1-positive cells (left), Blimp1 KO cells (middle), and Blimp1-negative somatic cells (right) at the E/MS (top), LS/0B (middle), and E/MB (bottom) stages are shown according to the color code at the bottom.
In contrast, in the Blimp1 mutant PGC-like cells, although the selected genes showed considerable degrees of expression frequencies, their coordinated activation was severely impaired. They did, however, exhibit better coordination than in somatic neighbors. Therefore, the coordinated and ordered activation of the specification genes occurs successfully only in the presence of Blimp1, suggesting that there would be no Blimp1-independent, specific factors that function to induce multiple specification genes with high specificity in the incipient germ cell lineage.
In summary, combined evidence indicates that Blimp1 is not a single initiator but a dominant—perhaps the most robust—coordinator for the establishment of the germ cell fate in mice.
Discussion
Using a highly representative single-cell microarray technology and Q-PCR analysis, we uncovered complex genome-wide expression dynamics that specify the germ cell fate from the epiblast in mice, both in wild-type (initial 48 h) and Blimp1-deficient (two critical stages) conditions. With the expression of key markers identified in our previous studies (Saitou et al. 2002; Yabuta et al. 2006), we preclassified wild-type Blimp1-positive cells from all the developmental stages. We then analyzed the genome-wide expression profiles associated with representative classes, establishing the developmental traits of these classes and a pathway for germ cell specification. For Blimp1-deficient cells, based on the knowledge from the wild-type analysis, we were able to identify those acquiring relatively proper PGC characteristics. Combined data thus revealed not only the dominant function of Blimp1 for repressing “somatic genes,” but also the presence of Blimp1-independent mechanisms, although relatively weak, for PGC specification, leading to a synthetic and comprehensive view of the specification of germ cell fate (Fig. 7). This is the first study to define the genome-wide dynamics for the specification of a key lineage in development at single-cell resolution.
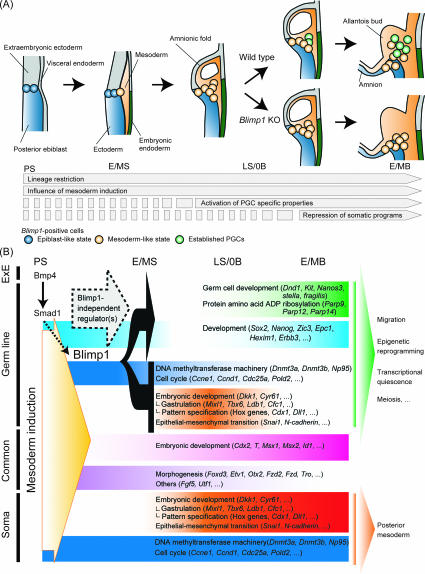
Figure 7.
A model of the global transcriptional dynamics for the specification of germ cell fate in mice. (A) Schematic representation of germ cell specification in mice. The most proximal posterior epiblast cells are determined as Blimp1-positive, lineage-restricted PGC precursors at the PS stage (blue circle). These cells then exhibit a mesoderm-like state under the influence of the mesoderm induction (orange circle). In the presence of Blimp1, the PGC precursors start to activate PGC-specific properties and then repress somatic programs, hence becoming established PGCs (green circle). In the absence of Blimp1, the mesoderm-like-state cells are propelled toward a somatic mesodermal fate as the embryo develops. (B) Summary of genes associated with germ cell specification. We identified 493 “specification” genes and 330 “somatic” genes, representatives of which are shown with the GO functional classifications. Genes that represent common developmental programs between germ and somatic cells are also shown. Blimp1 functions as a dominant repressor of somatic genes and as a positive regulator of the specification genes with high specificity. Blimp1-independent mechanisms are involved in the induction of approximately half of the specification genes. (ExE) Extraembryonic ectoderm.
We isolated single cells from embryos as quickly as possible for cDNA synthesis: Embryo isolation, dissection of an embryonic fragment bearing developing germline cells, dissociation of the fragment into single cells, and the picking up of single cells for cDNA synthesis were all completed in less than ∼1.5 h. We strictly evaluated the quality of all the amplifications by assessing the quantitative amplification of spike RNAs and some housekeeping genes, eliminating amplification-by-amplification variations (Kurimoto et al. 2006, 2007). We defined differential expression based on rigorous statistical tests that have been shown to identify expression differences between single-cell populations with high confidence (Kurimoto et al. 2006). Our extensive Q-PCR analyses of expression levels and frequencies validated essentially all the microarray expression data. On the other hand, we performed statistical analysis so stringently that some genes with genuine biologically relevant difference would not have been detected. Indeed, our present analysis detected with high accuracy the expression of all known genes associated with germ cell specification, underscoring the reliability of the single-cell microarray data.
Defined pathway and developmental time window for the specification of germ cell fate
The established PGCs have been defined as stella-positive and Hoxb1-negative cells (Saitou et al. 2002; Yabuta et al. 2006). Indeed, >80% of the Blimp1-positive cells we isolated from E8.25 hindgut endoderm were stella-positive, Sox2-positive, and Hoxb1-negative (Fig. 1). However, at earlier stages, we showed clearly that a majority of the Blimp1-positive cells were Hoxb1-positive: At the LS/0B stage, as many as 86% of the Blimp1-positive cells expressed Hoxb1. On the other hand, genetic lineage tracing experiments have shown that all the Blimp1-expressing cells at early stages (∼LS/0B stages) contribute almost invariably to stella-positive PGCs (Ohinata et al. 2005). Therefore, it appears that all the lineage-restricted PGC precursors express Hoxb1 initially, but subsequently repress it in order to be established as PGCs. This is considered to be a reflection of the signaling that PGC precursors unavoidably receive at the posterior proximal epiblast, from which they originate. This notion is supported by two lines of evidence: First, the Group V somatic genes that cosegregate with Hoxb1 and show transient up-regulation in Blimp1-positive cells at the LS/0B stage are highly enriched with genes associated with mesoderm induction, such as those for “embryonic development,” “gastrulation,” “pattern specification,” and “morphogenesis” (Fig. 3). Moreover, genes commonly up-regulated from the epiblast in the established PGCs and their somatic neighbors are also enriched with those for “embryonic development” and “morphogenesis” (Fig. 3). Second, in the Blimp1 mutants, all the Blimp1Δ-positive cells that are destined for a PGC fate become entirely sensitive to mesoderm-inducing signals and express Hoxb1 and almost the entire somatic program at the E/MB stage (Fig. 4). Initial reduction of Sox2 in the Blimp1-positive cells (Fig. 1B–D, panels iii, iv; Wood and Episkopou 1999) would also be an effect of mesoderm-inducing activities. The evidence presented by this study thus is a clear demonstration of the induction of PGC fate from the epiblast and its segregation from the somatic mesoderm.
On the other hand, our data showed that a majority of E/MB- and EHF-stage Blimp1-positive and Hoxb1-positive cells exhibit expression profiles similar to their somatic neighbors (Fig. 2). However, the clustering of all the isolated Blimp1-positive cells by the Q-PCR data on the selected genes showed that the E/MB-stage Blimp1-positive, Hoxb1-positive cells with higher Blimp1 levels were, indeed, grouped with established PGCs (Supplemental Fig. S10). Consistent with this, many specification genes exhibited positive level-to-frequency and level-to-level correlations with Blimp1 transcript levels (Fig. 5; Supplemental Figs. S7–S9); furthermore, the expression coverage analysis indicated that repression of Hoxb1 is one of the final events in germ cell specification (Fig. 6). Collectively, these findings suggest that after the E/MB stage, although cells expressing Hoxb1 with low Blimp1 levels may no longer contribute to the germ cell lineage, cells with Hoxb1 with high Blimp1 levels will still do so. Indeed, Blimp1 continues to be expressed in the most proximal posterior epiblast cells and some mesodermal cells that appear to originate from these epiblast cells at least until E8.25 (data not shown). The competence of a given cell to express high levels of Blimp1 would depend on both intrinsic factors (e.g., expression of signaling components for Blimp1 induction) and extrinsic factors (e.g., distance from signal centers ≈ signal concentration) that might gradually change along with dynamic morphogenetic processes during gastrulation. These factors would thus restrict the time window for germ cell induction in mice. It remains to be determined until what stage all the Blimp1-positive epiblast cells contribute exclusively to the germ cell lineage and how long the Blimp1-positive epiblast cells can produce PGCs.
Defined ‘building blocks’ of the established PGCs and the role of Blimp1 for PGC specification
Our study has defined, for the first time, the genome-wide expression properties of the established PGCs in comparison to the proximal epiblast and somatic neighbors (Fig. 3; Supplemental Fig. S3), and the full gene set that is under the influence of Blimp1 activity (Fig. 4; Supplemental Fig. S5). Since the identification of founder PGCs as alkaline phosphatase-positive cells in the extraembryonic mesoderm (Ginsburg et al. 1990), only a few genes expressed specifically in these cells have been identified (Hayashi et al. 2007). The specification and somatic genes, both with three major expression dynamics identified in this study, will serve as a novel foundation to explore the mechanism of germ cell specification at the molecular and genome-wide levels.
One of the events that has been proposed to be associated with, and be critical for, PGC specification is the repression of a “somatic program” represented by the Hox gene expression (Saitou et al. 2002). Remarkably, the genome-wide analysis we presented here revealed that the expression dynamics group represented by Hoxb1 (Group V), which is highly enriched with genes associated with mesodermal development, is a relatively minor group in the somatic genes. Instead, those genes that exhibit consistent down-regulation from the epiblast (Group IV) constitute the majority of the somatic genes (Fig. 3). Therefore, it is critical to note that PGCs indeed repress somatic mesodermal genes, but it is only a part of their repression program. We found that the Group IV genes included those for “cell cycle” involved in the promotion of the S phase and a number of critical epigenetic modifiers including machineries both for de novo and maintenance DNA methylation. These findings are consistent with the previous notion that PGCs lengthen their cell cycle time upon their lineage restriction from the epiblast (Tam and Snow 1981; Lawson and Hage 1994), and undergo genome-wide epigenetic reprogramming in their prolonged G2 phase (Seki et al. 2007). The potential extinction of all the DNA methyltransferase activity from PGCs argues for the involvement of a passive mechanism for the genome-wide DNA demethylation in PGCs (Seki et al. 2005). This will require further careful investigation.
Regarding the specification genes, we showed that Blimp1 is not required for the activation of approximately half of them (Fig. 4). However, we provided multiple lines of evidence that Blimp1 positively influences the proper up-regulation of many specification genes with high specificity, and hence regulates PGC properties in a dose-dependent manner (Figs. 5, 6; Supplemental Figs. S7–S9). This is consistent with the fact that Blimp1+/− embryos bear reduced numbers of PGCs (Ohinata et al. 2005; Vincent et al. 2005; Robertson et al. 2007) and findings that Blimp1 expression levels control multiple cell differentiation pathways (Kallies et al. 2004; Robertson et al. 2007).
In contrast, Blimp1 is essential for the repression of almost entire somatic genes of all expression dynamics groups (Fig. 4). How could Blimp1 exert these functions? Blimp1 has been known to function as a potent transcriptional repressor by binding to a consensus A/C-AG-T/C-GAAAG-T/C-G/T sequence (Kuo and Calame 2004) and recruiting several factors including Groucho, HDAC2 (Ren et al. 1999; Yu et al. 2000), G9a (Gyory et al. 2004), and Prmt5 (Ancelin et al. 2006), presumably in a context-dependent manner. Therefore, for the repression of the somatic genes, Blimp1 might function similarly, and those that were affected as early as the LS/0B stage (∼12 h after the onset of Blimp1 expression) might be its direct targets. From the studies of Blimp1 function in plasma cell and, more recently, epidermal differentiation, several direct targets of Blimp1 have been identified, including c-Myc, CIITA, Pax5, Spi-B, Id3 (Lin et al. 1997, 2002; Piskurich et al. 2000; Shaffer et al. 2002; Kuo and Calame 2004; Sciammas and Davis 2004; Horsley et al. 2006), Nfat5, Fos, Dusp16, and Blimp1 itself (Magnusdottir et al. 2007). Among these, c-Myc is repressed in PGCs and may thus be a direct target of Blimp1 in germ cell specification. However, we found that there is only a small overlap (∼5%) between the genes affected by the loss of Blimp1 in germ cell specification and in plasma cell (Supplemental Fig. S11; Shaffer et al. 2002; Sciammas and Davis 2004) and epidermal differentiation (Magnusdottir et al. 2007). We analyzed the distribution of a consensus sequence for Blimp1 binding throughout the genome and found that it is distributed equally in the upstream region of the specification (both with or without correlation to Blimp1), somatic, and unrelated genes (data not shown). Therefore, for a more integrated analysis of Blimp1 function, it is critical to perform a careful system-by-system analysis. Since the number of PGCs in vivo, especially at early stages, is too small for any biochemical analysis, it will be a critical challenge to establish an appropriate in vitro system to recapitulate germ cell specification, which may clarify the mechanism of Blimp1 function both in the activation of specification genes and the repression of somatic genes.
Blimp1-independent mechanism for germ cell specification and the potential function of the specification genes
There are at least two critical questions that require clarification: First, is there any factor that works independently from Blimp1 and has a comparably profound role in germ cell specification? Second, are there any specific factors working upstream of Blimp1?
In regard to the first question, the presence of Blimp1-independent mechanisms that confer PGC properties became evident from the microarray study of Blimp1-deficient cells (Fig. 4C,D). However, the specification genes that were properly acquired in the absence of Blimp1 were mostly those with low specificity (Fig. 4C,D). Moreover, the expression coverage analysis of the multiple specification genes with high specificity both in the wild-type and Blimp1 mutant cells (Fig. 6) indicated that Blimp1 is an absolute requirement for the coordinated activation of these specification genes. Therefore, Blimp1 is not a single initiator but a major—presumably the most robust—coordinator for the specification of germ cell fate. To address the second question, using the array data, we screened for genes that could potentially work upstream of Blimp1. However, we failed to identify such candidates, at least among the genes on the Affymetrix slide (Supplemental Fig. S12). Blimp1 expression is absent in the Bmp4 and Smad1 mutants (Ohinata et al. 2005; Saitou et al. 2005; Y. Ohinata and M. Saitou, unpubl.). A small number of the initial Blimp1-positive cells undergo transient up-regulation of Hoxb1 (and the Group V genes) and down-regulation of Sox2 (and the Group II genes), as do the somatic neighbors (Figs. 1, 3), indicating that initial Blimp1-positive cells are indeed nascent PGC precursors. The present microarray data, showing the tight clustering of the individual PS-stage PrE cells (three Blimp1-positive and five Blimp1-negative) (Fig. 2), demonstrated that Blimp1 expression is not yet a discriminating property among the Oct4-positive, T-positive posterior PrE cells. This provides further evidence that germ cell fate is induced in the PrE cells from around E6.25 onward, and that the top of the hierarchy of specific transcriptional regulators is most likely Blimp1.
Based on this evidence, we propose that the Bmp4–Smad signaling directly or indirectly activates, in parallel, Blimp1 and a number of the specification genes, especially those that show higher coverage in the Blimp1-positive cells at earlier stages (and those with low specificity). Currently, it is unknown why only a restricted number of proximal epiblast cells activate Blimp1 and are allocated as PGC precursors, a matter that deserves further investigation. Blimp1 is essential for the coordinated, efficient induction and up-regulation of the high-specificity specification genes and is the dominant factor for the repression of virtually all somatic genes (Fig. 7). The microarray and extensive Q-PCR analysis have identified genes including Prdm14, Elf3, Dnd1, Tdrd5, Parp9, Tcfap2c, and Zic3 as specification genes exhibiting early onset of expression and high coverage in the Blimp1-positive cells. Genes such as Klf2, Klf5, Fiat, Gadd45a, Kit, Sox2, stella, Nanos3, Zfp462, and Ddit4 showed relatively late onset of expression and low coverage (Fig. 6). It has so far been known that at least three signals—Bmp4, Bmp8b, and Bmp2—apparently operating at different times, are required for the full activation of PGC fate (Lawson et al. 1999; Ying et al. 2000; Ying and Zhao 2001). Therefore, a combination of the responses to the signal inputs and transcriptional regulator cascades would be critical for the generation of the full germ cell program. In this sense, it will be critical to evaluate how the expression of the specification genes would be affected in the absence of the relevant signaling molecules.
Among the specification genes, Dnd1, Kit, and Nanos3 are known to be critical for early germ cell development, although the causes and the mechanisms of early PGC death in these mutants have been elusive (Mintz and Russell 1957; Buehr et al. 1993; Tsuda et al. 2003; Youngren et al. 2005). The comprehensive expression profiles we presented, as well as future analysis of the aberrant transcriptomes of these mutants and those of the other key specification genes, should provide an essential foundation upon which to construct a molecular network that guarantees the establishment and propagation of the germ cell lineage in mice.
Materials and methods
Embryo isolation, single-cell cDNA preparation, and Q-PCR analysis
All the animals were treated with appropriate care according to the RIKEN ethics guidelines. Noon of the day when the vaginal plugs of the mated females were identified was scored as E0.5. Isolation of embryos (C57BL/6 background), dissection of embryonic fragments, and single-cell cDNA preparations were as described (Kurimoto et al. 2006; Yabuta et al. 2006). In brief, fragments of wild-type and Blimp1 mutant embryos (the Blimp1-knockout strain of C57BL/6 background is a kind gift from A. Tarakhovsky and D. O’Carroll [Ohinata et al. 2005]) containing developing PGCs or PGC-like cells were cut out and dissociated with 0.05% trypsin and 0.5 mM EDTA into single cells. These cells were randomly picked up with mouth pipettes into tubes containing cell lysis buffer and incubated for 90 sec at 70°C. First-strand cDNAs were synthesized by RT using oligo (dT)-tagged primer. Remnant primer after RT was degraded by exonuclease. The cDNAs were then tailed with poly(dA) by terminal deoxynucleotidyl transferase. The second strand of the poly(dA)-tailed cDNAs were synthesized with another oligo(dT)-tagged primer and subjected to 20-cycle PCR using the same two primers. The PCR products were subjected to another nine-cycle PCR to tail with T7 promoter, gel purification, and final one-cycle PCR.
Q-PCR analysis was performed as described (Kurimoto et al. 2006; Yabuta et al. 2006). Transcript copy numbers were estimated using spiked RNAs (1000, 100, 20, five copies) as a standard, and transformed into a log10 scale. Expression levels lower than 10 copies per cell were considered as low expression. The level-to-level correlation coefficient (R) and linear regression coefficients were calculated using Microsoft Excel, and the P-value of R was calculated against the null hypothesis R = 0. Level-to-frequency logistic regression was performed using the program glm in R (R Development Core Team 2005), and the P-value is calculated against the null hypothesis that the regression coefficient equals zero.
Whole-mount immunofluorescence analysis
Blimp1-mVenus mice are essentially identical to Blimp1-mEGFP mice (Ohinata et al. 2005) except that EGFP is replaced with Venus, a brighter mutant of YFP (Nagai et al. 2002). A more detailed characterization of Blimp1-mVenus mice will be described elsewhere. For whole-mount immunofluorescence analysis, isolated embryos were fixed in 4% paraformaldehyde in PBS for 2 h at 4°C, washed three times with PBS–0.2% Triton X-100 (PBS-T), and blocked with PBS-T with 1% normal goat serum (Vector Laboratories) overnight. Embryos were then incubated with primary antibodies (anti-Oct4, rabbit polyclonal, Santa Cruz Biotechnology; anti-Sox2, rabbit polyclonal, a kind gift from S. Yamanaka [Maruyama et al. 2005]; anti-Kit, rat monoclonal, a kind gift from S. Nishikawa [Nishikawa et al. 1991]; anti-Sox3 [Abcam]; anti-AP2γ [Santa Cruz Biotechnology]; and anti-GFP, rat monoclonal, Nakarai) in blocking solution for 96 h, washed eight times with PBS-T, incubated with appropriate secondary antibodies (Invitrogen) and DAPI for 48 h in blocking solution, washed eight times with PBS-T, and mounted in Vectasheild (Vector Laboratories) for observation by confocal microscopy.
Microarray data analysis
Microarray hybridization to an Affymetrix GeneChip Mouse Genome 430.2 Array was performed according to the manufacturer’s instructions. The microarray data were processed using the dChip software, with log2 transformation and zero truncation as described (Kurimoto et al. 2006). For ANOVA, the 134 single-cell cDNAs were prospectively classified into 13 groups based on representative gene expressions detected in microarrays as detailed in Supplemental Table S3. ANOVA was performed for probes with absolute call “Present” at least 40% in at least one group using the TMEV4 software (Saeed et al. 2003), and 5694 genes were found significant (false-positive rate <0.05 for 1000 permutations). The gene expression levels were averaged, essentially based on the clustering analysis (Fig. 2), as detailed in Supplemental Table S4. Based on our previous study (Kurimoto et al. 2006), we defined genes as differentially expressed between two cell types using three criteria: (1) the log-averaged expression level (absence of expression is assumed as log2 expression level of zero) is greater than five copies of transcript per cell in either cell type; (2) the expression level difference is >3.5-fold; and (3) at least one of the three statistical tests are satisfied—(i) t-test, (ii) t-test for cells calling Present only, or (iii) χ2 test for independence between cell types and absolute calls (P < 0.05). The complete list of the specification and somatic genes is shown in Supplemental Table S1. Unsupervised hierarchical clustering and PCA were performed using the TMEV4 software and R. Functional classification of genes was performed using software Expression Analysis Systematic Explorer (EASE) (Hosack et al. 2003), and significant enrichment was defined based on an EASE score <0.05.
The accession number for the microarray data presented in this study (Gene Expression Omnibus database) is GSE11128.
Acknowledgments
We dedicate this work to Anne McLaren, who has made enormous contributions to mammalian germ cell biology. We are also grateful to M. Azim Surani for his encouragement throughout this study. We thank the members of the Functional Genomics Subunit, Center for Developmental Biology, RIKEN Kobe Institute, especially Junko Nishio for her excellent help in the microarray experiments, S. Nishikawa for the anti-Kit antibody (ACK2), S. Yamanaka for the anti-Sox2 antibody, A. Miyawaki for the Venus plasmid, and A. Tarakhovsky and D. O’Carroll for the Blimp1 knockout mice. K.K. and Y.O. are fellows in the Special Postdoctoral Researchers Program of RIKEN. This study was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a PRESTO project grant from the Japan Science and Technology Agency.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1649908.
References
- Ancelin K., Lange U.C., Hajkova P., Schneider R., Bannister A.J., Kouzarides T., Surani M.A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A., Eggan K., Skaletsky H., Akutsu H., Berry D.L., Yanagimachi R., Page D.C., Jaenisch R. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Buehr M., McLaren A., Bartley A., Darling S. Proliferation and migration of primordial germ cells in We/We mouse embryos. Dev. Dyn. 1993;198:182–189. doi: 10.1002/aja.1001980304. [DOI] [PubMed] [Google Scholar]
- Chang H., Matzuk M.M. Smad5 is required for mouse primordial germ cell development. Mech. Dev. 2001;104:61–67. doi: 10.1016/s0925-4773(01)00367-7. [DOI] [PubMed] [Google Scholar]
- Chiquoine A.D. The identification, origin and migration of the primordial germ cells in the mouse embryo. Anat. Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- Chu G.C., Dunn N.R., Anderson D.C., Oxburgh L., Robertson E.J. Differential requirements for Smad4 in TGFβ-dependent patterning of the early mouse embryo. Development. 2004;131:3501–3512. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- Ciruna B., Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour C.G., Akam M. Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Ginsburg M., Snow M.H., McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Gyory I., Wu J., Fejer G., Seto E., Wright K.L. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- Hacohen N., Kramer S., Sutherland D., Hiromi Y., Krasnow M.A. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Kobayashi T., Umino T., Goitsuka R., Matsui Y., Kitamura D. SMAD1 signaling is critical for initial commitment of germ cell lineage from mouse epiblast. Mech. Dev. 2002;118:99–109. doi: 10.1016/s0925-4773(02)00237-x. [DOI] [PubMed] [Google Scholar]
- Hayashi K., de Sousa Lopes S.M., Surani M.A. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- Horsley V., O’Carroll D., Tooze R., Ohinata Y., Saitou M., Obukhanych T., Nussenzweig M., Tarakhovsky A., Fuchs E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack D.A., Dennis Jr G., Sherman B.T., Lane H.C., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hasbold J., Tarlinton D.M., Dietrich W., Corcoran L.M., Hodgkin P.D., Nutt S.L. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 2004;200:967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.C., Calame K.L. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J. Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y., Ono Y., Uno K.D., Yamada R.G., Ueda H.R., Saitou M. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y., Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat. Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- Lawson K.A., Hage W.J. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found. Symp. 1994;182:68–84. doi: 10.1002/9780470514573.ch5. [DOI] [PubMed] [Google Scholar]
- Lawson K.A., Dunn N.R., Roelen B.A., Zeinstra L.M., Davis A.M., Wright C.V., Korving J.P., Hogan B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes & Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Dedhar S., Kalluri R., Thompson E.W.2006. . The epithelial–mesenchymal transition: New insights in signaling, development, and disease J. Cell Biol. 172973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wong K., Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- Lin K.I., Angelin-Duclos C., Kuo T.C., Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol. Cell. Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusdottir E., Kalachikov S., Mizukoshi K., Savitsky D., Ishida-Yamamoto A., Panteleyev A.A., Calame K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc. Natl. Acad. Sci. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama M., Ichisaka T., Nakagawa M., Yamanaka S. Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells. J. Biol. Chem. 2005;280:24371–24379. doi: 10.1074/jbc.M501423200. [DOI] [PubMed] [Google Scholar]
- Mintz B., Russell E.S. Gene-induced embryological modifications of primordial germ cells in the mouse. J. Exp. Zool. 1957;134:207–237. doi: 10.1002/jez.1401340202. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Kusakabe M., Yoshinaga K., Ogawa M., Hayashi S., Kunisada T., Era T., Sakakura T. In utero manipulation of coat color formation by a monoclonal anti-c-kit antibody: Two distinct waves of c-kit-dependency during melanocyte development. EMBO J. 1991;10:2111–2118. doi: 10.1002/j.1460-2075.1991.tb07744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y., Payer B., O’Carroll D., Ancelin K., Ono Y., Sano M., Barton S.C., Obukhanych T., Nussenzweig M., Tarakhovsky A., et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Peinado H., Olmeda D., Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Piskurich J.F., Lin K.I., Lin Y., Wang Y., Ting J.P., Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat. Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2005. [Google Scholar]
- Ren B., Chee K.J., Kim T.H., Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes & Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson E.J., Charatsi I., Joyner C.J., Koonce C.H., Morgan M., Islam A., Paterson C., Lejsek E., Arnold S.J., Kallies A., et al. Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development. 2007;134:4335–4345. doi: 10.1242/dev.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Saitou M., Payer B., O’Carroll D., Ohinata Y., Surani M.A. Blimp1 and the emergence of the germ line during development in the mouse. Cell Cycle. 2005;4:1736–1740. doi: 10.4161/cc.4.12.2209. [DOI] [PubMed] [Google Scholar]
- Sato M., Kimura T., Kurokawa K., Fujita Y., Abe K., Masuhara M., Yasunaga T., Ryo A., Yamamoto M., Nakano T. Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech. Dev. 2002;113:91–94. doi: 10.1016/s0925-4773(02)00002-3. [DOI] [PubMed] [Google Scholar]
- Sciammas R., Davis M.M. Modular nature of Blimp-1 in the regulation of gene expression during B cell maturation. J. Immunol. 2004;172:5427–5440. doi: 10.4049/jimmunol.172.9.5427. [DOI] [PubMed] [Google Scholar]
- Seki Y., Hayashi K., Itoh K., Mizugaki M., Saitou M., Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Seki Y., Yamaji M., Yabuta Y., Sano M., Shigeta M., Matsui Y., Saga Y., Tachibana M., Shinkai Y., Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Braun R.E. Pathway to totipotency: Lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K., et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tam P.P., Snow M.H. Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J. Embryol. Exp. Morphol. 1981;64:133–147. [PubMed] [Google Scholar]
- Tanaka S.S., Matsui Y. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech. Dev. 2002;119 (Suppl. 1):S261–S267. doi: 10.1016/s0925-4773(03)00126-6. [DOI] [PubMed] [Google Scholar]
- Tanaka S.S., Yamaguchi Y.L., Tsoi B., Lickert H., Tam P.P. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev. Cell. 2005;9:745–756. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Tremblay K.D., Dunn N.R., Robertson E.J. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Sasaoka Y., Kiso M., Abe K., Haraguchi S., Kobayashi S., Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Turner C.A., Mack D.H., Davis M.M. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Vincent S.D., Dunn N.R., Sciammas R., Shapiro-Shalef M., Davis M.M., Calame K., Bikoff E.K., Robertson E.J. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Wood H.B., Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Yabuta Y., Kurimoto K., Ohinata Y., Seki Y., Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol. Reprod. 2006;75:705–716. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- Yeung K.Y., Ruzzo W.L. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- Ying Y., Zhao G.Q. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev. Biol. 2001;232:484–492. doi: 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- Ying Y., Liu X.M., Marble A., Lawson K.A., Zhao G.Q. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol. Endocrinol. 2000;14:1053– 1063. doi: 10.1210/mend.14.7.0479. [DOI] [PubMed] [Google Scholar]
- Youngren K.K., Coveney D., Peng X., Bhattacharya C., Schmidt L.S., Nickerson M.L., Lamb B.T., Deng J.M., Behringer R.R., Capel B., et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Angelin-Duclos C., Greenwood J., Liao J., Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 2000;20:2592–2603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]