Abstract
Background
Gene-based vaccine delivery is an important strategy in the development of a preventive vaccine for acquired immunodeficiency syndrome (AIDS). Vaccine Research Center (VRC) 004 is the first phase 1 dose-escalation study of a multiclade HIV-1 DNA vaccine.
Methods
VRC-HIVDNA009-00-VP is a 4-plasmid mixture encoding subtype B Gag-Pol-Nef fusion protein and modified envelope (Env) constructs from subtypes A, B, and C. Fifty healthy, uninfected adults were randomized to receive either placebo (n = 10) or study vaccine at 2 mg (n = 5), 4 mg (n = 20), or 8 mg (n = 15) by needle-free intramuscular injection. Humoral responses (measured by enzyme-linked immunosorbant assay, Western blotting, and neutralization assay) and T cell responses (measured by enzyme-linked immunospot assay and intracellular cytokine staining after stimulation with antigen-specific peptide pools) were measured.
Results
The vaccine was well tolerated and induced cellular and humoral responses. The maximal CD4+ and CD8+ T cell responses occurred after 3 injections and were in response to Env peptide pools. The pattern of cytokine expression by vaccine-induced HIV-specific T cells evolved over time, with a diminished frequency of interferon-γ–producing T cells and an increased frequency of interleukin-2–producing T cells at 1 year.
Conclusions
DNA vaccination induced antibody to and T cell responses against 3 major HIV-1 subtypes and will be further evaluated as a potential component of a preventive AIDS vaccine regimen.
More than 25 million people have died since HIV/AIDS was identified in 1981, and an estimated 14,000 new infections occur daily [1]. Development of a globally relevant HIV-1 vaccine is critical for controlling this pandemic. The combination of a high transcriptional error rate and frequent recombination results in a remarkable amount of genetic diversity among HIV-1 strains and presents a challenge for selecting vaccine antigens. A joint meeting between the Vaccine Research Center (VRC) (National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH], Department of Health and Human Services) and the Joint United Nations Programme on HIV/AIDS concluded that testing of multiclade candidate vaccines is a high international scientific priority [2]. Sequences encoding gene products from 3 high-incident HIV subtypes were used to produce the candidate vaccine evaluated in the present study [3, 4]. Multiple antigens expressed by the vaccine elicit responses against multiple epitopes and may diminish the chances for immune escape.
Delivering antigens by DNA plasmids has potential advantages over other vector delivery systems, notably the lack of antivector immunity. Despite examples of vaccine-induced protection in mice and nonhuman primates [5, 6], DNA immunization has shown limited immunogenicity in humans [7–11]. Here, we report the findings from a phase 1 clinical trial of a multigene, multiclade HIV-1 DNA candidate vaccine and demonstrate the induction of HIV-1–specific T cell and antibody responses.
METHODS
Study design
VRC 004 (NIH 03-I-0022) was conducted at the NIH by the VRC. It was a randomized, double-blind, placebo-controlled phase 1 dose-escalation study. A randomization sequence was obtained by use of computer-generated random numbers, and blocking ensured balance across groups. The study was opened to accrual on 13 November 2002 and was unblinded on 24 September 2004. Vaccine safety and immunogenicity were the primary and secondary objectives, respectively. Eligibility criteria included HIV-seronegative status; age 18–40 years; being amenable to HIV risk-reduction counseling; having good general health as determined by medical history, physical exam, and laboratory tests; and having no prior exposure to investigational HIV vaccines. Fifty subjects were randomized to receive placebo (n = 10) or vaccine at doses of 2 mg (n = 5), 4 mg (n = 20), or 8 mg (n = 15). Safety reviews were conducted in both the 2-mg and 4-mg groups (5 vaccine recipients and 2 placebo recipients) before randomizing the remaining 36 subjects to the 4-mg, 8-mg, or placebo groups. The NIAID Intramural Data and Safety Monitoring Board conducted safety reviews for the dose escalation from 4 to 8 mg as well as at 6-month intervals throughout the study. Injections (1 mL/injection) were administered on day 0 and at weeks 4 and 8. Arms were alternated for sequential vaccinations, except for the delivery of the 8-mg dose of vaccine, which required 1-mL injections of 4 mg into both arms. Evaluations included laboratory tests, physical assessments by clinicians, and self-assessment for local and systemic symptoms recorded on 7-day diary cards. Adverse events were graded for severity by use of a preapproved table that incorporated a 5-point scale and were coded by use of Medical Dictionary for Regulatory Activities terminology. HIV testing was done by RNA polymerase chain reaction (Roche Amplicor HIV-1 Monitor Test) and ELISA (Abbott HIVAB HIV-1/HIV-2 rDNA); Western blotting (Genetic Systems HIV Western blot kit; BioRad Laboratories; performed at the Mayo Laboratory, Rochester, MN) was done if ELISA results were positive. The social impact of participating in an HIV vaccine study was monitored.
Vaccine
The vaccine, VRC-HIVDNA009-00-VP, was developed by the VRC and is manufactured by Vical; it is composed of 4 closed, circular, DNA plasmids at a concentration of either 2 mg/mL or 4 mg/mL (figure 1). The plasmid expressing clade B HIV-1 Gag-Pol-Nef fusion polyproteins comprised 50% of the vaccine by weight. The plasmids expressing Env glycoprotein from clades A, B, and C each comprised 16.67% of the vaccine by weight. Before formulation of the vaccine product, expression levels of individual plasmids were assessed semiquantitatively by Western blot densitometry and were compared with standards run under the same conditions. Preclinical testing demonstrated the product to have an acceptable safety profile [13, 14].
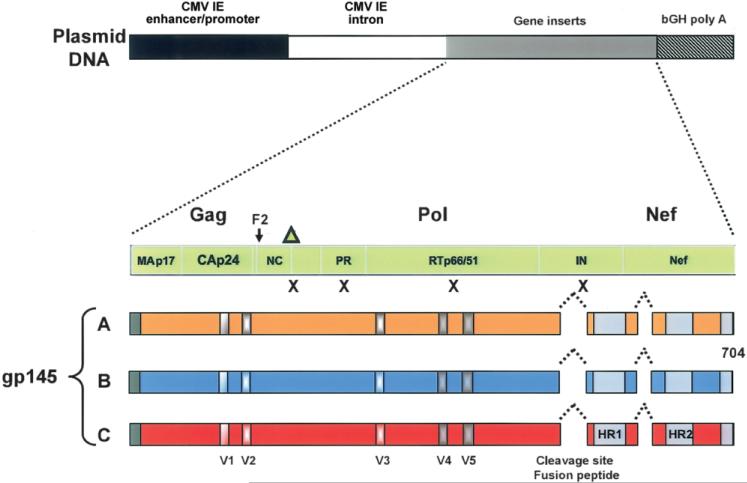
Figure 1.
Schematic of the DNA vaccine design. Four separate DNA plasmids were produced by inserting individual HIV-1 gene constructs into the pVR1012 backbone under the control of the cytomegalovirus (CMV) immediate-early (IE) promoter, followed by the bovine growth hormone polyadenylation (bGH poly A) sequence [3, 12]. The synthetic gag gene is from the clade B strain HXB2 (GenBank accession no. K03455), the synthetic pol gene (pol/h) is from the clade B strain NL4-3 (GenBank accession no. M19921), and the synthetic nef gene (nef/h) is from the clade B strain PV22 (GenBank accession no. M19921). Mutations (indicated by Xs), including the deletion of the carboxy-terminus of Gag (indicated by the triangle), were introduced in the protease and reverse-transcriptase genes to prevent processing of the pol gene products and to reduce the potential for functional enzymatic activity. This resulted in a fusion protein that directly reads through the frame shift in Gag (F2) through Pol and into Nef. This gene product is not able to assemble or produce pseudoparticles. To create synthetic gp145, versions of the envelope genes were truncated immediately downstream of the transmembrane domain of gp41. In each construct, the cleavage site and fusion peptide at the junction of gp120 and gp41 were deleted, and a portion of the interspace between the 2 heptad-repeat regions in gp41 was deleted. The Env gene products are primarily cell associated rather than secreted. The EnvA sequence is from 92rw020 (CCR5 tropic; GenBank accession no. U08794), the EnvB sequence is from HXB2 (CXCR4 tropic; GenBank accession no. K03455), and the EnvC sequence is from 97ZA012 (CCR5 tropic; GenBank accession no. AF286227). HR1–HR2, heptad-repeat regions in gp41; IN, integrase; NC, nucleocapsid; PR, protease; V1–V5, variable regions in envelope.
The placebo used was calcium and magnesium free (Vical). All injections were administered with a needle-free injection device into the deltoid muscle by use of a Biojector 2000 (Bioject).
Antibodies
Unconjugated mouse anti–human CD28, unconjugated mouse anti–human CD49d, allophycocyanin (APC)–conjugated mouse anti–human CD3, fluorescein isothiocyanate (FITC)–conjugated mouse anti–human CD8, peridinin chlorophyll protein–conjugated mouse anti–human CD4, and a mixture of phycoerythrin (PE)–conjugated mouse anti–human interferon (IFN)–γ and interleukin (IL)–2 monoclonal antibodies were obtained from Becton Dickinson Immunocytometry Systems (BDIS). Independent evaluation of IFN-γ and IL-2 required CD4cy5.5PE and CD8QDot655 conjugation (for details, see http://drmr.com/abcon) in combination with CD3cy7APC, IFN-γ FITC, and IL-2 APC (BDIS). To determine the optimum concentration for staining, reagents were independently titrated.
Cell preparation
Peripheral-blood mononuclear cells (PBMCs) were prepared by Ficoll-Paque density gradient centrifugation (Pharmacia). PBMCs were frozen in heat-inactivated fetal calf serum containing 10% dimethylsulfoxide in a Forma CryoMed cell freezer. Cells were stored at −180°C. Immunogenicity assays were performed on thawed specimens; average viability was >95%.
Peptides
Peptides (15 aa in length, overlapping by 11 aa, and corresponding to the vaccine inserts) were synthesized at >85% purity as confirmed by high-performance liquid chromatography. Peptides were pooled for each protein (EnvA, EnvB, EnvC, Gag, Nef, and Pol [2 pools]) and were used at a final concentration of 2.5 μg/mL.
Cell stimulation
PBMCs (106) in 200 μL of R-10 medium (RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin G, 100 μg/mL streptomycin sulfate, and 1.7 mmol/L sodium glutamate) were incubated with costimulatory anti-CD28 and anti-CD49d monoclonal antibodies (1 μg/mL each) each and with each peptide (2.5 μg/mL) in 96-well V-bottom plates. Cells incubated with costimulatory antibodies only were included in every experiment, to control for spontaneous production of cytokine and activation of cells before addition of peptides. Staphylococcal enterotoxin B (SEB; 1 μg/mL; Sigma-Aldrich) was used as a positive control for lymphocyte activation. Cultures were incubated at 37°C in a 5% CO2 incubator for 6 h in the presence of brefeldin A (10 μg/mL; Sigma).
Intracellular cytokine staining (ICS) assays
ICS assays for IFN-γ and/or IL-2 in CD4+ and CD8+ T cells were performed after stimulation with overlapping peptide pools representing the vaccine antigens. PBMCs were permeabilized for 7 min in 200 μL of a solution containing 100 μL of Tween 20 (Sigma), 160 μL of deionized water, and 40 μL of 10× FACS-Lyse solution (BDIS) at room temperature; were washed twice in cold Dulbecco's PBS containing 1% fetal bovine serum and 0.02% sodium azide (FACS buffer); and were stained with directly conjugated anti-human antibodies for 20 min on ice. Stained cells were then immediately washed twice with cold FACS buffer. The cells were resuspended in Dulbecco's PBS containing 1% paraformaldehyde (Electron Microscopy Systems) and then stored at 4°C until analysis.
Flow-cytometric analysis
Flow-cytometric analysis was performed on a FACSCalibur flow cytometer (BDIS). Between 50,000 and 250,000 events, gated on small lymphocytes, were acquired and analyzed using FlowJo software (version 8.1; Tree Star Software). The same cytokine, CD4, and CD8 gates were used for the entire trial.
Enzyme-linked immunospot (ELISpot) assays
ELISpot assays were performed using a commercially available ELISpot kit (BD Biosciences) [3]. PBMCs were stimulated overnight at 37°C in triplicate wells at a density of 2 × 105 cells/well for all stimulations other than SEB, which was conducted at 5 × 104 cells/well. After incubation, cells were lysed, and the wells were washed and incubated for 2 h at room temperature in the presence of biotinylated IFN-γ detection antibodies. Subsequently, the wells were incubated with an avidin–horseradish peroxidase solution for 1 h at room temperature, followed by incubation for 20 min with 3-amino-9-ethylcarbazole substrate solution. The plate was air-dried for a minimum of 2 h before spot quantitation on a CTL ELISpot image analyzer (Cellular Technology). Results are expressed as the mean number of spot-forming cells per 106 PBMCs.
Measurement of antibody responses
Research ELISAs were performed to delineate the antibody response to individual viral antigens encoded in the vaccine. End-point titers of antibodies directed against HIV antigens were determined using 96-well Immulon 2 plates (Dynex Technologies) coated with a preparation of purified recombinant HIV proteins, in accordance with methods adapted from those described elsewhere [15]. Analysis by immunoprecipitation followed by Western blotting (IP–Western blotting) was performed as described else-where [3]. Screening for HIV-1 neutralization activity was performed on 1:5 dilutions of serum in a flow cytometry–based assay that measured levels of intracellular p24 after a single round of HIV-1 infection of PBMCs, as described elsewhere [16].
Statistical methods
T cell data are summarized by rates of positive response to the individual peptide pools and exact, 2-sided 95% confidence intervals (CIs). Positivity criteria consisted of a statistical hypothesis test for a difference between stimulated and unstimulated wells and a minimal level of response requirement, as described elsewhere [17]. The minimum threshold for background-corrected positive-response percentage was 0.0241% for CD4+ T cells and 0.0445% for CD8+ T cells for the ICS assays and was 50 sfc/106 PBMCs for the ELISpot assays. SAS (version 8.2; SAS Institute) and Splus (version 6.0; Insightful) were used for analyses.
RESULTS
Subject population
Subjects had a mean age of 29 years, and 60% were men. Vaccine and placebo recipients had similar demographic characteristics (table 1). Vaccinations were completed in 9 of the 10 subjects in the placebo group, in 5 of the 5 subjects in the 2-mg group, in 20 of the 20 subjects in the 4-mg group, and in 14 of the 15 subjects in the 8-mg group. All subjects completed 52 weeks of follow-up.
Table 1.
Subject demographics and vaccine reactogenicity.
| Category, parameter | Placebo recipients (n = 10) | Vaccine recipients (n = 40) |
|---|---|---|
| Demographics | ||
| Sex | ||
| Male | 8 (80) | 22 (55) |
| Female | 2 (20) | 18 (45) |
| Age | ||
| 18-20 years | 1 (10) | 2 (5) |
| 21-30 years | 4 (40) | 22 (55) |
| 31-40 years | 5 (50) | 16 (40) |
| Mean ± SD, years | 30 ± 7 | 29 ± 5 |
| Range, years | 20-39 | 18-38 |
| Race | ||
| White | 7 (70) | 34 (85) |
| Black or African American | 1 (10) | 4 (10) |
| Asian/Pacific Islander | 2 (20) | 1 (2.5) |
| American Indian/Alaskan Native | 0 (0) | 0 (0) |
| Multiracial | 0 (0) | 1 (2.5) |
| Ethnicity | ||
| Non-Hispanic/Latino | 9 (90) | 39 (98) |
| Hispanic/Latino | 1 (10) | 1 (2.5) |
| Reactogenicity summarya | ||
| Local | ||
| None | 2 (20) | 1 (2.5) |
| Mild | 7 (70) | 30 (75) |
| Moderate | 1 (10) | 9 (22.5) |
| Severe | 0 (0) | 0 (0) |
| Systemic | ||
| None | 3 (30) | 10 (25) |
| Mild | 5 (50) | 22 (55) |
| Moderate | 2 (20) | 8 (20) |
| Severe | 0 (0) | 0 (0) |
NOTE. Data are no. (%) of subjects, unless otherwise indicated.
Each subject was counted once for his or her worst severity score during the 7 days after each vaccination.
Vaccine safety
Three adverse events that were possibly related to vaccination were notable. They included grade 3 asymptomatic neutropenia with onset 27 days after the third vaccination (4-mg group), grade 3 urticaria with onset 4 days after the third vaccination (4-mg group), and grade 2 maculopapular rash with onset 27 days after the second vaccination (8-mg group); the last subject did not receive the third vaccination. All resolved without sequelae and had alternative explanations. Vaccinations were well tolerated, and there were no episodes of severe reactogenicity (table 1). The placebo group reported an incidence of pain similar to that of the vaccine groups but reported less induration and erythema; there was no suggestion of a dose effect.
Fourteen (40% [95% CI, 24%–58%]) of the 35 subjects who received 4- or 8-mg injections had vaccine-induced positive diagnostic ELISA results, and 7 (20% [95% CI, 8%–37%]) had indeterminate Western blot results (table 2). Most commonly, the first positive ELISA result occurred at week 12, with a return to background levels by week 52; there were 6 subjects with a positive ELISA result at week 52.
Table 2.
Frequency of vaccine-induced antibody responses.
| Group | Commercial ELISA | Commercial Western blot | ELISA EnvC | IP–Western blot |
|---|---|---|---|---|
| 2 mg (n = 5) | 0 (0) | 0 (0) | 1 (20) | 0 (0) |
| 4 mg (n = 20) | 11 (55) | 5 (25) | 12 (60) | 8 (40) |
| 8 mg (n = 15) | 3 (20) | 2 (13) | 11 (73) | 12 (80) |
| Placeboa (n = 8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
NOTE. Data are no. (%) of subjects. IP, immunoprecipitation.
Two subjects who became infected with HIV, both in the placebo group, are not included in this table. All other subjects (n = 48) remained polymerase chain reaction (PCR) negative throughout the study. Aside from the results for the 2 HIV-infected subjects, the Western blot results were all in the indeterminate category. A positive Western blot result required a band at p24 in addition to a band for at least 1 of the envelope glycoproteins (gp41, gp120, or gp160). If there were bands present that did not meet the positivity criteria, the result was reported as indeterminate.
Vaccine-specific antibody responses
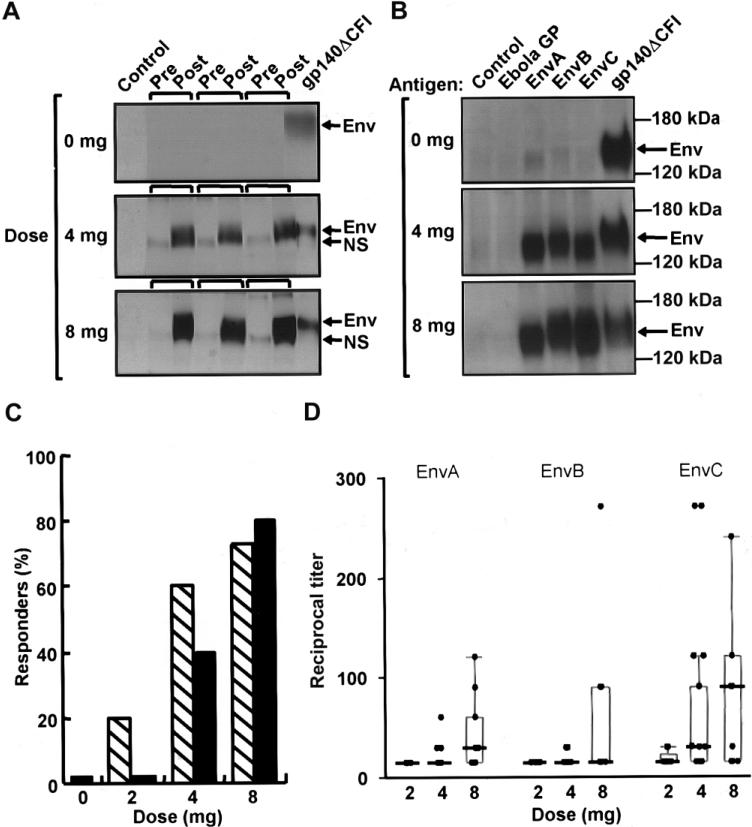
Env-specific antibody responses were observed by IP-Western blotting in postimmunization serum only (figure 2A). EnvA-, EnvB-, and EnvC-specific antibody responses were observed (figure 2B). The frequency of antibody responses increased with the dose; there were none observed at the 2-mg dose, 8 (40%) of 20 at the 4-mg dose, and 12 (80%) of 15 at the 8-mg dose (figure 2C, black bars).
Figure 2.
Induction, specificity, and dose response of the vaccine-induced antibody response to clades A, B, and C Env proteins. A, Western blot analysis of Env antigens captured by immunoprecipitation using prevaccination (pre) and 12-week (post) serum samples from representative placebo recipients (0 mg) and vaccine recipients (4- or 8-mg doses). Arrows indicate the EnvB-specific band distinguished from a small nonspecific (NS) background band. B, Specific reactivity of antibody induced by vaccination to EnvA, EnvB, and EnvC but not to an unrelated Ebola virus glycoprotein–negative control (Ebola GP). All proteins were produced in the supernatants of 293T cells transfected with the vaccine plasmids. Arrows indicate the Env-specific band. Results are shown for representative subjects from the placebo group (0 mg) and from the 4- and 8-mg groups. No positive bands were detected for any of the placebo recipients at any time point in the study. C, Frequency of positive antibody responders to purified proteins at week 12 as measured by end-point titration ELISA (hatched bars) or by immunoprecipitation followed by Western blotting (black bars), as a function of dose. No antibody was detected by these methods for placebo recipients or for vaccine recipients before immunization. D, Peak response to purified proteins for EnvA, EnvB, and EnvC as measured by end-point titration ELISA for each subject, by dose group. The peak response (reciprocal titer) occurred between weeks 8 and 24. The box plots indicate the median, 25th, and 75th percentiles for each dose level, and the error bars show the 5th and 95th percentile.
A dose effect for antibody response was also suggested by the results of an end-point titration ELISA for EnvA, EnvB, and EnvC (figure 2C and 2D). A positive result for at least 1 Env protein at week 12 was observed in 24 (60% [95% CI, 43%–75%]) of 40 subjects. All 24 positive subjects had detectable antibody against EnvC, 12 had detectable antibody against EnvA, and 9 had detectable antibody against EnvB. At week 12, 1 (20%) of 5 recipients of 2 mg, 12 (60%) of 20 recipients of 4 mg, and 11 (73%) of 15 recipients of 8 mg had a positive response (figure 2C, hatched bars). The peak antibody titer for each of the envelope antigens occurred between weeks 8 and 24 and was greatest for EnvC in the 8-mg group (figure 2D). Vaccine-induced HIV antibody was observed in only 1 subject by 1 test method in the 2-mg group. Serum neutralizing activity against the MN strain of HIV-1 was <80% in all subjects, indicating that no substantial neutralizing activity was present.
Vaccine-induced T cell responses
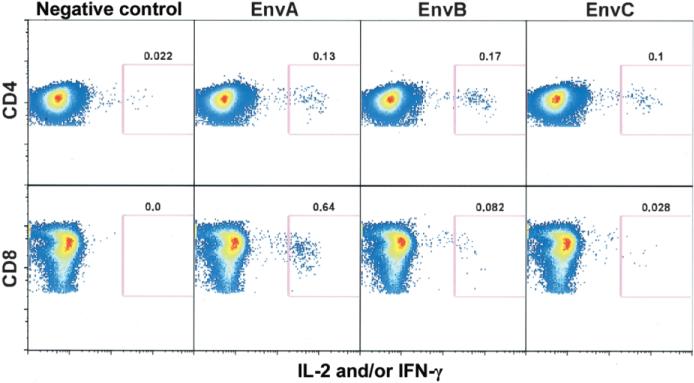
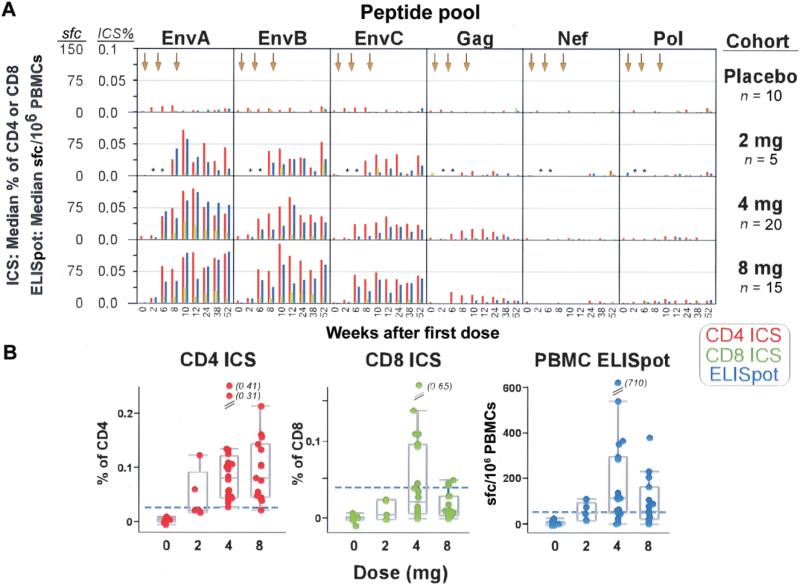
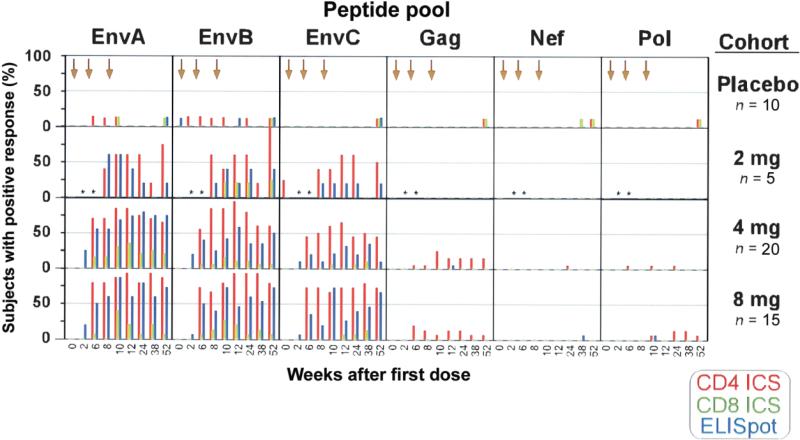
T cell responses were detected by both ELISpot and ICS assay and were most frequent to Env antigens. The ICS cytokine gating strategy is shown by a representative analysis at week 12, which demonstrates responses to EnvA, EnvB, and EnvC peptide pools in CD4+ and CD8+ T cells (figure 3). EnvA peptides elicited responses of the greatest magnitude (figure 4A and 4B). Responses were detected after 2 immunizations in most vaccine recipients, with the highest response rate observed at week 12 (4 weeks after the third immunization) (figure 5 and table 3). The median magnitude of the T cell response as measured by both ELISpot and ICS assay generally peaked at week 10 or 12 for all peptide pools and persisted through week 52 (figure 4A). For EnvA, week 12 T cell responses ranged from 0.024% to 0.41% of total CD4+ T cells (as measured by ICS assay), from 0.045% to 0.65% of total CD8+ T cells (as measured by ICS assay), and from 50 to 710 sfc/106 PBMCs (as measured by ELISpot assay) (figure 4B). False-positive results occurred, with a magnitude just over the threshold, at a frequency of 1 of 48 for the ICS assay for CD4+ T cells, 0 of 48 for the ICS assay for CD8+ T cells, and 2 of 48 for the ELISpot assay (figure 5).
Figure 3.
Measurement of HIV-specific T cell responses. The T cell response measurements were largely based on flow-cytometric detection of intracellular cytokine production in peripheral-blood mononuclear cells (PBMCs) stimulated with peptide pools representing the vaccine antigen. Because this is a relatively new approach for determining immunogenicity in clinical trials of preventive vaccines, an example of the primary data and gating strategy for this method is demonstrated for a representative subject. CD4+ and CD8+ T cell responses are shown for a single recipient of the 4-mg dose at week 12 (4 weeks after completion of the vaccination schedule). PBMCs were incubated either with costimulatory antibodies only (negative control) or with EnvA, EnvB, or EnvC peptide pools, and production of interferon (IFN)–γ and/or interleukin (IL)–2 was measured on the same wavelength. Gating (pink box) reflects cells producing higher levels of cytokine. The numeric value above each gate represents the percentage of total CD4+ or CD8+ T cells producing cytokine.
Figure 4.
Magnitude of T cell responses to specific vaccine components. T cell responses were measured by intracellular cytokine staining (ICS) assay to detect interferon (IFN)–γ and/or interleukin-2 and by IFN-γ enzyme-linked immunospot (ELISpot) assay for all placebo and vaccine recipients. A, Median magnitudes of peptide pool–specific responses, shown as a percentage of total CD4+ or CD8+ T cells for the ICS assay (scale 0–0.1) and as the no. of spot-forming cells per 106 peripheral-blood mononuclear cells (PBMCs) for the ELISpot assay (scale 0–150), for all subjects, by dose group. The time course for each subject is shown by study week on the X-axis; arrows along the top of the graph designate time points for immunizations (study weeks 0, 4, and 8). Within each of the 24 boxes, the entire time course of the study is represented. Each column shows the responses to a peptide pool representing the respective vaccine antigens (EnvA, EnvB, EnvC, Gag, Nef, and Pol). Each row represents a dose cohort (placebo recipients, 2-mg recipients, 4-mg recipients, and 8-mg recipients). Red bars represent CD4+ T cell responses as measured by ICS assay, green bars represent CD8+ T cell responses as measured by ICS assay, and blue bars represent CD4+ or CD8+ T cell responses as measured by ELISpot assay. Time points without samples for analysis are represented by asterisks. B, Magnitudes of specific T cell responses to a peptide pool for EnvA at study week 12, by dose group. EnvA-specific responses are shown for each subject as measured by 3 assays; shown are the percentages of CD4+ or CD8+ T cells producing cytokine as measured by ICS assay and the no. of spot-forming cells per 106 PBMCs as measured by ELISpot assay. The left plot shows EnvA-specific CD4+ T cell responses, the middle plot shows EnvA-specific CD8+ T cell responses, and the right plot shows EnvA-specific CD4+ or CD8+ responses. The box plots indicate the median, 25th, and 75th percentiles for each dose level, and the error bars show the 5th and 95th percentiles. The horizontal dashed line on each plot indicates the laboratory threshold of positivity for each assay (for the ICS assay, 0.0241% for CD4+ T cells and 0.0445% for CD8+ T cells; for the ELISpot assay, 50 sfc/106 PBMCs).
Figure 5.
Frequencies of subjects with detectable T cell responses. All T cell responses for each antigen are shown for all subjects at all time points. Each of the 24 boxes shows the frequency of response to a different antigen in a different dose group for the entire time course of the study. The Y-axis of each of the 24 boxes shows the frequency of response as the percentage of subjects in a dose group with a positive response to the respective peptide pool for each assay. The X-axis shows the time course of the study by study week (0–52) in each of the 24 plots. Each row represents a dose cohort (placebo recipients, 2-mg recipients, 4-mg recipients, and 8-mg recipients). Each column represents the peptide pool used for stimulation of peripheral-blood mononuclear cells (EnvA, EnvB, EnvC, Gag, Nef, and Pol). Red bars represent CD4+ T cell responses as measured by intracellular cytokine staining (ICS) assay, green bars represent CD8+ T cell responses as measured by ICS assay, and blue bars represent CD4+ or CD8+ T cell responses as measured by enzyme-linked immunospot (ELISpot) assay. No week 2 data and only 1 sample from week 6 are included for the 2-mg cohort (time points without samples for analysis are represented by asterisks).
Table 3.
Frequency of T cell responses at weeks 12 and 52, as assessed by intracellular cytokine staining assay.
| Week 12 frequency | Week 52 frequency | |||
|---|---|---|---|---|
| T cell subset, peptide pool | 4-mg group (n = 20) | 8-mg group (n = 15) | 4-mg group (n = 20) | 8-mg group (n = 15) |
| CD4+ T cells | ||||
| EnvA | 17 (85) | 14 (93) | 13 (65) | 13 (87) |
| EnvB | 19 (95) | 14 (93) | 12 (60) | 12 (80) |
| EnvC | 13 (65) | 11 (73) | 9 (45) | 11 (73) |
| Gag | 3 (15) | 2 (13) | 3 (15) | 1 (7) |
| Nef | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Pol-1a | 0 (0) | 0 (0) | 0 (0) | 1 (7) |
| Pol-2a | 0 (0) | 0 (0) | 1 (5) | 2 (13) |
| Any | 20 (100) | 14 (93) | 14 (70) | 14 (93) |
| CD8+ T cells | ||||
| EnvA | 7 (35) | 3 (20) | 4 (20) | 1 (7) |
| EnvB | 2 (10) | 3 (20) | 1 (5) | 1 (7) |
| EnvC | 0 (0) | 1 (7) | 0 (0) | 0 (0) |
| Gag | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Nef | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Pol-1a | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Pol-2a | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Any | 7 (35) | 5 (33) | 4 (20) | 1 (7) |
NOTE. Data are no. (%) of subjects.
Pol protein is too large to include all peptides in 1 pool; therefore, the Pol peptides were divided into 2 pools.
As measured by ELISpot assay, there was a statistically significant difference in the number of positive EnvA, EnvB, and EnvC responses in vaccine recipients (n = 39) at week 12, compared with that at baseline (P < .0001, P < .0001, and P = .001, respectively, exact McNemar tests). Gag, Pol, and Nef responses among vaccine recipients were not significantly different between baseline and week 12.
Positive CD4+ T cell responses as measured by ICS assay for at least 1 of the vaccine-specific antigens were detected in 39 (97.5%) of the 40 vaccine recipients for at least 1 time point by week 12. At week 12, compared with that at baseline, there was a statistically significant difference (P ≤ .0001 for all, exact McNemar tests) in the number of vaccine recipients with a CD4+ T cell response to EnvA, EnvB, and EnvC but not to Gag, Pol, or Nef.
Positive CD8+ T cell responses as measured by ICS assay for peptides representing at least 1 of the vaccine antigens were detected in 16 (40%) of the 40 vaccine recipients by week 12. At week 12, compared with that at baseline, there was a statistically significant difference (P = .002 and P = .0313, respectively, exact McNemar tests) in the number of vaccine recipients with a CD8+ T cell response to EnvA and EnvB but not to EnvC; no vaccine recipients had a CD8+ T cell response to Gag, Pol, or Nef.
Although there was a trend toward a greater magnitude (figure 4) and frequency (figure 5) of T cell responses in recipients of the 4- or 8-mg dose than in the recipients of the 2-mg dose, this difference was not statistically significant at week 12. The pattern of responses as measured by ELISpot assay throughout the study was consistent with the pattern as measured by ICS assay. The frequency and magnitude were greatest for EnvA and EnvB, with weaker responses to EnvC, few responses to Gag, and no responses to Nef or Pol (figure 5).
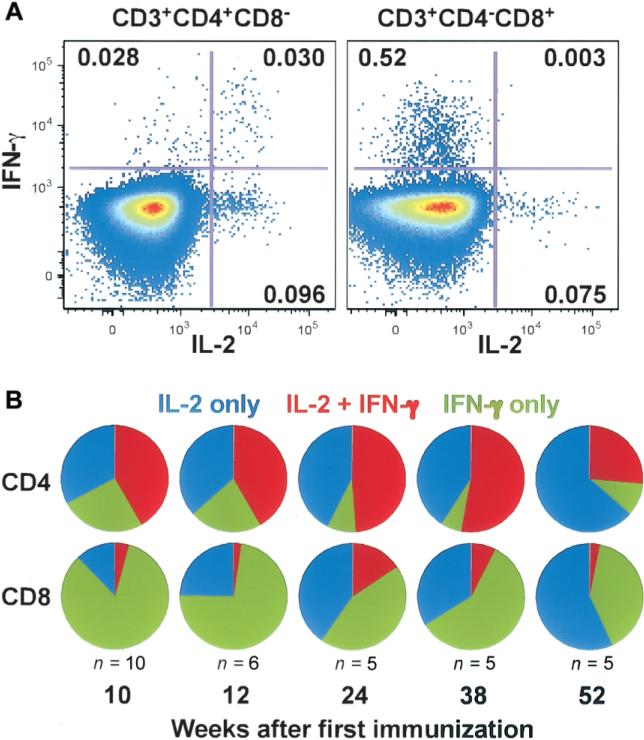
Once T cell responses were detected, they remained detectable in the majority of subjects for the 52 weeks of follow-up (figures 4A and 5 and table 3). The functional properties of the vaccine-induced T cell response changed qualitatively over time. A representative T cell cytokine-production profile for IL-2 and IFN-γ at week 12 is shown in figure 6A. The CD4+ and CD8+ T cell subsets contain cells that make IL-2 only, IFN-γ only, or both. At week 12, CD4+ T cells tended to make IL-2 only or in combination with IFN-γ, whereas the great majority of CD8+ T cells typically produced IFN-γ only (figure 6A). At week 52, both T cell subsets tended to have a higher frequency of IL-2–producing cells and a lower frequency of IFN-g–producing cells (figure 6B).
Figure 6.
Evolution of cytokine-expression patterns in vaccine-induced T cells over time. Multiparameter flow cytometry was performed in a subset of subjects to define the relative frequency of interferon (IFN)–γ and interleukin (IL)–2 production in T cells stimulated by the vaccine-specific peptide pools. A, Response of either CD4+ or CD8+ T cells from a representative vaccine recipient is shown at study week 12 after stimulation with the EnvA peptide pool. For each of the 2 plots, the Y-axis shows CD3+ (CD4+ or CD8+) T cells producing IFN-γ, whereas the X-axis shows IL-2 production. The plot on the left demonstrates CD4+ T cell production of IL-2 alone (in the bottom right quadrant; 0.096%), IFN-γ alone (in the top left quadrant; 0.028%), or both (in the top right quadrant; 0.030%). The plot on the right demonstrates CD8+ T cell production of IFN-γ alone (in the top left quadrant; 0.52%), IL-2 alone (in the bottom right quadrant; 0.075%), or both (in the top right quadrant; 0.003%). Over time, the relative frequency of cells producing IFN-γ appeared to diminish, and the relative frequency of vaccine-specific IL-2–producing cells appeared to increase, as shown in panel B by use of pie charts demonstrating the average cytokine-expression pattern from a subset of vaccine recipients with the highest initial responses to the EnvA peptide pool over the 52-week time course of the study. There are 2 sets of pie charts: the top row shows CD4+ T cells, and the bottom row shows CD8+ T cells, both as measured by intracellular cytokine staining assay. Five time points of the study are represented (week 10, 12, 24, 38, and 52), and the no. of representative subjects included at each time point is listed. The pie charts show the distribution of cytokine-expression patterns of each T cell responding to the EnvA peptide pool: the blue segments represent T cells producing IL-2 only, the red segments represent T cells producing both IL-2 and IFN-γ, and the green segments represent T cells producing IFN-γ only.
DISCUSSION
This 4-plasmid HIV-1 DNA candidate vaccine was assessed as safe and well tolerated in healthy uninfected adults. Importantly, DNA immunization induced both HIV-1–specific antibody and T cell responses to the Env proteins of the 3 major subtypes of HIV-1. We have shown that DNA as a gene-delivery platform for HIV-1 vaccine antigen expression can induce significant immune responses that are sustained for at least 1 year in the majority of antigen-naive subjects. The concept of using bacteria-derived plasmid DNA to deliver vaccine antigens has many attractive features, including ease and flexibility of construction, scalable manufacturing capacity, stability, intracellular production of vaccine antigen, transient expression, no induction of antivector immunity, induction of a balanced CD4+ and CD8+ T cell response as well as of antibody, and lack of significant local or systemic reactogenicity. The broad immunogenicity of the VRC HIV-1 DNA candidate vaccine probably reflects a combination of factors, including optimization of vector design, manufacturing methods, delivery, sample processing, and immunological end-point measurements.
A notable feature of the immunogenicity analysis was the response to Env antigens relative to Gag, Pol, and Nef. The dominant antigenicity of Env was observed for both antibody and T cell responses, with EnvA peptides eliciting the greatest T cell responses and EnvC binding the most vaccine-induced antibody. All the genes encoding the vaccine antigens were codon modified to improve expression. The Env constructs received additional modification to remove the cleavage site, the fusion peptide of gp41, a region of the interspace between the 2 gp41 heptad repeats, and the cytoplasmic domain [3], but there is no indication that these changes were responsible for liberating additional T cell epitopes. The gag/pol/nef construct was designed to remove the frame shift, resulting in improved expression of Pol and Nef, at the expense of 66 aa from the carboxy-terminus of Gag [18]. This construct was immunogenic in preclinical testing and exhibited strong protein expression in vitro [4]. Here, the Gag, Pol, and Nef antigens elicited only a modest induction of CD4+ T cell responses despite representing 50% of the plasmid mixture by weight. It is not known why the induction of Gag-, Pol-, and Nef-specific responses were lower than Env-specific responses, but there are several possible explanations for increased Env immunogenicity. First, since Env is a membrane glycoprotein, its intracellular trafficking and mode of antigen presentation differs from those of the internal viral proteins. Second, Gag, Pol, and Nef are not glycosylated, are synthesized in the cytoplasm, and localize to a subcellular compartment different from that of Env. Therefore, it is possible that the steps involved in antigen processing and the kinetics of antigen presentation may differ. Finally, our experiments in animal models indicate that Env contains more CD4+ T cell epitopes than does Gag or other proteins [19]. Thus, the increased immunogenicity of Env is potentially an intrinsic property of the expressed antigen.
Antibody responses induced by this 4-plasmid combination vaccine were detected by IP-Western blotting and research ELISA for vaccine-specific antigens as well as by commercial ELISA and Western blotting for native viral proteins (table 2). Analysis of neutralization did not reveal significant activity. On the basis of animal studies, high levels of binding antibody and neutralizing activity are not anticipated after DNA immunization alone but can be achieved after subsequent boosting with replication-defective recombinant adenovirus vectors [20]. The induction of measurable antibody to clades A, B, and C Env proteins with DNA alone in humans is encouraging and suggests that DNA vaccination is a viable strategy for priming both cellular and humoral responses. Whether the response to all the Env proteins indicates cross-reactivity between clades or between clade-specific antibodies is not known. As multivalent vaccines become more immunogenic and clinical evaluation expands, the complexity of interpreting routine commercial diagnostic tests will increase. Alternative approaches to serological diagnosis, such as gene-based diagnostic approaches, will be needed in the future.
This DNA vaccine consistently induced T cell responses. Envspecific CD4+ T cell responses were present in nearly all subjects. Detection of this response may be attributed to the development of a highly sensitive flow cytometry–based assay that detects IFN-γ and/or IL-2 on the same channel. Other studies have shown that most of the CD4+ T cell response can be captured by the combination of these 2 cytokines [21, 22]; if IFN-γ alone is used as the end point, a portion of the CD4+ T cell response is missed. CD8+ T cells are key effectors for clearing virus-infected cells [23] and have been associated with control of lentivirus replication in both nonhuman primates (NHPs) and humans [24–32]. Twelve (34%) of the 35 vaccine recipients who received the 4-mg or 8-mg doses had detectable HIV Env–specific CD8+ T cell responses at week 12. The magnitude of positive responses ranged from just above background to 0.65% of total CD8+ T cells. When detected, the magnitude of the CD8+ T cell response is comparable to that seen in NHP studies of similar vaccines, although typically CD8+ T cell responses can be detected in 100% of the animals [33, 34]. An interesting feature of the T cell response was that, over the course of a year, a higher frequency of EnvA-specific CD4+ and CD8+ T cells made IL-2 only. Reports that show that IFN-γ–producing CD4+ T cells are not long lived [35] and that IL-2–producing CD8+ T cells have a greater proliferative capacity in HIV-1–infected patients with nonprogressive disease [36] suggest that these IL-2–producing T cells may have the capacity for expansion after subsequent antigen exposure.
Demonstrating the safety and immunogenicity of a multigene, multiclade HIV DNA vaccine is an important step in the development of a globally relevant vaccination strategy. The capacity to make precise measurements of vaccine-induced T cell responses will improve the chances of defining correlates of immune protection in future trials.
Acknowledgments
We thank the study volunteers and the many community organizations that support the importance of being involved in the clinical evaluation of candidate HIV vaccines. We also thank the National Institutes of Health Clinical Center staff, the Clinical Center Pharmacy staff (Judith Starling, Hope Decederfelt, and others), the National Institute of Allergy and Infectious Diseases (NIAID) staff, the Patient Recruitment and Public Liaison and Office of Communications and Public Liaison staff, the members of the NIAID Intramural Data and Safety Monitoring Board, the Capital Area Vaccine Effort members, EMMES Corporation (Phyllis Zaia, Lihan Yan, and others), Vical (Kevin Bracken, Jennifer Meek, and others), and other supporting staff (Richard Jones, Nancy Barrett, Bryan Glover, Katina Bryan, Monique Young, and Ariella Blejer) who made this work possible. We are grateful as well for the advice and important preclinical contributions of Vaccine Research Center investigators and key staff, including Daniel Douek, Doria (Woody) Dubois, Jennifer Fischer, Yue Huang, Wing-Pui Kong, Peter Kwong, Laurence Lemiale, Norman Letvin, Abraham Mittelman, Steve Perfetto, Srini Rao, John Rathmann, Rebecca Sheets, Robert Seder, Judith Stein, Ellen Turk, Jessica Wegman, Richard Wyatt, Ling Xu, and Zhi-yong Yang.
Financial support: National Institute of Allergy and Infectious Diseases intramural research program.
Footnotes
Potential conflicts of interest: G.J.N. and B.K.C. are named on patent applications for the vaccine concept presented in this work. All other authors report no potential conflicts of interest.
VACCINE RESEARCH CENTER (VRC) 004 STUDY TEAM
Brenda Larkin, Michael Scott, Colleen Thomas, Sarah Hubka, Ingelise Gordon, Pamela Edmonds, Steve Rucker, Laura Novik, Tiffany Alley, Lin Gu, Lewis McCurdy, Andrew Catanzaro, Janie Parrino, and Joseph Casazza of the VRC, National Institute of Allergy and Infectious Diseases (NIAID), and Jorge Flores of the Division of AIDS, NIAID.
Clinicaltrials.gov registry number: NCT00047931.
References
- 1.UNAIDS/WHO . AIDS epidemic update: December 2005. UNAIDS; Geneva: 2005. [Google Scholar]
- 2.Nabel G, Makgoba W, Esparza J. HIV-1 diversity and vaccine development. Science. 2002;296:2335. doi: 10.1126/science.296.5577.2335. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti BK, Kong WP, Wu BY, et al. Modifications of human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76:5357–68. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong WP, Huang Y, Yang ZY, Chakrabarti BK, Moodie Z, Nabel GJ. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J Virol. 2003;77:12764–72. doi: 10.1128/JVI.77.23.12764-12772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rollman E, Hinkula J, Arteaga J, et al. Multi-subtype gp160 DNA immunization induces broadly neutralizing anti-HIV antibodies. Gene Ther. 2004;11:1146–54. doi: 10.1038/sj.gt.3302275. [DOI] [PubMed] [Google Scholar]
- 6.Donnelly JJ, Friedman A, Martinez D, et al. Preclinical efficacy of a prototype DNA vaccine: enhanced protection against antigenic drift in influenza virus. Nat Med. 1995;1:583–7. doi: 10.1038/nm0695-583. [DOI] [PubMed] [Google Scholar]
- 7.Wang R, Doolan DL, Le TP, et al. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science. 1998;282:476–80. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- 8.Roy MJ, Wu MS, Barr LJ, et al. Induction of antigen-specific CD8+ T cells, T helper cells, and protective levels of antibody in humans by particle-mediated administration of a hepatitis B virus DNA vaccine. Vaccine. 2000;19:764–78. doi: 10.1016/s0264-410x(00)00302-9. [DOI] [PubMed] [Google Scholar]
- 9.MacGregor RR, Boyer JD, Ugen KE, et al. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J Infect Dis. 1998;178:92–100. doi: 10.1086/515613. [DOI] [PubMed] [Google Scholar]
- 10.MacGregor RR, Boyer JD, Ciccarelli RB, Ginsberg RS, Weiner DB. Safety and immune responses to a DNA-based human immunodeficiency virus (HIV) type I env/rev vaccine in HIV-infected recipients: follow-up data. J Infect Dis. 2000;181:406. doi: 10.1086/315199. [DOI] [PubMed] [Google Scholar]
- 11.MacGregor RR, Ginsberg R, Ugen KE, et al. T-cell responses induced in normal volunteers immunized with a DNA-based vaccine containing HIV-1 env and rev. AIDS. 2002;16:2137–43. doi: 10.1097/00002030-200211080-00005. [DOI] [PubMed] [Google Scholar]
- 12.Chakrabarti BK, Ling X, Yang ZY, et al. Expanded breadth of virus neutralization after immunization with a multiclade envelope HIV vaccine candidate. Vaccine. 2005;23:3434–45. doi: 10.1016/j.vaccine.2005.01.099. [DOI] [PubMed] [Google Scholar]
- 13.Sheets RL, Stein J, Manetz TS, et al. Toxicological safety evaluation of DNA plasmid vaccines against HIV-1, Ebola, severe acute respiratory syndrome, or West Nile virus is similar despite differing plasmid backbones or gene-inserts. Toxicol Sci. 2006;91:620–30. doi: 10.1093/toxsci/kfj170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheets RL, Stein J, Manetz TS, et al. Biodistribution of DNA plasmid vaccines against HIV-1, Ebola, severe acute respiratory syndrome, or West Nile virus is similar, without integration, despite differing plasmid backbones or gene inserts. Toxicol Sci. 2006;91:610–9. doi: 10.1093/toxsci/kfj169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malenbaum SE, Yang D, Cheng-Mayer C. Evidence for similar recognition of the conserved neutralization epitopes of human immunodeficiency virus type 1 envelope gp120 in humans and macaques. J Virol. 2001;75:9287–96. doi: 10.1128/JVI.75.19.9287-9296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mascola JR, Louder MK, Winter C, et al. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J Virol. 2002;76:4810–21. doi: 10.1128/JVI.76.10.4810-4821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catanzaro AT, Koup RA, Roederer M, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J Infect Dis. 2006;194:1638–49. doi: 10.1086/509258. in this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Kong WP, Nabel GJ. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J Virol. 2001;75:4947–51. doi: 10.1128/JVI.75.10.4947-4951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L, Kong WP, Nabel GJ. Enhanced breadth of CD4 T-cell immunity by DNA prime and adenovirus boost immunization to human immunodeficiency virus Env and Gag immunogens. J Virol. 2005;79:8024–31. doi: 10.1128/JVI.79.13.8024-8031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascola JR, Sambor A, Beaudry K, et al. Neutralizing antibodieselicited by immunization of monkeys with DNA plasmids and recombinant adenoviral vectors expressing human immunodeficiency virus type 1 proteins. J Virol. 2005;79:771–9. doi: 10.1128/JVI.79.2.771-779.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Rosa SC, Lu FX, Yu J, et al. Vaccination in humans generates broad T cell cytokine responses. J Immunol. 2004;173:5372–80. doi: 10.4049/jimmunol.173.9.5372. [DOI] [PubMed] [Google Scholar]
- 23.Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–2. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- 24.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–10. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barouch DH, Santra S, Schmitz JE, et al. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–92. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 26.Barouch DH, Kunstman J, Kuroda MJ, et al. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature. 2002;415:335–9. doi: 10.1038/415335a. [DOI] [PubMed] [Google Scholar]
- 27.Koup RA, Safrit JT, Cao Y, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–5. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitz JE, Kuroda MJ, Santra S, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 29.Pinto LA, Sullivan J, Berzofsky JA, et al. ENV-specific cytotoxic T lymphocyte responses in HIV seronegative health care workersoccupationally exposed to HIV-contaminated body fluids. J Clin Invest. 1995;96:867–76. doi: 10.1172/JCI118133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogg GS, Jin X, Bonhoeffer S, et al. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–6. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 31.Rowland-Jones S, Sutton J, Ariyoshi K, et al. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 32.Rowland-Jones SL, Nixon DF, Aldhous MC, et al. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet. 1993;341:860–1. doi: 10.1016/0140-6736(93)93063-7. [DOI] [PubMed] [Google Scholar]
- 33.Seaman MS, Xu L, Beaudry K, et al. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J Virol. 2005;79:2956–63. doi: 10.1128/JVI.79.5.2956-2963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santra S, Seaman MS, Xu L, et al. Replication-defective adenovirus serotype 5 vectors elicit durable cellular and humoral immune responses in nonhuman primates. J Virol. 2005;79:6516–22. doi: 10.1128/JVI.79.10.6516-6522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu CY, Kirman JR, Rotte MJ, et al. Distinct lineages of TH1 cells have differential capacities for memory cell generation in vivo. Nat Immunol. 2002;3:852–8. doi: 10.1038/ni832. [DOI] [PubMed] [Google Scholar]
- 36.Zimmerli SC, Harari A, Cellerai C, Vallelian F, Bart PA, Pantaleo G. HIV-1-specific IFN-gamma/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc Natl Acad Sci USA. 2005;102:7239–44. doi: 10.1073/pnas.0502393102. [DOI] [PMC free article] [PubMed] [Google Scholar]