Abstract
The mating-type determination circuit in Saccharomyces yeast serves as a classic paradigm for the genetic control of cell type in all eukaryotes. Using comparative genetics, we discovered a central and conserved, yet previously undetected, component of this genetic circuit: active repression of α-specific genes in a cells. Upon inactivation of the SUM1 gene in Saccharomyces bayanus, a close relative of Saccharomyces cerevisiae, a cells acquired mating characteristics of α cells and displayed autocrine activation of their mating response pathway. Sum1 protein bound to the promoters of α-specific genes, repressing their transcription. In contrast to the standard model, α1 was important but not required for α-specific gene activation and mating of α cells in the absence of Sum1. Neither Sum1 protein expression, nor its association with target promoters was mating-type-regulated. Thus, the α1/Mcm1 coactivators did not overcome repression by occluding Sum1 binding to DNA. Surprisingly, the mating-type regulatory function of Sum1 was conserved in S. cerevisiae. We suggest that a comprehensive understanding of some genetic pathways may be best attained through the expanded phenotypic space provided by study of those pathways in multiple related organisms.
Keywords: Sum1, natural variation, cell-type determination, gene regulation
Owing to three decades of intensive genetic, molecular, and biochemical analysis, the genetic circuit responsible for determining mating type in Saccharomyces cerevisiae stands as the most thoroughly characterized cell-type regulatory pathway in eukaryotes. The two haploid yeast mating types, a and α, are distinguished by their ability to mate with each other to form the third cell type, the a/α diploid. Two sets of genes, the a-specific genes and the α-specific genes, are differentially transcribed in a cells and α cells, respectively, to determine these distinct mating phenotypes.
The mating-type-specific patterns of gene expression are dictated by the allele present at the mating-type (MAT) locus. The MATα allele encodes two transcription factors: α1, which activates α-specific genes, and α2, which represses a-specific genes. The MATa allele encodes only the a1 protein, which forms a heterodimer with α2 in MATa/α diploids to repress a third set of genes, the haploid-specific genes. In the standard model, known as the α1–α2 hypothesis (Strathern et al. 1981), expression of a-specific genes in a cells is a default state, resulting merely from the absence of the α1 and α2 proteins. Two transcription factors common to both a and α cells, Ste12 and Mcm1, are also necessary for proper expression of mating-type genes. Ste12 works at two regulatory levels in both mating types to activate transcription: It is required for basal transcription of a-specific and α-specific genes in the absence of the mating pheromones, a-factor and α-factor, and for their further induction in response to pheromones (Fields and Herskowitz 1985; Kirkman-Correia et al. 1993). Mcm1, a MADS-box transcription factor similar to mammalian Serum Response Factor, is required for the activation of α-specific genes and for both the activation and the repression of a-specific genes (Jarvis et al. 1989; Elble and Tye 1991; Hwang-Shum et al. 1991; Bruhn and Sprague 1994). In α cells, Mcm1 homodimers interact directly with α1 to activate α-specific genes; similarly, Mcm1 homodimers interact with α2 to repress a-specific genes (Smith and Johnson 1992). Ste12 and Mcm1 themselves interact to activate transcription, forming complexes with α1 at α-specific gene promoters, or acting on their own at a-specific gene promoters in a cells (Fig. 1A; Sengupta and Cochran 1990; Yuan et al. 1993; Bruhn and Sprague 1994).
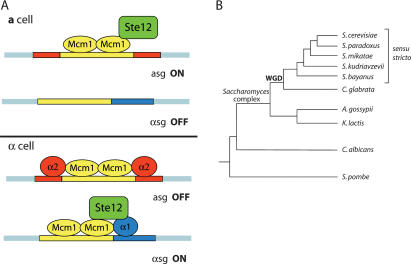
Figure 1.
(A) The standard model for the transcriptional control of haploid mating-type genes in S. cerevisiae. The operator elements, contained within a-specific gene (asg) and α-specific gene (αsg) promoters, are schematized as colored boxes, which correspond to binding sites for α1 (blue), α2 (red), and Mcm1 (yellow). (B) Simplified phylogenetic tree, based on concatenated sequences of 153 genes present in all species shown, depicts the sensu stricto yeasts and some related yeast species. “WGD” denotes the whole-genome duplication that occurred along the branch leading to C. glabrata and the Saccharomyces species. Modified from Fitzpatrick et al. (2006).
Together, Mcm1 and Ste12 can activate a-specific genes, yet they require α1 to activate α-specific genes. This additional coactivator requirement has been explained solely by differential activator–DNA affinities encoded in distinct classes of Mcm1-binding sites. Whereas the highly conserved, palindromic sequence found in a-specific gene promoters, known as the P element, binds Mcm1 tightly, the related sequence known as the P′Q element, which confers α-cell specificity, binds Mcm1 with reduced affinity. Thus, the standard model holds that cooperative DNA binding by α1 and Mcm1 is necessary for α-specific gene expression. Despite extensive mutational and in vitro binding analyses of P′Q elements, no evidence for repression of α-specific gene expression has emerged (Bender and Sprague 1987; Flessel et al. 1989; Ganter et al. 1993; Hagen et al. 1993).
Recently, the full force of modern genomics analysis has been brought to bear on the yeast mating-type determination circuit, including microarray analyses of mating-type-specific gene expression, genome-wide location assays of mating-type regulatory proteins, and computational analysis of DNA regulatory sites (Zeitlinger et al. 2003; Galgoczy et al. 2004). These studies revealed additional mating-type-regulated genes and a novel mating-type-regulated dimension to osmolarity tolerance, but no substantial alterations were made to the mating-type regulatory circuit itself. However, these efforts, and similar studies in other organisms, are potentially limited by the amount of phenotypic information accessible within a single species.
Interspecies genetic analysis may allow deeper probing into gene circuits because of mutations that strengthen or weaken known genetic interactions, or that bring about novel interactions. Other comparative studies have attempted to understand how genetic variation between species creates phenotypic variation. For example, the varying shapes and sizes of beaks in Galapagos finches can be explained by changes in BMP4 expression during development (Abzhanov et al. 2004). Here, we attempt to harness the natural variation present in related species to identify components of gene regulatory pathways that have eluded phenotypic detection in single-species analyses. Indeed, the distinction between these goals can sometimes blur: This study was inspired by differences in phenotype caused by the same mutation in different species, yet led to discovery of a common regulatory component of both species.
The power of comparative genetic analysis is greatly expanded in taxa in which the genomes of multiple related species have been sequenced (Stark et al. 2007). The sensu stricto yeasts comprise several closely related Saccharomyces species—five of which have published genome sequences—that afford an excellent opportunity for studying gene regulatory pathways and their evolution (Cliften et al. 2003; Kellis et al. 2003). S. bayanus is the most evolutionarily distant of the sensu stricto yeasts from S. cerevisiae (Fig. 1B). Thus, comparisons between these two species are useful for making inferences about the sensu stricto ancestral state. The nucleotide substitution level between S. bayanus and S. cerevisiae is approximately equivalent to that between mouse and human (Kellis et al. 2003), offering a calibration for how that amount of variation affects function.
Analysis of the Saccharomyces bayanus genome sequence suggested that transcriptional repression by the Silent Information Regulator (Sir) proteins had changed in that lineage (Kellis et al. 2003; J. Babiarz and L. Teytelman, unpubl.). While examining transcriptional silencing in S. bayanus, we discovered an unexpected species-specific and mating-type-specific colony-wrinkling phenotype in a cells lacking the SUM1 gene. SUM1 encodes a transcriptional repressor of several dozen genes, many of which are expressed only during meiosis. Similar to silencing by the Sir proteins, Sum1 uses a NAD+-dependent histone deacetylase, called Hst1, to effect transcriptional repression. Sum1 recruits Hst1 via a protein called Rfm1, with these three proteins constituting a transcriptional repressor complex (McCord et al. 2003). A gain-of-function allele, SUM1-1, can restore silencing to the HMRa mating type locus in sir mutants (Klar et al. 1985; Laurenson and Rine 1991; Rusche and Rine 2001). The fortuitous phenotype of S. bayanus sum1Δ mutants suggested either that some dimension of mating type control had changed in S. bayanus, or that a conserved regulatory mechanism had been missed in studies of in S. cerevisiae. The results presented here revealed the existence of a conserved repression mechanism operating on α-specific genes in a cells of both species, uncovering the first new central component of the mating type circuit in 20 years.
Results
S. bayanus MATa sum1Δ had mating-type-specific and species-specific phenotypes
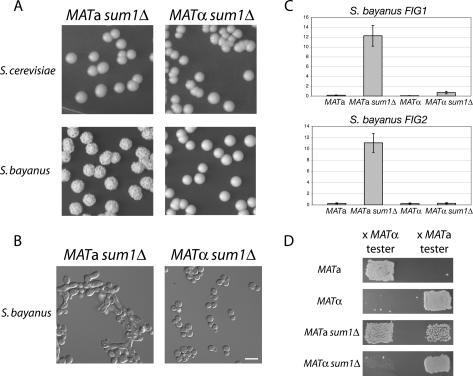
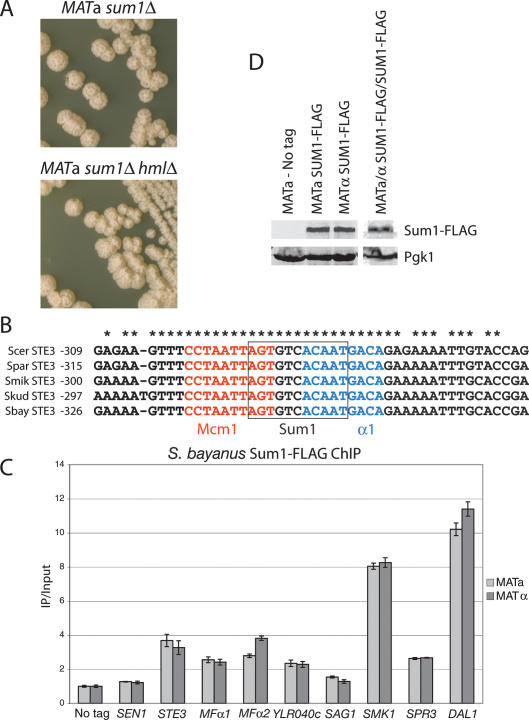
Upon targeted inactivation of the SUM1 gene in S. bayanus, we observed a dramatic mating-type-specific change in colony morphology. MATa sum1Δ colonies were distinctly wrinkled, whereas MATα sum1Δ colonies were smooth and identical to wild type (Fig. 2A; see Fig. 4B, below, for wild-type comparison). Microscopic inspection of MATa sum1Δ cells revealed that some cells had elongated cell morphology reminiscent of the shmoon (plural of shmoo) formed in response to mating pheromone (Fig. 2B). The mutant cells also formed lightly flocculent clumps in liquid culture (indicated by their faster sedimentation than wild type), suggesting that up-regulation of adhesion molecules in sum1Δ mutants altered colony morphology (Supplemental Fig. S1). These phenotypes were especially intriguing given that S. cerevisiae sum1Δ mutants of both mating types have no discernable cell or colony phenotypes (Fig. 2A) and their mating behavior is identical to wild type (Klar et al. 1985; Laurenson and Rine 1991). Given the shmoon-like cells, we asked whether the mating response in S. bayanus MATa sum1Δ cells was activated even in the absence of a mating partner. Indeed, the pheromone-induced genes FIG1 and FIG2 were highly up-regulated in MATa sum1Δ cells relative to wild type (Fig. 2C).
Figure 2.
Species-specific and MATa-specific phenotypes in sum1Δ mutants. (A, bottom panels) S. bayanus MATa sum1Δ and MATα sum1Δ colonies grown for 5 d on YPD (for wild-type comparison see Fig. 4B). (Top panels) S. cerevisiae sum1Δ colonies (W303 background) of both mating types grown for 5 d on YPD. (B) DIC microscopy of S. bayanus sum1Δ cells from colonies grown on YPD. Bar, 10 μm. (C) Quantitative RT–PCR analysis of the S. bayanus mating pheromone-induced genes FIG1 and FIG2 in wild-type and sum1Δ cells of both mating types. (D) Patch mating assays of S. bayanus wild-type and sum1Δ cells of both mating types. Growth of a patch is approximately proportional to the mating ability of the strain indicated to the left of the picture. Note that a subset of MATa sum1Δ cells mated with a cells, while most cells mated with α cells.
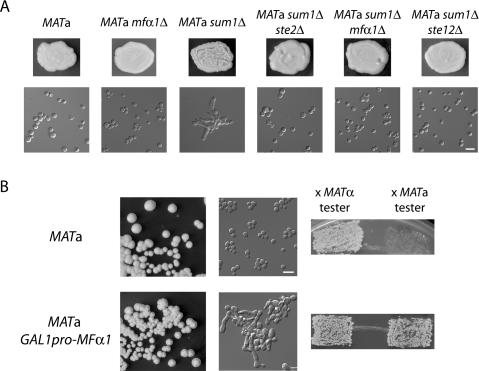
Figure 4.
Sum1 prevented autostimulation of a cells by α-factor. (A) Inactivation of the mating pheromone signaling pathway suppressed MATa sum1Δ phenotypes. Top panels depict patches of each indicated genotype growing on YPD. Bottom panels show DIC microscopy of cells taken from the above patches. (B, left panels) MATa wild-type and GAL1pro-MFα1 colonies grown for 5 d on YPG. Cells taken from the same colonies at left were photographed (middle panels) and tested in patch mating assays (right panels). On YPD, the GAL1pro-MFα1 strain behaved identically to the wild-type MATa strain (data not shown). Bars, 10 μm.
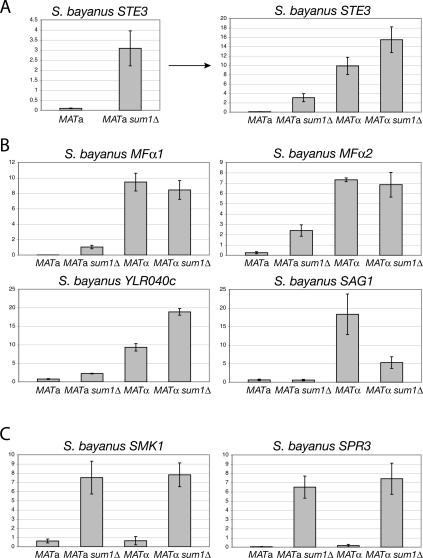
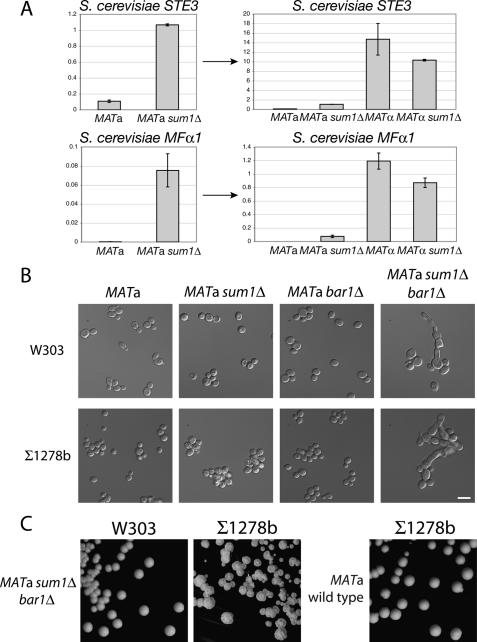
Mating tests on sum1Δ mutants of both mating types revealed that the activated mating response indeed reflected a change in mating behavior. A subset of MATa sum1Δ mutant cells mated with the MATa tester strain, with more cells retaining the ability to mate with the MATα strain (Fig. 2D). This bimating ability of the population suggested that the expression of mating genes normally expressed only in α cells was not properly regulated. This prediction was born out in quantitative RT–PCR analysis: The expression of MFα1 and MFα2, the two genes encoding α-factor, and STE3, the a-factor receptor gene, were increased 90-fold, 10-fold, and 30-fold, respectively, in MATa sum1Δ cells compared with wildtype (Fig. 3A,B). The expression of YLR040c, an α-specific gene of unknown function, also was increased by threefold. However, expression of SAG1, which encodes the α-specific agglutinin involved in cell–cell adhesion during mating, was unaffected in mutant a cells but decreased threefold in MATα sum1Δ mutants relative to wild-type α cells (Fig. 3B; discussed further below).
Figure 3.
α-Specific genes were up-regulated in S. bayanus MATa sum1Δ cells analysis compared with wild type. (A) Expression of the α-specific gene STE3 was assayed by quantitative RT–PCR. MATa wild type versus sum1Δ are shown on separate plot at left as an example of this two-way comparison for all α-specific genes. These same data are replotted at the right for four-way comparison with wild-type and mutant α cells. (B) Expression of the α-specific genes MFα1, MFα2, YLR040c, and SAG1 in wild-type and sum1Δ cells of both mating types. (C) Expression of the meiotic genes SMK1 and SPR3, which are targets of Sum1 repression in S. cerevisiae, in wild-type and sum1Δ cells of both mating types.
Two other features of α-specific gene expression were noteworthy. First, for STE3 and YLR040c, a small increase in gene expression was observed in MATα sum1Δ mutants compared with wild type (discussed further below). Second, although substantial α-specific gene expression occurred in MATa sum1Δ cells, the levels reached only 10%–30% of those in wild-type α cells. This difference in expression was potentially due to the absence of the α1 activator in a cells. Nonetheless, the expression of most α-specific genes in MATa sum1Δ cells did not fit the standard model for mating type determination developed in S. cerevisiae, and implied that Sum1 was a novel repressor of α-specific genes in S. bayanus.
In principle, the gene sets repressed by Sum1 could have completely changed—from meiotic genes to α-specific genes—between the two species. To ask whether Sum1 repression of meiotic genes was conserved in S. bayanus, the expression of two meiotic genes that are repressed by Sum1 in S. cerevisiae, SMK1 and SPR3, was analyzed in S. bayanus sum1Δ cells. The increase in expression observed for these genes in sum1Δ mutants of both mating types (Fig. 3C) indicated that Sum1 retained its role as a repressor of meiotic genes in S. bayanus.
Sum1 prevented autostimulation of a cells by α-factor
The gene expression data suggested that S. bayanus MATa sum1Δ mutants expressed and responded to α-factor, producing shmoon and the bimating phenotype. However, it remained possible that stimulation by a-factor through Ste3, the a-factor receptor, also contributed to the mutant phenotypes, as loss of Sum1 led to expression of STE3 in a cells. To distinguish between autostimulation of MATa sum1Δ cells by misexpression of α-factor or by misexpression of the a-factor receptor, we generated double mutants in the MATa sum1Δ background with genes required for pheromone signaling, inactivating different steps in the mating pathway. Strikingly, inactivation of STE2, which encodes the α-factor receptor and is expressed only in a cells, completely suppressed sum1Δ colony wrinkling, cell elongation, and flocculation phenotypes (Fig. 4A). MFα1 produces the majority of α-factor, whereas MFα2 produces a smaller fraction (Kurjan 1985). Consistent with these studies, the S. bayanus MATa sum1Δ mfα1Δ double mutants had very little flocculence and no apparent cell elongation (Fig. 4A), whereas MATa sum1Δ mfα2Δ double mutants had phenotypes indistinguishable from MATa sum1Δ single mutants (data not shown). Similar to STE2 deletion, inactivation of STE12 completely suppressed the MATa sum1Δ phenotypes (Fig. 4A). In contrast, deletion of STE3 enhanced wrinkling, bimating, and expression of FIG2 (data not shown). This result was consistent with expression of STE3 in a cells dampening the pheromone-signaling response (Roth et al. 2000; Rivers and Sprague 2003). Taken together, these data established that the cell and colony morphology defects in MATa sum1Δ mutants were caused by production of α-factor and subsequent stimulation by the same or neighboring cells within a growing colony.
To test whether the α-factor response was sufficient for the MATa sum1Δ phenotypes, the galactose-inducible GAL1 promoter was introduced into the genome of an otherwise wild-type S. bayanus MATa strain so as to drive expression of the MFα1 gene. In contrast to wild-type a cells, MATa GAL1pro-MFα1 cells grown on galactose-containing medium formed wrinkled colonies that contained cells with enhanced flocculence and a high percentage of shmoon (Fig. 4B). Additionally, when assayed on galactose medium, the inducible MFα1-expressing cells mated robustly with both a cells and α cells (Fig. 4B). Thus, expression of MFα1 in S. bayanus a cells was both necessary and sufficient to produce the flocculation, shmooing, and bimating phenotypes observed in the MATa sum1Δ mutant. Furthermore, the cell elongation was classic “shmooing” behavior caused by derepression of α-specific genes in a cells that, by definition, lack the α1 activator protein.
As Sum1 had been implicated in repression of the silent mating type loci, HML and HMR, it was possible that induction of α-specific genes and associated phenotypes in the absence of Sum1 were indirect effects of derepression of the silenced α1 gene at HMLα (Irlbacher et at. 2005). However, MATa sum1Δ hmlΔ double mutants possessed phenotypes identical to those of MATa sum1Δ single mutants (Fig. 5A). Therefore, the sum1Δ phenotypes did not depend on HMLα, ruling out this indirect-effect explanation.
Figure 5.
Sum1 repressed α-specific genes directly by binding to their promoters. (A) S. bayanus MATa sum1Δ and MATa sum1Δ hmlΔ colonies grown on YPD for 5 d. (B) Sum1-binding site predictions for the STE3 promoters from five sensu stricto yeasts. A predicted Sum1 site partially overlapped the known Mcm1 and α1 sites in the P′Q element of the STE3 promoter. The sequence encompassing all three binding sites was 100% conserved across all five species. (Scer) S. cerevisiae; (Spar) S. paradoxus; (Smik) S. mikatae; (Skud) S. kudriavzevii; (Sbay) S. bayanus. Numbers indicate the nucleotide position relative to the start codon. (C) ChIP of Sum1-Flag was performed, followed by quantitative PCR using primers corresponding to select promoter regions in the S. bayanus genome. IP amplification values were first normalized to input amplification values. These data were then normalized to a representative “No Tag” amplification value. (D) Sum1 protein levels were assayed by immunoblotting for Flag-tagged Sum1p (top row) and phosphoglucokinase (Pgk1p, bottom row), a loading control, using S. bayanus whole-cell extracts. Lanes from two separate blots are shown, with protein levels in haploid cells shown in lanes 1–3, and in diploid cells in lane 4. Lysate from the same haploid cells’ samples was run on both gels.
Sum1 bound to and repressed α-specific gene promoters
In the standard model of mating-type determination, no repressor of α-specific genes exists; merely the absence of the α1 activator protein prevents their expression. To explore the possibility that Sum1 directly repressed α-specific genes, we scanned their promoters (defined here as 1 kb of sequence 5′ of the start codon) for consensus Sum1-binding sites derived from detailed analyses of Sum1’s specific DNA sequence binding in vitro and its repression activity in vivo (Pierce et al. 2003). Using the consensus sequence AGYGWCACAAA, a near-perfect match in the STE3 promoter was detected that was 100% conserved across the sensu stricto yeast species (Fig. 5B). With a less stringent consensus, DSYGWCAYWDW, at least one match was found in the STE3, MFα1, and YLR040c promoters, each of which was highly conserved across the sensu stricto. Intriguingly, the sequence matches in the STE3 and MFα1 promoters occurred within the previously characterized P′Q elements, which in S. cerevisiae direct their α-specific expression through binding α1 and Mcm1 (for the YLR040c promoter, the match occurred 100 base pairs [bp] 5′ of the predicted P′Q box). None of the matches found in the SAG1 promoter were well conserved; however, this promoter was less conserved than the other α-specific gene promoters.
Sum1 was Flag-tagged at its C terminus in S. bayanus haploid strains and shown to provide Sum1 function in a sum1Δ mutant. Chromatin immunoprecipitation (ChIP) was used to test whether Sum1 protein bound the promoters of α-specific genes. Quantitative PCR analysis of Sum1-Flag precipitates revealed two- to fourfold enrichment of the STE3, MFα1, MFα2, and YLR040c promoters relative to precipitates from no-tag control strains (Fig. 5C). A negative control primer set amplified a region within the SEN1 ORF. As positive controls, we examined S. bayanus Sum1 binding at three loci to which the S. cerevisiae Sum1 protein binds (Lee et al. 2002). Strong enrichment (more than eightfold) of both the SMK1 and DAL1 promoters was observed (Fig. 5C), but only 2.5-fold enrichment of the SPR3 promoter was observed (comparable with the MFα1 level), despite this gene being a confirmed Sum1 target in S. cerevisiae (Xie et al. 1999). A simple model consistent with the ChIP and genetic analysis was that, at least in S. bayanus, Sum1 bound directly to α-specific gene promoters to repress their transcription.
Sum1 repression of α-specific genes was conserved in S. cerevisiae
The conserved Sum1-binding site in the S. cerevisiae STE3 promoter suggested that similar repression might occur throughout the Saccharomyces clade, although Sum1 had not previously been implicated in control of mating type in S. cerevisiae. Data from genome-wide localization studies (Lee et al. 2002; Harbison et al. 2004; Robert et al. 2004) identified highly reproducible binding of both Sum1 and Hst1 at the STE3 promoter in S. cerevisiae a cells. Furthermore, microarray data indicated that the expression of STE3, YLR040c, and FIG1 increased in S. cerevisiae MATa sum1Δ cells compared with wild-type a cells (Pierce et al. 2003), although the biological impact of this increase was unknown. Thus, we tested whether Sum1 repressed α-specific genes in S. cerevisiae despite the absence of gross phenotypes in MATa sum1Δ mutants of this species. In fact, quantitative RT–PCR analysis of α-specific genes revealed an expression profile similar to that of S. bayanus. Expression of S. cerevisiae STE3 and MFα1 increased 10-fold and 50-fold, respectively, in MATa sum1Δ cells compared with wild type (Fig. 6A).
Figure 6.
Sum1-mediated repression of α-specific genes was conserved in S. cerevisiae. (A) Expression of STE3 and MFα1 in S. cerevisiae was assayed by quantitative RT–PCR. MATa wild-type versus sum1Δ cells are shown on separate plots at left to allow two-way comparison. These same data are replotted at right for four-way comparison with wild-type and mutant α cells. (B) DIC microscopy of S. cerevisiae MATa sum1Δ bar1Δ double-mutant cells plus wild-type and single mutant controls of W303 (top row) and Σ1278b (bottom row) backgrounds. Bar, 10 μm. (C) Σ1278b MATa sum1Δ bar1Δ colonies grown on YPD for 5 d (middle panel); compare with wild-type colonies (right panel). W303 sum1Δ bar1Δ colonies, which had only a subtle change in morphology, are shown at the left for comparison (cf. Fig. 2A).
These data presented a conundrum: If Sum1 repressed α-specific genes in S. cerevisiae, then why did MATa sum1Δ mutants of this species appear normal? We inspected the S. bayanus orthologs of S. cerevisiae mating genes to ask whether changes in mating genes might contribute to the S. bayanus MATa sum1Δ phenotype. Pairwise alignment of the S. cerevisiae and S. bayanus a-specific gene BAR1, which encodes a protease that degrades α-factor, revealed a single base-pair deletion (ΔT741) in the S. bayanus ortholog. This frameshift mutation was predicted to generate an inactive protein lacking the C-terminal catalytic site. Although this mutation had been annotated as a sequencing error (Kellis et al. 2003), we confirmed its presence by resequencing. This loss-of-function bar1 allele likely rendered S. bayanus a cells hypersensitive to α-factor. Mating pheromone halo assays confirmed that S. bayanus a cells were, in fact, hypersensitive to α-factor, but α cells were not hypersensitive to a-factor (data not shown). Transformation of S. bayanus MATa sum1Δ cells with a plasmid bearing the S. cerevisiae BAR1 gene suppressed the colony wrinkling phenotype, confirming that the bar1 mutation contributed to the phenotypic difference between species (Supplemental Fig. S2).
To test whether Bar1 function masked some of the phenotypic consequences of loss of Sum1, BAR1 was deleted in a S. cerevisiae MATa sum1Δ strain (W303 background). Intriguingly, MATa sum1Δ bar1Δ double-mutant cells had readily observable shmooing behavior, recapitulating much of the S. bayanus sum1Δ phenotype in S. cerevisiae. However, the S. cerevisiae mutant colonies were not wrinkled and the cells were not flocculent (Fig. 6B,C), suggesting that interspecies differences in adhesion molecules, or in their ability to be induced by α-factor, might account for the rest of the phenotype. A clue to this discrepancy is that S. cerevisiae a cells undergo pseudohyphal-like growth upon low-level stimulation by α-factor (Erdman and Snyder 2001). In the S. cerevisiae Σ1278b strain, which is competent to form pseudohyphae, the MATa sum1Δ bar1Δ double mutant fully recapitulated the shmooing and colony wrinkling phenotypes seen in the orthologous S. bayanus mutant (Fig. 6C). Thus, some difference between W303 and Σ1278b affected the ability of pheromone signaling to trigger flocculence (Liu et al. 1996; Guo et al. 2000).
α1 was required to overcome repression by Sum1 in α cells, but Mcm1 and Ste12 could activate transcription in the absence of Sum1 and α1
As Sum1 blocked α-specific gene expression, some mechanism must prevent it from repressing these genes in α cells. Sum1 protein was present at equivalent levels in both a cells and α cells (Fig. 5D). Sum1 bound equivalently in vivo to all promoters assayed in a cells and α cells (Fig. 5C). Therefore, neither the synthesis of nor the DNA-binding activity of Sum1 was mating-type-regulated.
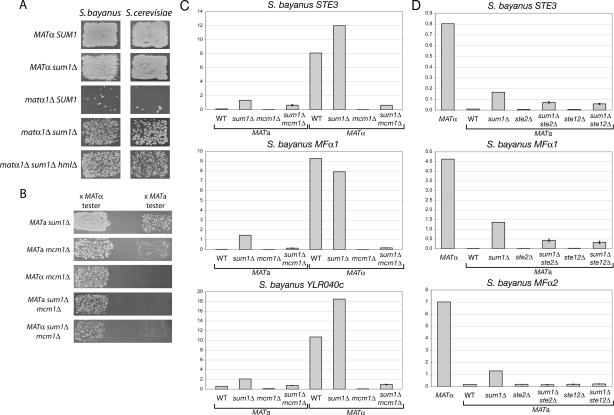
A simple model to explain how α-specific genes were expressed despite the presence of a repressor on their promoters was that α1 protein somehow counteracted Sum1’s repressive function, and contributed to transcription activation together with Mcm1 and Ste12. If α1 were required to overcome Sum1 repression in α cells, and were not completely required for α-specific gene expression, then deletion of SUM1 should suppress the nonmating phenotype of matα1Δ mutants. Indeed, whereas both S. cerevisiae and S. bayanus matα1Δ mutants were almost completely sterile, matα1Δ sum1Δ double mutants of both species mated substantially better (Fig. 7A). Furthermore, matα1Δ sum1Δ hmlΔ triple mutants mated equivalently to matα1Δ sum1Δ double mutants, demonstrating that removing Sum1 restored mating ability to α cells that were otherwise sterile because they lacked the α1 activator. Thus, Sum1 was active in both mating types of both species, and α1 was required to overcome Sum1 repression, in addition to providing a transcriptional activation function.
Figure 7.
Sum1 was a general repressor of mating-type-specific genes. (A) Patch mating assays (using only the MATa tester) were performed on MATα, MATα sum1Δ, matα1Δ, matα1Δ sum1Δ, and matα1Δ sum1Δ hmlΔ strains in both S. bayanus and S. cerevisiae (W303). (B) Patch mating assays were performed on S. bayanus sum1Δ, mcm1Δ, and sum1Δ mcm1Δ strains of the indicated mating types. (C) Expression of α-specific genes in cells of indicated genotypes was assayed by quantitative RT–PCR (WT, wild type). Expression of STE3 is reduced 10-fold in mcm1Δ cells (columns 3 and 7) compared with wild-type a cells (column 1). It should be noted that ACT1 (actin) RNA levels in mcm1Δ mutant cells were reduced twofold, causing the normalized expression values for sum1Δ mcm1Δ cells to be inflated. (D) Expression of α-specific genes in cells of indicated genotypes was assayed by quantitative RT–PCR (WT, wild type).
The restoration of mating in matα1Δ sum1Δ double mutants argued that Mcm1 and Ste12 could activate α-specific genes on their own, in contrast to the need for α1 as in the α1–α2 hypothesis. To test whether Mcm1 and Ste12 were necessary for α-specific gene expression in the absence of Sum1, mating ability and α-specific gene expression were assayed in sum1Δ mcm1Δ and sum1Δ ste12Δ double mutants. (The MCM1 gene was not essential in S. bayanus, in contrast to S. cerevisiae, although some lethality was observed in germinating spores from mcm1Δ/+ diploids and some viable cells had a growth defect [data not shown].) sum1Δ ste12Δ mutant cells of both mating types were completely sterile (data not shown); likewise, MATα mcm1Δ strains were unable to mate as α cells (Fig. 7B). The complete loss of mating with a cells by MATα mcm1Δ cells was not surprising, given that Mcm1 is required for activation of α-specific genes and repression of a-specific genes (Passmore et al. 1988; Elble and Tye 1991). However, unexpectedly, an equivalent small fraction of both mcm1Δ and sum1Δ mcm1Δ mutant cells mated as a cells, regardless of the allele present at MAT. Although most mcm1Δ mutant cells in the population were unable to mate, the a-like mating ability in a subset of cells demonstrated that at least some a-specific gene expression was Mcm1-independent (Kronstad et al. 1987). As suggested by the mating assays, Mcm1 was necessary for α-specific gene expression in the presence or absence of Sum1 (Fig. 7C). Indeed, in MATa mcm1Δ cells, STE3 expression was down 10-fold from its level in wild-type a cells.
Ste12, on the other hand, was partially required for α-specific gene expression, with the exception of MFα2 (Fig. 7D). The expression of STE3 and MFα1 remained severalfold increased in both MATa sum1Δ ste12Δ cells and MATa sum1Δ ste2Δ cells compared with wild-type, ste2Δ, or ste12Δ controls, yet neither gene was expressed in the double mutants to the level of the sum1Δ single mutant. It appeared that the level of expression of α-specific genes was a composite of two effects: derepression in the absence of Sum1, and autostimulation by α-factor. Specifically, expression of MFα2 in both MATa sum1Δ ste12Δ and MATa sum1Δ ste2Δ cells was equivalent to that in wild-type a cells, consistent with this gene being completely Ste12-dependent (Fields et al. 1988). The remaining STE3 and MFα1 expression in the absence of Sum1 and Ste12 implied that Mcm1 could activate some α-specific genes, albeit weakly, without other known coactivators. Interestingly, purified recombinant Mcm1 can bind P′Q elements in vitro, although it does so with low affinity relative to its binding of P elements from a-specific genes (Grayhack 1992).
The mechanism of repression of α-specific genes
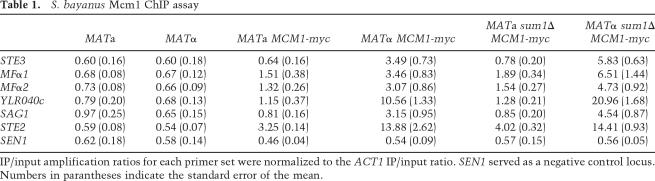
The above results pointed to a simple mechanism for repression of α-specific genes by Sum1: preventing Mcm1 from binding to DNA. To test this model, ChIP assays were performed on Myc-tagged Mcm1 in wild-type and sum1Δ cells. Consistent with this model, ChIP revealed enhanced association of Mcm1 with all five α-specific gene promoters in MATα sum1Δ cells compared with MATα wild type (Table 1). STE2, an a-specific gene, served as a control, as Mcm1 bound this gene’s promoter in both cell types, and its binding was unaffected by deletion of SUM1 (as predicted) (see Fig. 1). The greater enrichment of STE2 in α cells versus a cells may reflect conformational changes in Mcm1’s association with DNA introduced by the α2 protein. Intriguingly, Mcm1 precipitates from MATa sum1Δ cells did not show a substantial enrichment of α-specific gene promoter sequences compared with MATa wild type. However, although the standard model suggests that Mcm1 does not bind to α-specific gene promoters in the absence of α1 protein, we observed association of Mcm1 with the MFα1, MFα2, and YLR040c promoters in MATa cells. Thus, models to explain the mechanism of α-specific gene activation in sum1Δ cells need not require enhanced occupancy by Mcm1 (discussed further below).
Table 1.
S. bayanus Mcm1 ChIP assay
IP/input amplification ratios for each primer set were normalized to the ACT1 IP/input ratio. SEN1 served as a negative control locus. Numbers in parantheses indicate the standard error of the mean.
To explore whether the mechanism of repression of α-specific genes involved the proposed Sum1–Hst1–Rfm1 repressor complex, the HST1 and RFM1 genes were inactivated. S. bayanus MATa hst1Δ and MATa rfm1Δ mutants had colony and cellular phenotypes similar, although not identical, to MATa sum1Δ mutants (Supplemental Fig. S3). These results suggested that histone deacetylation by Hst1 was required for repression of α-specific genes, and that the interactions between Sum1, Rfm1, and Hst1 were conserved between S. bayanus and S. cerevisiae.
Discussion
The standard model of mating-type control in Saccharomyces cerevisiae posits that a is the default mating type, with the α mating type requiring the activation of α-specific gene expression by α1 (in combination with Mcm1 and Ste12), and the repression of a-specific gene expression by α2 (also in combination with Mcm1) (Strathern et al. 1981; Sprague 1990). We found that the Sum1 protein acted as a direct repressor of α-specific genes in both S. bayanus and S. cerevisiae (Fig. 8A). At least two differences between these two species conferred phenotypic differences that allowed the discovery of this conserved aspect of mating type determination, which has been overlooked for 26 years. Part of the phenotypic difference was explained by S. bayanus a cell hypersensitivity to α-factor due to a bar1 mutation. (This mutation was probably a polymorphism among S. bayanus strains [M. Dunham, A. Tsong, pers. comm.].) However, another factor, present in both S. bayanus and S. cerevisiae strain Σ1278b, caused sum1Δ bar1 mutant a cells to flocculate in response to α-factor. A good candidate for this “flocculence factor” was FLO8, which is required for flocculation in Σ1278b and has inactivating mutations in both W303 and S288C strains of S. cerevisiae (Liu et al. 1996).
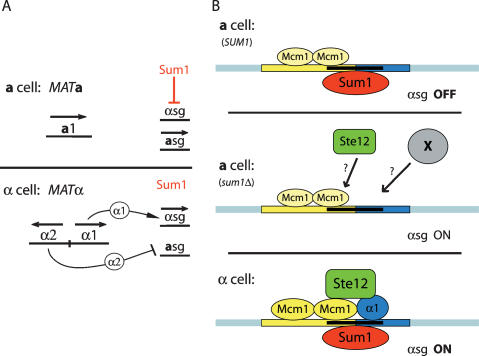
Figure 8.
Models for Sum1-mediated repression of α-specific genes. (A) Model for Sum1 role in repression of α-specific genes and mating type determination. In contrast to the standard model, active repression of α-specific genes by Sum1 was required for proper a cell identity. (B) Molecular mechanism for α-specific gene regulation. In a cells, Sum1 repressed α-specific genes by binding either to sites directly overlapping the α1 and Mcm1-binding sites, or to other nearby sites. Mcm1 bound some, but not all, α-specific gene promoters in a cells. Without repression by Sum1, Mcm1 and Ste12 were able to activate α-specific genes in a cells to a level that permitted α-like mating. In α cells, α1 induced high levels of α-specific gene expression and was required to overcome the block imposed by Sum1. Despite their high levels of transcription, Sum1 remained associated with α-specific genes in α cells.
Modifying the model for control of α-specific gene expression
Our results demonstrated two roles for the α1 protein: counteracting Sum1 repression, and inducing the maximal expression of α-specific genes (Fig. 3A,B). MATa sum1Δ and matα1Δ sum1Δ cells, both of which lack α1, were capable of α cell mating behavior (Fig. 7A,B). Consistent with the standard model, Mcm1 and Ste12 were necessary for mating of α cells and for robust α-specific gene expression in the absence of Sum1 (Fig. 7B–D). As Mcm1 and Ste12 were able to activate transcription in the absence of α1 and Sum1, the α1/Mcm1 cooperative binding model cannot fully explain α-specific gene activation (Fig. 8). Indeed, Mcm1 was associated with the MFα1, MFα2, and YLR040c promoters in MATa cells (Table 1). Mcm1’s occupancy of α-specific gene promoters was enhanced in the absence of Sum1, but only in MATα cells (when α1 is present). Surprisingly, Mcm1 ChIP of α-specific gene promoters was not substantially enhanced in MATa sum1Δ cells compared with MATa wild-type cells. This result forced us to consider two additional models for the interplay between Sum1 and the coactivator proteins at α-specific genes: (1) Sum1 interfered with transcriptional activation at a step downstream from Mcm1 binding, or (2) a novel activator was recruited to α-specific genes in the absence of Sum1. Formally, Sum1 might have blocked DNA binding by Ste12 in MATa cells. However, some expression of α-specific genes was observed in MATa sum1Δ ste12Δ cells (Fig. 7D), and Ste12-binding sites in most α-specific gene promoters did not overlap predicted Sum1-binding sites, making increased promoter occupancy by Ste12 in sum1Δ cells seem a less likely explanation.
Our data also highlighted gene-specific aspects of α-specific gene regulation. One exception to the model that Sum1 repressed α-specific genes was SAG1, whose expression in α cells was promoted by SUM1 (Fig. 3B). These data raised the possibility that some other factor represses SAG1, implying that there may be yet another dimension to α-specific gene regulation. The expression of MFα2 increased in a cells lacking Sum1, however, this increase was completely abolished in sum1Δ ste2Δ and sum1Δ ste12Δ double mutants (Fig. 7D). SAG1 and MFα2 thus appear to be regulated differently than MFα1, STE3, and YLR040c, which may represent unanticipated complexity in α-cell mating behavior.
Sum1 repressed α-specific genes yet was expressed and active in both a cells and α cells (Fig. 8B). Therefore, some mechanism involving α1, Mcm1, and Ste12 must exist to prevent Sum1 from repressing α-specific genes in α cells. One possible mechanism is DNA bending by α1 and Mcm1 (Hagen et al. 1993; Carr et al. 2004), which may alter the physical conformation of the promoter, preventing Sum1 from making protein–DNA or protein–protein interactions critical for repression. It is also possible that α1 and Mcm1 out-compete Sum1 for contacts with general transcription factors, Mediator, or RNA Polymerase holoenzyme. Finally, it is possible that α-specific gene activators directly contact Sum1 to prevent repression. Notably, some mcm1 and ste12 alleles are specifically defective in activating expression of α-specific genes in α cells (Passmore et al. 1988; La Roche et al. 1995). These mutant proteins may fail to oppose Sum1 repression, or they may be unable to interact with α1.
Sum1 binding to α-specific gene promoters in their active state could be explained if this repressor bound to the minor groove of DNA while the α1 and Mcm1 activators bound the major groove. This scenario seemed plausible given that Sum1 contains two “AT-hooks”—short, positively charged motifs of thirteen amino acids—in its N-terminal half. These moieties often mediate binding to the minor groove of AT-rich DNA sequences (Reeves and Nissen 1990; Huth et al. 1997). Regardless of the specific mechanism involved, our results suggest a more complex model of α-specific promoter function than previously appreciated.
Control of cell-type-determining genes during differentiation
Why should α-specific genes have required repression by Sum1? As in the development of distinct cell types in multicellular eukaryotes, proper yeast cell identity must be ensured as soon as a differentiation event occurs; in this case, a mating-type switch. The most straightforward role for mating-type regulation by Sum1 was preventing α-mating behavior in a cells, particularly when cells switch from MATα to MATa. In such a cell, α-specific genes would continue to be expressed if there were any residual α1 protein. Without active repression of α-specific genes, this newly formed a cell might mate with another a cell, generating a MATa/a diploid unable to undergo meiosis and destined for meiotic oblivion should it mate with an α cell to form a triploid. A complementary role for Sum1 was maintenance of robust a cell mating ability, as expression of α-specific genes in a cells causes decreased mating ability (Roth et al. 2000; Rivers and Sprague 2003). Furthermore, our results show that α-specific genes can be expressed at a substantial level even in the absence of their primary activator. For a cells that secreted even a low level of α-factor, their adaptation and subsequent desensitization (Dietzel and Kurjan 1987) would likely make them less sensitive to signals from bona fide mating partners. (Indeed, Sum1 was necessary for preventing autostimulation of a cells with α-factor. We note that our data did not formally distinguish between autocrine stimulation of an a cell by α-factor produced by that same a cell versus stimulation occurring primarily in trans, by α-factor secreted from neighboring cells. As a substantial amount of α-factor was likely present within growing MATa sum1Δ colonies, some level of trans-stimulation undoubtedly occurred.)
Further insight into why this function of Sum1 evolved might be gained by asking when it evolved. Given the evolutionary distance between S. cerevisiae and S. bayanus, our results suggest that the Sum1 protein likely performed at least two functions in the sensu stricto ancestor: repression of α-specific genes and repression of meiotic genes. The conservation of the Sum1 protein and predicted Sum1-binding sites in α-specific gene promoters in Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces kudriavzevii support this idea. α-Specific gene activation by α1 appears to be conserved throughout Ascomycete fungi (Tsong et al. 2003). However, Sum1 orthologs are present in Ashbya gossypii and Klyuveromyces lactis, but are not in species more distant from S. cerevisiae. Therefore, it would be interesting to ask how α-specific genes were regulated in ancestral Ascomycetes. It is possible either that a different repression mechanism is used outside of the Saccharomyces complex, or that Sum1-based repression is unique to these species, possibly to accomplish a unique aspect of their mating behavior (Fig. 1B).
Advantages of comparative genetic analysis
In retrospect, active repression of α-specific genes has evaded detection for so long largely because conventional biochemical and single-species genetic approaches were not sufficiently sensitive. One apparent complication was overlap of the α1/Mcm1 activators’ binding sites by the putative Sum1-binding site in the promoters of some α-specific genes. Previously characterized mutations in the STE3 promoter that abolished activator binding (Hagen et al. 1993) also ablated the Sum1-binding site, obscuring the contribution of Sum1 to repression of STE3 expression. Similarly, gel-shift analyses of the STE3 and MFα1 operators largely failed to detect a complex containing anything other than α1 and Mcm1 (Bender and Sprague 1987; Jarvis et al. 1988). However, reaction conditions optimized for α1/Mcm1 binding or low Sum1 protein abundance relative to that of α1 and Mcm1 may have confounded the ability of such experiments to detect Sum1 binding. Sum1 recruitment to α-specific genes might involve chromatin or other protein–protein interactions, rather than simply site-specific DNA binding.
The perspective of evolutionary biology offers the ability to infer conservation of functions between organisms, and the ability to ask how alterations in genetic circuits generate novel forms. Current large-scale biochemical, genetic, and computational efforts seek to identify novel genetic pathways and novel components of known pathways (Tong et al. 2004; Krogan et al. 2006; Collins et al. 2007). However, the recognition of a genetic pathway’s components may depend on the range of phenotypes produced by common perturbations in multiple species. As we have shown, comparative genetic analysis in closely related species can provide the phenotypic depth sometimes needed for ascribing novel functions to specific genes. In this case, adventitious variation in phenotypes between species revealed a regulatory circuit conserved among species. Because nature provides variation not captured by model organisms, more detailed pictures of shared and unique regulatory pathways should emerge from a montage of multiple species’ genetic interaction networks.
Materials and methods
Yeast strains, culture, and genetic manipulations
Yeast strains are listed in Table 1. Heterothallic S. bayanus wild-type strains were derived from CB54001, obtained from Ed Louis. The HO gene was inactivated in this diploid strain and haploid MATa and MATα hoΔ strains were isolated by sporulation and tetrad dissection of the HO/hoΔ heterozygotes. Auxotrophic markers ade2-1 and his3-1 were generated in haploid prototrophic strains by EMS mutagenesis followed by screening for colonies unable to grow on minimal media. For lys2-5 and ura3-1, mutagenized haploids were plated on α-aminoadipate and 5-FOA media, respectively, to select for resistant mutants. Strains JRY8729 and JRY8730 were generated by sequential crosses of individual auxotrophic mutants and dissecting tetrads for multiple-marker mutants. All growth of S. bayanus was performed using standard conditions for S. cerevisiae, except that plate and liquid culturing was performed at 25°C for both species. S. cerevisiae wild-type strain W303-1a has been described previously (Thomas and Rothstein 1989). The MATa Σ1278b strain, JRY6896, was originally designated 10560-4A (G. Fink), and was obtained from Jeremy Thorner (JTY2560). One-step gene replacement, C-terminal 13xMyc tag integration (Longtine et al. 1998; Goldstein and McCusker 1999) and C-terminal 3xFlag tag integration (Gelbart et al. 2001) have been described previously, and these genetic manipulations were performed identically for both S. bayanus and S. cerevisiae. All gene disruptions in both species (sum1Δ∷Hyg, etc.) were confirmed using PCR to examine the 5′ and 3′ ends of targeted ORFs. To construct strain JRY8736, the GAL1 promoter (Longtine et al. 1998) was integrated by homologous recombination immediately upstream of the ATG of the MFα1 gene, removing 50 bp of the native promoter.
Microscopy and mating assays
DIC microscopy was performed using a Nikkon Eclipse E600 microscope (100× objective). Cells were gently dispersed in complete synthetic media and spotted onto 2% agarose pads mounted on slides. Multiple fields were observed to score cellular morphology. Patch-mating assays were performed by mixing approximately equal amounts of query strain and tester strain, each obtained from individual colonies, on YPD (glucose) or YPG (galactose) and incubating overnight. The following day, patches were replica plated onto minimal media to select for diploids. Patch mating assays shown in Figure 4 (galactose induction of MATa wild-type and GAL1pro-MFα1 strains) were performed on rich medium containing 2% galactose as the carbon source before replica plating to minimal media containing 2% glucose and no galactose. The tester strains used in all mating assays were JRY2726 (MATa) and JRY2728 (MATα), both of which are S. cerevisiae his4. All S. bayanus strains described in the text mated equivalently with these tester strains and with S. bayanus tester strains.
RNA and protein analysis
RNA isolation was performed using the hot-phenol method (Schmitt et al. 1990). Total RNA was digested with Amplification-grade DNase I (Invitrogen) and purified using the RNeasy Minelute kit (Qiagen). cDNA was synthesized using the SuperScript III First-Strand Synthesis System for RT–PCR and oligo(dT) primer (Invitrogen). Quantitative PCR on cDNA was performed using an MX3000P machine (Stratagene) and the DyNAmo HS SYBR Green qPCR kit (NEB). Amplification values for all primer sets were normalized to actin (ACT1) cDNA amplification values. Samples were analyzed in triplicate for two or three independent RNA preparations. For Figure 7, C and D, analysis, duplicate mcm1Δ, sum1Δ mcm1Δ, ste2Δ, ste12Δ, sum1Δ ste2Δ, and sum1Δ ste12Δ samples were processed in parallel to single wild-type and sum1Δ control samples from identical strains to those analyzed in Figure 3.
Yeast whole-cell extracts were prepared using 20% TCA and solubilization in SDS loading buffer plus 100 mM Tris base. SDS-PAGE and immunoblotting were performed using standard procedures and the LiCOR imaging system. Anti-Flag M2 antibody from mouse (Sigma) was used to detect Flag-tagged Sum1 protein. Rabbit anti-Pgk1 antibody (Baum et al. 1978), a kind gift of Jeremy Thorner, was used to verify equal loading.
ChIP analysis
ChIPs were performed as described (Davies et al. 2005) using formaldehyde cross-linking of log-phase cultures for 1 h at room temperature. IPs were performed using anti-Flag M2-agarose from mouse and anti-c-Myc agarose from rabbit (Sigma). Quantitative PCR was performed on precipitated DNA fragments as described above. The “Reference” primer set, which amplifies a region within the SEN1 ORF, corresponded to a locus with a minimal level of Sum1 binding in the S. cerevisiae genome. Amplification values for the precipitated DNA were normalized to the values for the input DNA for all primer sets. Samples were analyzed in triplicate for three independent ChIPs and one no-tag control for each mating type.
Binding site predictions
To detect putative Sum1-binding sites, 1 kb of sequence 5′ of the ATG of each α-specific gene and predefined consensus binding sites (described in the text) were submitted to the SCPD database (M. Zhang laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY; http://rulai.cshl.edu/SCPD). To account for site degeneracy, sequential consensus searches that permitted zero or one mismatch were performed.
Acknowledgments
We thank Ed Louis for the sequenced S. bayanus strain, and Annie Tsong and members of the Rine laboratory for helpful discussions and insights. We especially thank Jeremy Thorner for many helpful discussions, and for gifts of the Pgk1 antibody and yeast strains. This work was supported by an NSF predoctoral fellowship to O.A.Z. and by a grant from the National Institutes of Health (NIGMS 31105) to J.R. We dedicate this paper to the memory of our friend, Daniel E. Koshland Jr.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1640008.
References
- Abzhanov A., Protas M., Grant B.R., Grant P.R., Tabin C.J. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Baum P., Thorner J., Honig L. Identification of tubulin from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 1978;75:4962–4966. doi: 10.1073/pnas.75.10.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Sprague G.F. MATα1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell-type-specific genes. Cell. 1987;50:681–691. doi: 10.1016/0092-8674(87)90326-6. [DOI] [PubMed] [Google Scholar]
- Bruhn L., Sprague G.F. MCM1 point mutants deficient in expression of α-specific genes: Residues important for interaction with α1. Mol. Cell. Biol. 1994;14:2534–2544. doi: 10.1128/mcb.14.4.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr E.A., Mead J., Vershon A.K. α1-induced DNA bending is required for transcriptional activation by the Mcm1–α1 complex. Nucleic Acids Res. 2004;32:2298–2305. doi: 10.1093/nar/gkh560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Davies B.S., Wang H.S., Rine J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: Similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 2005;25:7375–7385. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel C., Kurjan J. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: A model for desensitization to pheromone. Mol. Cell. Biol. 1987;7:4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R., Tye B.K. Both activation and repression of a-mating-type-specific genes in yeast require transcription factor Mcm1. Proc. Natl. Acad. Sci. 1991;88:10966–10970. doi: 10.1073/pnas.88.23.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S., Snyder M. A filamentous growth response mediated by the yeast mating pathway. Genetics. 2001;159:919–928. doi: 10.1093/genetics/159.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Herskowitz I. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell. 1985;42:923–930. doi: 10.1016/0092-8674(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Fields S., Chaleff D.T., Sprague G.F. Yeast STE7, STE11, and STE12 genes are required for expression of cell-type-specific genes. Mol. Cell. Biol. 1988;8:551–556. doi: 10.1128/mcb.8.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick D.A., Logue M.E., Stajich J.E., Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flessel M.C., Brake A.J., Thorner J. The MFα1 gene of Saccharomyces cerevisiae: Genetic mapping and mutational analysis of promoter elements. Genetics. 1989;121:223–236. doi: 10.1093/genetics/121.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgoczy D.J., Cassidy-Stone A., Llinas M., O’Rourke S.M., Herskowitz I., DeRisi J.L., Johnson A.D. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2004;101:18069–18074. doi: 10.1073/pnas.0407611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter B., Tan S., Richmond T.J. Genomic footprinting of the promoter regions of STE2 and STE3 genes in the yeast Saccharomyces cerevisiae. J. Mol. Biol. 1993;234:975–987. doi: 10.1006/jmbi.1993.1652. [DOI] [PubMed] [Google Scholar]
- Gelbart M.E., Rechsteiner T., Richmond T.J., Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: Analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 2001;21:2098–2106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grayhack E.J. The yeast α1 and MCM1 proteins bind a single strand of their duplex DNA recognition site. Mol. Cell. Biol. 1992;12:3573–3582. doi: 10.1128/mcb.12.8.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Styles C.A., Feng Q., Fink G.R. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D.C., Bruhn L., Westby C.A., Sprague G.F. Transcription of α-specific genes in Saccharomyces cerevisiae: DNA sequence requirements for activity of the coregulator α1. Mol. Cell. Biol. 1993;13:6866–6875. doi: 10.1128/mcb.13.11.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth J.R., Bewley C.A., Nissen M.S., Evans J.N., Reeves R., Gronenborn A.M., Clore G.M. The solution structure of an HMG-I(Y)–DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- Hwang-Shum J.J., Hagen D.C., Jarvis E.E., Westby C.A., Sprague G.F. Relative contributions of MCM1 and STE12 to transcriptional activation of a- and α-specific genes from Saccharomyces cerevisiae. Mol. Gen. Genet. 1991;227:197–204. doi: 10.1007/BF00259671. [DOI] [PubMed] [Google Scholar]
- Irlbacher H., Franke J., Manke T., Vingron M., Ehrenhofer-Murray A.E. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes & Dev. 2005;19:1811–1822. doi: 10.1101/gad.334805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E.E., Hagen D.C., Sprague G.F. Identification of a DNA segment that is necessary and sufficient for α-specific gene control in Saccharomyces cerevisiae: Implications for regulation of α-specific and a-specific genes. Mol. Cell. Biol. 1988;8:309–320. doi: 10.1128/mcb.8.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E.E., Clark K.L., Sprague G.F. The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes & Dev. 1989;3:936–945. doi: 10.1101/gad.3.7.936. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kirkman-Correia C., Stroke I.L., Fields S. Functional domains of the yeast STE12 protein, a pheromone-responsive transcriptional activator. Mol. Cell. Biol. 1993;13:3765–3772. doi: 10.1128/mcb.13.6.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A.J., Kakar S.N., Ivy J.M., Hicks J.B., Livi G.P., Miglio L.M. SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics. 1985;111:745–758. doi: 10.1093/genetics/111.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kronstad J.W., Holly J.A., MacKay V.L. A yeast operator overlaps an upstream activation site. Cell. 1987;50:369–377. doi: 10.1016/0092-8674(87)90491-0. [DOI] [PubMed] [Google Scholar]
- Kurjan J. α-Factor structural gene mutations in Saccharomyces cerevisiae: Effects on α-factor production and mating. Mol. Cell. Biol. 1985;5:787–796. doi: 10.1128/mcb.5.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Roche S.D., Shafer B.K., Strathern J.N. A ste12 allele having a differential effect on a versus α cells. Mol. Gen. Genet. 1995;246:80–90. doi: 10.1007/BF00290136. [DOI] [PubMed] [Google Scholar]
- Laurenson P., Rine J. SUM1-1: A suppressor of silencing defects in Saccharomyces cerevisiae. Genetics. 1991;129:685–696. doi: 10.1093/genetics/129.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C.A., Fink G.R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- McCord R., Pierce M., Xie J., Wonkatal S., Mickel C., Vershon A.K. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 2003;23:2009–2016. doi: 10.1128/MCB.23.6.2009-2016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore S., Maine G.T., Elble R., Christ C., Tye B.K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 1988;204:593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- Pierce M., Benjamin K.R., Montano S.P., Georgiadis M.M., Winter E., Vershon A.K. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol. Cell. Biol. 2003;23:4814–4825. doi: 10.1128/MCB.23.14.4814-4825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., Nissen M.S. The A.T–DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- Rivers D.M., Sprague G.F. Autocrine activation of the pheromone response pathway in matα2-cells is attenuated by SST2- and ASG7-dependent mechanisms. Mol. Genet. Genomics. 2003;270:225–233. doi: 10.1007/s00438-003-0914-3. [DOI] [PubMed] [Google Scholar]
- Robert F., Pokholok D.K., Hannett N.M., Rinaldi N.J., Chandy M., Rolfe A., Workman J.L., Gifford D.K., Young R.A. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A.F., Nelson B., Boone C., Davis N.G. Asg7p–Ste3p inhibition of pheromone signaling: Regulation of the zygotic transition to vegetative growth. Mol. Cell. Biol. 2000;20:8815–8825. doi: 10.1128/mcb.20.23.8815-8825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L.N., Rine J. Conversion of a gene-specific repressor to a regional silencer. Genes & Dev. 2001;15:955–967. doi: 10.1101/gad.873601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.E., Brown T.A., Trumpower B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P., Cochran B.H. The PRE and PQ box are functionally distinct yeast pheromone response elements. Mol. Cell. Biol. 1990;10:6809–6812. doi: 10.1128/mcb.10.12.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.L., Johnson A.D. A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an α2 dimer. Cell. 1992;68:133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- Sprague G.F., Jr. Combinatorial associations of regulatory proteins and the control of cell type in yeast. Adv. Genet. 1990;27:33–62. doi: 10.1016/s0065-2660(08)60023-1. [DOI] [PubMed] [Google Scholar]
- Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J., Hicks J., Herskowitz I. Control of cell type in yeast by the mating type locus. The α1–α2 hypothesis. J. Mol. Biol. 1981;147:357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tsong A.E., Miller M.G., Raisner R.M., Johnson A.D. Evolution of a combinatorial transcriptional circuit: A case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- Xie J., Pierce M., Gailus-Durner V., Wagner M., Winter E., Vershon A.K. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y.O., Stroke I.L., Fields S. Coupling of cell identity to signal response in yeast: Interaction between the α1 and STE12 proteins. Genes & Dev. 1993;7:1584–1597. doi: 10.1101/gad.7.8.1584. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C.T., Hannett N.M., Volkert T.L., Fink G.R., Young R.A. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]