Abstract
A pleiotropic cytokine, tumor necrosis factor-α (TNFα), regulates the expression of multiple macrophage gene products and thus contributes a key role in host defense. In this study, we have investigated the specificity and mechanism of activation of members of the c-Jun-NH2-terminal kinase/stress-activated protein kinase (JNK/SAPK) subfamily of mitogen-activated protein kinases (MAPKs) in mouse macrophages in response to stimulation with TNFα. Exposure of macrophages to TNFα stimulated a preferential increase in catalytic activity of the p46 JNK/SAPK isoform compared with the p54 JNK/SAPK isoform as determined by: (i) separation of p46 and p54 JNK/SAPKs by anion exchange liquid chromatography and (ii) selective immunodepletion of the p46 JNK/SAPK from macrophage lysates. To investigate the level of regulation of p46 JNK/SAPK activation, we determined the ability of MKK4/SEK1/JNKK, an upstream regulator of JNK/SAPKs, to phosphorylate recombinant kinase-inactive p46 and p54 JNK/SAPKs. Endogenous MKK4 was able to transphosphorylate both isoforms. In addition, both the p46 and p54 JNK/SAPK isoforms were phosphorylated on their TPY motif in response to TNFα stimulation as reflected by immunoblotting with a phospho-specific antibody that recognizes both kinases. Collectively, these results suggest that the level of control of p46 JNK/SAPK activation is distal not only to MKK4 but also to the p54 JNK/SAPK. Preferential isoform activation within the JNK/SAPK subfamily of MAPKs may be an important mechanism through which TNFα regulates macrophage phenotypic heterogeneity and differentiation.
Tumor necrosis factor-α (TNFα), a pleiotropic cytokine produced mainly by macrophages, has been shown to regulate the expression of many of the individual functional activities and gene products that collectively mediate the heterogeneous role of macrophages in inflammation, wound repair, and fibrosis (1, 2). Work reported by this laboratory has focused on delineating the mechanisms controlling the expression of insulin-like growth factor I (IGF-I), a macrophage-derived growth factor implicated in normal wound repair and in the pathologic processes involved in tissue fibrosis, especially pulmonary fibrosis (3), and for which TNFα has been shown to play an integral role (4, 5). In previously reported work, we showed that stimulation of mouse macrophages with TNFα led to an increase in IGF-I protein synthesis that was regulated at a pretranslational level following specific ligation of the TNFα receptor CD120a (p55) (6, 7). The 5′ flanking region of the IGF-I gene contains canonical cis-regulatory sequences for several transcription factors including AP- 1 (8). Because AP-1 has been shown to be activated by TNFα (9) and its DNA-binding activity is mediated by the homodimeric protooncogene product c-Jun or the heterodimeric c-Jun-c-Fos (10), the present study focuses on the signaling mechanisms utilized by TNFα that may contribute to the activation of AP-1.
In seeking to define these events, we have investigated the activation of members of the mitogen-activated protein kinase (MAPK) family of protein Ser/Thr kinases in mouse bone marrow-derived macrophages and whose downstream targets include both c-Jun and c-Fos. Currently, the MAPK family comprises three subfamilies, namely: (i) the extracellular signal-regulated kinases or ERK subfamily represented by p42mapk/erk2 and p44mapk/erk1 (11), (ii) the c-Jun NH2-terminal kinases/stress-activated protein kinases or JNK/SAPK subfamily, represented by the p46 JNK/SAPK and p54 JNK/SAPK isoforms and their variants (12–16), and (iii) the p38mapk subfamily (17). Previous work from this laboratory has shown that ligation of CD120a (p55) by TNFα in mouse macrophages stimulates a rapid, transient, and selective activation of the p42mapk/erk2 (18) and that ligation of CD 120a (p55) is rapidly (within 30 sec) followed by the activation of MEK kinase (MEKK) and by the c-Raf-1-independent activation of the specific MEK isoform, MEK1 (19, 20). Thus, unlike the responses of many other cell types in which dual activation of p42mapk/erk2 and p44mapk/erk1 occurs in a c-Raf-1-dependent fashion (21), the activation of the ERK family in macrophages in response to TNFα is restricted at the level of both the p42mapk/erk2 and the upstream kinases that regulate its activation. Although there are clear examples in which both p42mapk/erk2 and p44mapk/erk1 have been shown to be equally effective at phosphorylating and activating the same substrate, each enzyme has also been shown to display the ability to phosphorylate distinct substrates (22). Thus, the pattern of restricted activation of ERKs seen in macrophages is strongly suggestive that each kinase may phosphorylate a distinct group of substrates.
The JNK/SAPK subfamily of MAPKs has also been shown to be activated by TNFα (9, 13, 23) and has been defined principally on the basis of the ability of its members to activate the transcription factor c-Jun by phosphorylating residues Ser 63 and 73 located in the transactivation domain of the NH2 terminus of c-Jun (12). As with the ERK subfamily, emerging data suggest that p46 and p54 JNK/SAPKs, while showing some overlap in their catalytic activity, may also express distinct substrate preferences. For example, Kallunki et al. (24) have shown p54 JNK/SAPK to bind c-Jun with 25-fold greater affinity than p46 JNK/SAPK and have suggested that at the concentration of c-Jun encountered intracellularly, p54 JNK/SAPK is more likely to phosphorylate c-Jun than p46 JNK/SAPK, raising the question of alternative role(s) of p46 JNK/SAPK. Similarly, in COS cells expressing epitope-tagged p46 JNK/SAPK and p54 JNK/SAPK, it was shown that the specific activation of p54 JNK/SAPK was approximately 10-fold greater than that of p46 JNK/SAPK (23). Importantly, these investigators showed that in HOG1-deficient yeast (which are overly sensitive to osmotic stress), human p46 JNK/SAPK, but not p54 JNK/SAPK, was able to complement the deficiency in HOG1 expression, establishing that p46 and p54 JNK/SAPKs are not simply redundant kinases but can exhibit distinct functions (23). More recently, it has been suggested that in small cell lung cancer, activation of different JNK isoform activities may dictate the relative responsiveness of these tumor cells to the apoptotic effects of UV-irradiation with JNK1 activation signaling greater apoptosis than the signaling by the JNK2 isoform (25).
These issues thus raise the question of the role that signaling heterogeneity within the JNK/SAPK subfamily may play in the varied functional responses in macrophages (26), in particular with respect to the activation of AP-1. To address this question, we have investigated the restriction of JNK/SAPK activation in mouse bone marrow-derived macrophages following exposure to TNFα. In this report we show that although both major isoforms of JNK/SAPK are present in mouse macrophages, TNFα preferentially activates the p46 JNK/SAPK isoform. To establish the level of this isoform preference, we will show that endogenous macrophage MKK4/JNKK/SEK1 is able to transphosphorylate both p46 and p54 JNK/SAPK isoforms and that both isoforms are phosphorylated in response to stimulation with TNFα. Thus, these data suggest that the level of control of the preferential activation of p46 JNK/SAPK is likely to be distal to the p54 JNK/SAPK.
MATERIALS AND METHODS
Materials.
C3H/HeJ mice were used throughout the study and were bred at the National Jewish Center Biological Resource Center. Rabbit polyclonal anti-p46 JNK/SAPK, rabbit polyclonal anti-MKK4 (C-20), and phospho-specific JNK/SAPK antibodies were purchased from Santa Cruz Biotechnology. Anti-SAPK antibody raised against p54 SAPKβ in rabbits was a kind gift from John Kyriakis (Boston, MA). Anti-JNKK (MKK4, SEK1) antibody used for immunoprecipitation was purchased from PharMingen. Phospho-specific SEK1 (MKK4, JNKK) antibody was obtained from New England Biolabs. Recombinant c-Junl-79-GST and histidine-tagged kinase inactive p46 JNK/SAPK were generously provided by Gary Johnson (National Jewish Medical and Research Center, Denver, CO). Recombinant kinase inactive p54 SAPKβ-GST was a kind gift from James Woodgett (Princess Margaret Hospital, Toronto).
Macrophage Isolation and Culture.
Monolayers of mouse bone marrow-derived macrophages were prepared as previously described (6). On day 5, the colony-stimulating factor 1-containing medium was replaced with colony-stimulating factor 1-free medium [DMEM with penicillin (100 units/ml), streptomycin (100 μg/ml), and 0.1% (vol/vol) heat-inactivated FCS] for 18 hr prior to stimulation with TNFα.
Determination of JNK/SAPK Activity.
For measurement of JNK/SAPK activity, macrophage monolayers were stimulated as described (18) and then lysed at 4°C with 500 μl of an ice-cold lysis buffer [50 mM Tris⋅HCl buffer, pH 8.0, containing 137 mM NaCl, 10% glycerol, 1% (vol/vol) Nonidet P-40, 1 mM NaF, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 2 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride (PMSF)] (12). JNK/SAPK activity in each sample of lysate was bound to 15 μl of 1:1 slurry of lysis buffer:c-Junl-79-GST-Sepharose beads and incubated at 4°C for 2 hr. The beads were then washed twice with lysis buffer and twice with JNK/SAPK buffer (20 mM Hepes buffer, pH 7.2, containing 30 mM β-glycerophosphate, 10 mM p-nitrophenylphosphate, 10 mM MgCl2, 0.5 mM DTT, and 50 μM Na3VO4). The activity of JNK/SAPK was detected by phosphorylation of the c-Jun-GST fusion protein in an in vitro kinase assay and was assessed by incorporation of [γ-32P]ATP (10 μCi/sample; 1 Ci = 37 GBq) in JNK/SAPK buffer incubated at 30°C for 30 min. The phosphorylation reactions were terminated by the addition of an equal amount of 2× Laemmli sample buffer containing 20 mM DTT and boiled for 5 min. The proteins present in the supernatant were separated by SDS/PAGE and transferred onto nitrocellulose membranes, and 32P-labeled c-Jun-GST was detected by autoradiography. For immunoprecipitation of JNK/SAPK, the cell lysates were initially precleared with 15 μl of protein A-Sepharose beads for 10 min at 4°C. Then 1 μg of p46 JNK/SAPK antibody was added along with 15 μl of a 1:1 slurry of lysis buffer:protein A-Sepharose beads. The samples were incubated for 2 hr at 4°C. The JNK/SAPK assay was carried out in the presence of JNK/SAPK buffer, [γ-32P]ATP (10 μCi/sample), and 2 μg of soluble c-Jun1–79-GST per sample.
Resolution of JNK/SAPK Isoform Activities by Liquid Chromatography.
Unstimulated or TNFα-stimulated macrophage monolayers (≈11 × 106 cells) were scraped into 500 μl ice-cold lysis buffer [50 mM β-glycerophosphate buffer, pH 7.2, containing 1 mM EGTA, 2 mM MgCl2, 0.5% (vol/vol) Triton X-100, 1 mM DTT, 0.1 mM Na3VO4, 2 μg/ml leupeptin, and 2 μg/ml aprotinin]. Postnuclear supernatant was loaded onto an anion-exchange column (Mono Q, Biologic, Hercules, CA) equilibrated in 50 mM β-glycerophosphate, pH 7.2, containing 1 mM EGTA, 1 mM DTT, 0.1 mM Na3VO4, and 0.05 M NaCl and was eluted at a flow rate of 60 ml/hr for 40 ml with a linear gradient of 0.05–0.2 M NaCl in equilibration buffer. Two-milliliter fractions were collected, and 500-μl aliquots were tested for JNK/SAPK activity as described above. In addition, TCA-precipitated fractions were analyzed for JNK/SAPK proteins by SDS/PAGE, transferred to nitrocellulose membranes, and immunoblotted with p46 JNK/SAPK and p54 JNK/SAPK antibodies.
In Vitro Kinase Assay Of MKK4.
MKK4 catalytic activity was measured by immunoprecipitating MKK4 and subjecting it to an in vitro kinase assay using kinase-inactive p46 or p54 JNK/SAPKs as substrates in the presence of [γ-32P]ATP. Mouse macrophages were lysed with 1 ml EB lysis buffer [10 mM Tris⋅HCl buffer, pH 7.4, containing 5 mM EDTA, 50 mM NaCl, 0.1% (wt/vol) BSA, 1.0% (vol/vol) Triton X-100, 5 mM NaF, 1 mM PMSF, 2 mM Na3VO4, and 20 μg/ml aprotinin], and postnuclear supernatants were precleared with 15 μl of protein A-Sepharose beads. MKK4 was immunoprecipitated by incubation at 4°C for 2 hr with 0.5 μg of anti-JNKK (MKK4, SEK1) antibody and 15 μl of protein A-Sepharose beads. The beads were then washed twice with EB lysis buffer and twice with PAN buffer (10 mM Pipes buffer, pH 7.0, containing 21 μg/ml aprotinin, and 100 mM NaCl). MKK4 activity was assayed in a kinase reaction buffer [40 mm Hepes buffer, pH 7.2, containing 25 mM β-glycerophosphate, 2 mM Na3VO4, 20 mM MgCl2, 2 mM DTT, [γ-32P]ATP (20 μCi/sample)] and 1 μg of either kinase inactive p46 JNK/SAPK or kinase inactive p54 SAPKβ-glutathione S-transferase (GST) per sample for 30 min at 30°C. The phosphorylation reactions were terminated with an equal volume of 2× Laemmli sample buffer containing 20 mM DTT and boiled for 5 min. The phosphorylated proteins were separated by SDS/PAGE and electroblotted onto nitrocellulose membranes.
Western Blot Analysis.
Samples were separated by SDS/PAGE, transferred onto nitrocellulose membranes, and probed with the p46 JNK/SAPK, p54 JNK/SAPK, phospho-specific JNK/SAPK, or MKK4 antibodies as described (27). Bound antibodies were then detected by enhanced chemiluminescence (Amersham, Arlington Heights, IL) as previously described (18, 20).
RESULTS
Activation of JNK/SAPK by TNFα.
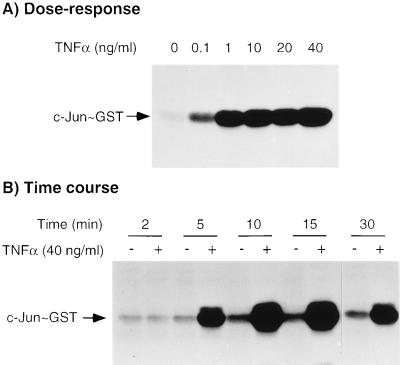
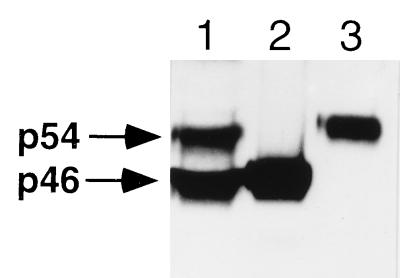
To investigate the activation of JNK/SAPK, monolayers of mouse bone marrow-derived macrophages were exposed to increasing concentrations of TNFα (0.1–40 ng/ml) for 10 min at 37°C before being lysed and analyzed for total JNK/SAPK activity. As illustrated in Fig. 1A, stimulation with TNFα resulted in a concentration-dependent activation of JNK/SAPK that peaked at a TNFα concentration of 1 ng/ml. The time course of activation of JNK/SAPK was investigated by incubating macrophage monolayers with 40 ng/ml of TNFα or with medium alone for time intervals up to 30 min before assaying for JNK/SAPK activity. As shown in Fig. 1B, peak activation of JNK/SAPK was detected approximately 10–15 min after the addition of TNFα, although the activity still remained elevated above that of unstimulated cells at 30 min. To verify that mouse macrophages expressed both the p46 and p54 JNK/SAPK isoforms, macrophage whole cell lysates, together with equal amounts (20 ng) of His-tagged recombinant p46 and p54 JNK/SAPK were separated by SDS/PAGE, transferred to nitrocellulose membranes, and immunoblotted with an antibody that recognizes both JNK/SAPK isoforms. As shown in Fig. 2, both p46 and p54 JNK/SAPK isoforms are expressed in roughly equivalent amounts in mouse macrophages. Using scanning densitometry to compare the p46 and p54 signals in the macrophage whole cell lysate with the standard recombinant proteins, we estimated that there were 21.2 ng of p46 JNK/SAPK per 106 cells and 28.1 ng of p54 JNK/SAPK per 106 cells.
Figure 1.
Concentration dependence and time course of JNK/SAPK activation by TNFα in mouse macrophages. (A) Bone marrow-derived macrophages were stimulated with the indicated concentrations of TNFα for 10 min. (B) Macrophages were stimulated with 40 ng/ml of TNFα for 2–30 min as shown. These results are representative of three experiments.
Figure 2.
Immunoblot of a macrophage whole cell lysate and recombinant histidine-tagged p46 and p54 JNK/SAPK isoforms showing that both p46 and p54 JNK/SAPKs are expressed in similar levels in mouse macrophages. Lanes: 1, macrophage whole cell lysate; 2, 20 ng His-tagged p46 JNK/SAPK; 3, 20 ng His-tagged p54 JNK/SAPK. Identical results were obtained in two independent experiments.
Separation of p46 JNK/SAPK and p54 JNK/SAPK by Anion-Exchange Liquid Chromatography.
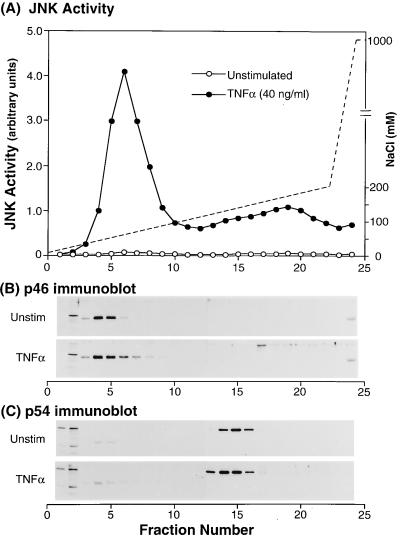
To investigate the relative contribution of p46 JNK/SAPK and p54 JNK/SAPK catalytic activity in response to TNFα, we separated the two kinases by anion-exchange liquid chromatography of detergent lysates of unstimulated and TNFα-stimulated mouse macrophages over a Mono-Q column and investigated the colocalization of JNK/SAPK activity with p46 and p54 JNK/SAPK proteins. Each fraction was assayed for: (i) JNK/SAPK activity and (ii) immunoreactive p46 JNK/SAPK and p54 JNK/SAPK following TCA precipitation and analysis by SDS/PAGE and immunoblotting of nitrocellulose blots with anti-p46 JNK/SAPK and anti-p54 JNK/SAPK antibodies. Following stimulation with TNFα under optimal conditions (40 ng/ml, 10 min), two major peaks of JNK/SAPK activity were detected in macrophage lysates (Fig. 3A). The first and most significant peak was eluted at a salt concentration of approximately 70 mM NaCl and colocalized with p46 JNK/SAPK (Fig. 3B). p54 JNK/SAPK was eluted at a NaCl concentration of approximately 140 mM (Fig. 3C) and was fully separated from p46 JNK/SAPK. p54 JNK/SAPK was associated with a low level of JNK/SAPK activity that comprised a shoulder of the second peak of activity. In addition, probing with an anti-phospho-JNK antibody showed that these same fractions also contained phosphorylated p54 JNK/SAPK (data not shown). Integration of the areas under the activity curves revealed that p46 JNK/SAPK represented 83% of the total JNK/SAPK activity. The second peak of JNK/SAPK activity eluted at approximately 180 mM NaCl and did not colocalize with either p46 JNK/SAPK or p54 JNK/SAPK (Fig. 3A). However, immunoblotting with anti-p42mapk/erk2, anti-p44mapk/erk1, and anti-p38mapk antibodies revealed the JNK/SAPK activity to be colocalized with p44mapk/erk1 but not with p38mapk or p42mapk/erk2 (data not shown). Although members of the ERK subfamily are not believed to phosphorylate intact c-Jun (28, 29), work reported by Pulverer et al. (30) has suggested that ERKs may phosphorylate Ser 63 and 73 of the truncated c-Junl-79-GST substrate used in these studies. However, ERK activation of c-Jun in vivo is unlikely because ERKs also phosphorylate c-Jun at Ser 243, which may negate any c-Jun activation (9, 31, 32).
Figure 3.
Separation of p46 JNK/SAPK and p54 JNK/SAPK by anion-exchange liquid chromatography. Macrophage monolayers were incubated in medium in the presence or absence of TNFα. (A) Mono-Q column assay of unstimulated (–○–) and TNFα-stimulated (–•–) whole cell lysates from a gradient of 0.05 M NaCl to 0.2 M NaCl at a flow rate of 60 ml/hr. JNK/SAPK activity was assayed by the solid phase in vitro kinase assay and was quantitated by PhosphoImager analysis measured in arbitrary units. (B) Immunoblot with the p46 JNK/SAPK antibody. (C) Immunoblot with the p54 JNK/SAPK antibody. These results are representative of three experiments.
Immunodepletion with the p46 JNK/SAPK Antibody.
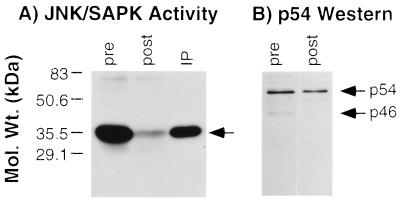
Confirmatory data that the p46 JNK/SAPK isoform was preferentially activated by TNFα were obtained by immunoprecipitating detergent lysates from TNFα-stimulated macrophages with the anti-p46 JNK/SAPK specific antibody and (i) measuring residual JNK/SAPK activity in the postimmunoprecipitation lysate and (ii) determining the extent of recovery of the kinase in the immunoprecipitate using soluble c-Junl-79-GST as substrate. As shown in Fig. 4A, immunoprecipitation with the anti-p46 JNK/SAPK antibody removed the majority of the catalytically active JNK/SAPK present initially in the whole cell lysate, and this was recovered in the immunoprecipitate. Quantification of the kinase activity by scanning densitometry before and after immunodepletion of p46 JNK/SAPK revealed that the p46 JNK/SAPK activity represented approximately 76% of total JNK/SAPK activity. Separation of the pre- and postimmunoprecipitation lysates by SDS/PAGE followed by immunoblotting with the anti-p54 SAPK antibody confirmed the specificity of the immunoprecipitation, in that equivalent amounts of the p54 JNK/SAPK were present before and after immunoprecipitation with the p46 JNK/SAPK antibody (Fig. 4B). These data thus support the conclusion that the p46 isoform is the predominant JNK/SAPK isoform activated by TNFα.
Figure 4.
Immunodepletion with the p46 JNK/SAPK antibody. (A) JNK/SAPK activity in the pre- and post-p46-immunoprecipitation lysates. JNK/SAPK activity in the immunoprecipitate (IP) was detected with soluble c-Jun-GST. (B) Immunoblot of the pre- and postimmunoprecipitated lysates with the p54 JNK/SAPK antibody. These results are representative of three experiments.
MKK4/JNKK/SEK1 Is Phosphorylated in a Time-Dependent Fashion by TNFα.
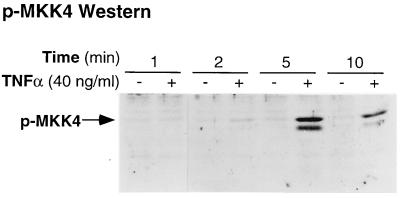
To further explore the mechanism underlying the preferential activation of p46 JNK/SAPK, we investigated the activation of MKK4/JNKK/SEK1, an upstream kinase implicated in JNK/SAPK activation in mouse macrophages stimulated with TNFα. Macrophage monolayers were stimulated with TNFα at 40 ng/ml for time intervals between 1 and 10 min and lysed in the presence of protease and phosphatase inhibitors. After separation by SDS/PAGE, immunoblotting of the whole cell lysates with a phospho-specific MKK4 antibody that detects MKK4/JNKK/SEK1 phosphorylated on residue Thr-223 revealed a time-dependent increase in MKK4/JNKK/SEK1 phosphorylation that was undetectable at 1 min of TNFα stimulation but that peaked at 5 min before returning toward baseline by 10 min (Fig. 5). Thus, MKK4/JNKK/SEK1 was rapidly and transiently phosphorylated in mouse macrophages in response to TNFα with a time course that is similar to the level of activity of JNK/SAPK.
Figure 5.
Time course of phosphorylation of MKK4/JNKK/SEK1 in response to TNFα. TNFα-stimulated macrophage whole cell lysates were immunoblotted with phospho-specific SEK1 antibody. These results are representative of three independent experiments.
MKK4/JNKK/SEK1 Is Catalytically Active in Mouse Macrophages.
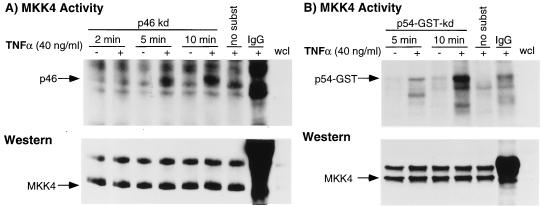
We next questioned whether the selectivity in p46 JNK/SAPK isoform activation may be due to a specificity in the catalytic activity of the MKK4/JNKK/SEK1. Activation of JNK/SAPK is mediated by dual phosphorylation of Thr-183 and Tyr-185 by MKK4/JNKK/SEK1 (33–35) and other less well characterized kinases (36). The approach used to investigate this issue was to immunoprecipitate MKK4/JNKK/SEK1 from TNFα-stimulated mouse macrophages and determine the ability of the kinase to transphosphorylate kinase-inactive p46 and p54 JNK/SAPKs in vitro. As shown in Fig. 6 (Upper), immunoprecipitation of MKK4/JNKK/SEK1, followed by an in vitro kinase assay using histidine-tagged kinase-inactive p46 and kinase-inactive p54-GST JNK/SAPK as substrates, revealed that in TNFα-stimulated cells, MKK4/JNKK/SEK1 exhibits similar catalytic activity toward both isoforms of JNK/SAPK. Immunoblotting with an MKK4/JNKK/SEK1 (C-20) antibody revealed that the kinase was immunoprecipitated in equal amounts from each sample (Fig. 6 Lower).
Figure 6.
Specific catalytic activity of endogenous MKK4 in mouse macrophages in response to TNFα. (A Upper) Immunoprecipitation of endogenous MKK4 followed by an in vitro kinase assay using kinase-inactive p46 JNK/SAPK showed that MKK4 is activated in a time-dependent fashion. (Lower) Immunoblot with the MKK4 (C-20) antibody showed equal loading of sample. (B Upper) Immunoprecipitation of MKK4 followed by an in vitro kinase assay with the kinase-inactive p54 SAPKβ-GST substrate revealed that MKK4 is able to phosphorylate p54 JNK/SAPK. (Lower) Immunoblot with the MKK4 (C-20) antibody. The no-substrate lane comprises macrophages stimulated with TNFα (40 ng/ml) for 10 min. These results are representative of three independent experiments.
Phosphorylation of p46 and p54 JNK/SAPK.
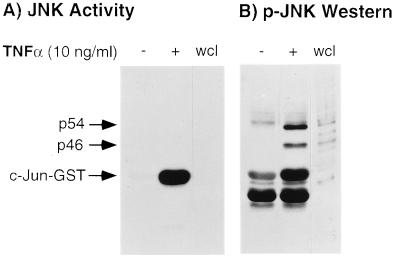
To investigate which JNK/SAPK isoform(s) were phosphorylated by TNFα, macrophage monolayers were stimulated with TNFα (10 ng/ml) for 10 min, lysed as described earlier, incubated with c-Junl-79-GST-Sepharose beads, and, following washing, the bound proteins were separated by SDS/PAGE. As shown previously, there was abundant JNK/SAPK activity upon TNFα stimulation (Fig. 7A). The nitrocellulose membranes were then immunoblotted with a phospho-specific JNK/SAPK antibody that detects phosphorylated Thr-183 and Tyr-185 residues on both JNK/SAPK isoforms. As shown in Fig. 7B, phosphorylated p46 and p54 JNK/SAPK were bound to the c-Jun1–79-GST-Sepharose beads following stimulation with TNFα with the p54 isoform showing greater binding of the phospho-specific antibody than the p46 isoform. Thus, although catalytic activity is preferentially exhibited by the p46 JNK/SAPK isoform, both isoforms are appropriately phosphorylated by their upstream kinase(s).
Figure 7.
JNK/SAPK activities in unstimulated and TNFα-stimulated mouse macrophages and corresponding immunoblots with the phospho-specific JNK/SAPK antibody. (A) JNK activity. (B) Immunoblot with phospho-specific JNK/SAPK antibody. wcl, whole cell lysate. These results are representative of three independent experiments.
DISCUSSION
As more information has become available, it has become increasingly apparent that differential activation of the three MAPK subfamilies and their individual subfamily members plays an important role in regulating distinct, and frequently opposing, cellular functions. For example, in L929 and U937 cells, exposure to TNFα results in the activation of the JNK/SAPK cascade and the initiation of programmed cell death (37, 38). However, as shown by Xia et al. (39), concomitant activation of ERKs during JNK/SAPK activation rescues PC12 cells from apoptosis following nerve growth factor withdrawal, suggesting that ERK activation can dramatically alter the consequences of JNK/SAPK activation. The results from the present study and from previous work from this laboratory suggest that differential and preferential activation of MAPK subfamily members in macrophages may also play an important role in the responses of these cells to TNFα. In previous work, we have shown that incubation with TNFα, which induces differentiative responses in macrophages (6, 26), initiates the selective activation of MEKK (19), MEK1 (20), and p42mapk/erk2 (18). In contrast, the macrophage-specific growth factor colony-stimulating factor 1 activates c-Raf-1 and p44mapk/erk1 (18, 19), suggesting that differential activation within the ERK subfamily may contribute to different functional and/or proliferative responses by these cells. The results from the present study, in which we have shown differential regulation of the p46 and p54 JNK/SAPK isoforms by TNFα, suggest that discrimination in the pattern of regulation of the JNK/SAPK subfamily may further contribute to the heterogeneity in functional responses in macrophages. In addition, the findings of preferential activation of p46 JNK/SAPK by TNFα support the notion that the p46 and p54 JNK/SAPKs are independently regulated. These findings may thus have broader implications on the heterogeneous activation of various transcription factor elements by members of the JNK/SAPK subfamily (16) and may partly explain the pleiotropic properties of TNFα.
Three approaches were used to determine how and where within the signaling cascade the preferential activation of p46 JNK/SAPK was regulated. First, we investigated the activation and role of the upstream kinase MKK4/JNKK/SEK1 in JNK/SAPK activation. Using a phospho-specific antibody that recognizes phosphorylated Thr-223, we have shown that stimulation of mouse macrophages with TNFα leads to a rapid and transient phosphorylation of MKK4/JNKK/SEK1. Thus, these data demonstrate the TNFα-dependent activation of MKK4/JNKK/SEK1 in primary cultures of mouse macrophages. In addition, they imply that this kinase may play a role in subsequent JNK/SAPK activation in these cells. Second, based on these findings, we questioned whether endogenous macrophage MKK4/JNKK/SEK1 was able to transphosphorylate p46 and p54 JNK/SAPKs using an immunoprecipitation/in vitro kinase assay employing kinase-inactive recombinant p46 and p54 JNK/SAPKs as substrates. The data clearly indicated that MKK4 was equally capable of phosphorylating both JNK/SAPK isoforms, suggesting that the level of regulation was distal to MKK4. Third, this conclusion was supported by the finding that both p46 and p54 JNK/SAPK isoforms were phosphorylated on Thr-183 and Tyr-185 using a phospho-specific antibody that recognizes this phosphorylated motif on both JNK/SAPK isoforms.
These collective findings suggest that the level of control is distal to the p54 JNK/SAPK isoform and is targeted at regulation of p54 catalytic activity. Potential mechanisms and their feasibility are as follows: (i) Activation of a p54 JNK/SAPK-specific phosphatase by TNFα. This possibility is supported by previous work showing that the protein synthesis inhibitor cycloheximide is a potent activator of p54 JNK/SAPK but not of p46 JNK/SAPK (14), suggesting that a labile phosphatase that requires translation to maintain its level may basally restrict p54 JNK/SAPK (15). However, based on the finding that both p46 and p54 JNK/SAPK were bound by c-Jun1–79-GST beads and phosphorylated on their respective TPY motif, as discussed above, this possibility would seem untenable. (ii) Although there may exist as yet uncharacterized JNKKs that preferentially activate p46 JNK/SAPK upon TNFα stimulation, that both isoforms were phosphorylated by MKK4/JNKK/SEK1 suggested to us that the level of regulation is not directed at MKK4/JNKK/SEK1. (iii) An inhibitor that specifically inactivates p54 JNK/SAPK catalytic activity. This possibility is supported by recent data showing that a nonenzymatic inhibitor of cyclin-dependent kinases inhibits JNK/SAPK activity (40). This p21 inhibitor is activated by DNA-damaging stimuli such as UV-irradiation, suggesting that it may serve to regulate JNK/SAPK activation. Although the p21 protein inhibited both p46 JNK/SAPK and p54 JNK/SAPK, there were clear differences in the level of inhibition of these different isoforms, raising the possibility of differential regulation of JNK/SAPK isoform activity by this inhibitor. (iv) Another potential mechanism for the p46 JNK/SAPK selectivity by TNFα may be due to differential effects of a scaffolding protein on the different isoforms of JNK/SAPKs. Although this has not been specifically reported in the MAPK family, it has been suggested (11) that an analogous mechanism may regulate differential activation of protein kinase C isozymes by the 14-3-3 zeta protein (41).
These findings raise a number of questions concerning the preferential activation of p46 JNK/SAPK by TNFα in macrophages. Could this be a cell-type-specific response that may be shared by other activators of the JNK/SAPK cascade? This possibility seems unlikely because previous studies have documented concurrent activation of both the p46 and p54 JNK/SAPK isoforms in mouse bone marrow-derived macrophages and RAW264.7 cells following stimulation with lipopolysaccharide (42) and in the human macrophage cell line U937 in response to stimulation with both cycloheximide and anisomycin (43). What are the functional consequences of a preferential activation of p46 JNK/SAPK? This issue currently remains enigmatic. Kallunki et al. (24) have suggested that p54 may preferentially activate c-Jun and AP-1. Perhaps the low level of p54 JNK/SAPK activity detected after stimulation with TNFα is sufficient to activate AP-1. Alternatively, perhaps the diminished activity of p54 JNK/SAPK serves to protect macrophages from apoptotic cell death in response to TNFα. Although these possibilities are currently speculative, the underlying questions clearly need to be addressed in the future.
In summary, we have shown that the p46 JNK/SAPK isoform is preferentially activated in mouse bone marrow-derived macrophages by TNFα and that this selectivity is not due to an inability of MKK4/JNKK/SEK1 to phosphorylate p54 JNK/SAPK but appears to be localized downstream of the kinase, possibly involving an inhibitor of p54 JNK/SAPK catalytic activity. The activation of p42mapk/erk2 (18) and p46 JNK/SAPK by TNFα occurred over a similar time course and, combined with the ability of ERKs to enhance the transcription of c-Fos and JNK/SAPK to activate c-Jun, suggests cooperation between these two MAPK pathways in mediating the effects of TNFα. A possible consequence of the activation of these two pathways by TNFα is the increased transcription of genes whose promoters contain a binding site for AP-1, such as IGF-I, which we have previously shown to be stimulated by TNFα (6). Furthermore, the results of this study reveal the complexities of the JNK/SAPK signal transduction pathways by TNFα and may provide additional insights as to how an array of genes may be regulated by TNFα.
Acknowledgments
We thank Linda Remigio for her expert technical assistance in this work. In addition, we thank Dr. Gary Johnson for the c-Jun-GST and kinase-inactive p46 JNK/SAPK constructs, Dr. James Woodgett for the p54 SAPKβ-GST construct, Dr. John Kyriakis for the p54 SAPK antibody and helpful discussions, and Drs. Anning Lin and Miguel Gijon for helpful discussions. We also thank Nadia deStackelburg, Barry Silverstein, and Leigh Landskroner for illustrative assistance. This work was supported by Public Health Service Grants RO1 HL55549 and SCOR HL55656 from the National Institutes of Health. E.D.C. is funded by a Clinical Investigator Development Award for the National Institutes of Health. B.W.W. was funded by a Medical Research Council of Canada Fellowship Grant.
ABBREVIATIONS
- TNFα
tumor necrosis factor-α
- IGF-I
insulin-like growth factor I
- MAPK
mitogen-activated protein kinase
- ERK
extracellular-signal-regulated kinase
- MEK
MAPK/ERK kinase
- MEKK
MEK kinase
- JNK
c-Jun amino terminal kinase
- SAPK
stress-activated protein kinase
- JNKK
JNK kinase
- MKK4
MAP kinase kinase 4
- SEK1
SAPK/ERK kinase 1
References
- 1.Nathan C F. J Clin Invest. 1987;79:319–326. doi: 10.1172/JCI112815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riches D W H. In: Macrophage Involvement in Wound Repair, Remodeling and Fibrosis. Clark R A F, editor. New York: Plenum; 1996. pp. 95–141. [Google Scholar]
- 3.Bitterman P B, Adelberg S, Crystal R G. J Clin Invest. 1983;72:1801–1813. doi: 10.1172/JCI111140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazaki Y, Araki K, Vesin C, Garcia I, Kapanci Y, Whitsett J A, Piguet P F, Vassalli P. J Clin Invest. 1995;96:250–259. doi: 10.1172/JCI118029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piguet P F, Collart M A, Grau G E, Kapanci Y, Vassalli P. J Exp Med. 1989;170:655–663. doi: 10.1084/jem.170.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lake F R, Noble P W, Henson P M, Riches D W H. J Clin Invest. 1994;93:1661–1669. doi: 10.1172/JCI117148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noble P W, Lake F R, Henson P M, Riches D W H. J Clin Invest. 1993;91:2368–2377. doi: 10.1172/JCI116469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kajimoto Y, Rotwein P. J Biol Chem. 1991;266:9724–9731. [PubMed] [Google Scholar]
- 9.Westwick J K, Weitzel C, Minden A, Karin M, Brenner D A. J Biol Chem. 1994;269:26396–26401. [PubMed] [Google Scholar]
- 10.Chiu R, Boyle W J, Meek J, Smeal T, Hunter T, Karin M. Cell. 1988;54:541–552. doi: 10.1016/0092-8674(88)90076-1. [DOI] [PubMed] [Google Scholar]
- 11.Su B, Karin M. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 12.Hibi M, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 13.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. Nature (London) 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 14.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 15.Cano E, Mahadevan L C. Trends Biochem Sci. 1995;20:117–122. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Derijard B, Davis R J. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 17.Han J, Lee J D, Bibbs L, Ulevitch R J. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 18.Winston B W, Riches D W H. J Immunol. 1995;155:1525–1533. [PubMed] [Google Scholar]
- 19.Winston B W, Lange-Carter C A, Gardner A M, Johnson G L, Riches D W H. Proc Natl Acad Sci USA. 1995;92:1614–1618. doi: 10.1073/pnas.92.5.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winston B W, Remigio L K, Riches D W H. J Biol Chem. 1995;270:27391–27394. doi: 10.1074/jbc.270.46.27391. [DOI] [PubMed] [Google Scholar]
- 21.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 22.Chuang C F, Ng S Y. FEBS Lett. 1994;346:229–234. doi: 10.1016/0014-5793(94)00480-3. [DOI] [PubMed] [Google Scholar]
- 23.Sluss H K, Barrett T, Derijard B, Davis R J. Mol Cell Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallunki T, Su B, Tsigelny I, Sluss H K, Derijard B, Moore G, Davis R, Karin M. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 25.Butterfield L, Storey B, Maas L, Heasley L E. J Biol Chem. 1997;272:10110–10116. doi: 10.1074/jbc.272.15.10110. [DOI] [PubMed] [Google Scholar]
- 26.Riches D W H. Semin Cell Biol. 1995;6:377–384. doi: 10.1016/s1043-4682(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 27.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minden A, Lin A, Smeal T, Derijard B, Cobb M, Davis R, Karin M. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cano E, Hazzalin C A, Mahadevan L C. Mol Cell Biol. 1994;14:7352–7362. doi: 10.1128/mcb.14.11.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pulverer B J, Hughes K, Franklin C C, Kraft A S, Leevers S J, Woodgett J R. Oncogene. 1993;8:407–415. [PubMed] [Google Scholar]
- 31.Davis R J. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 32.Chou S, Baichwal V, Ferrell J E., Jr Mol Biol Cell. 1992;3:1117–1130. doi: 10.1091/mbc.3.10.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derijard B, Raingeaud J, Barrett T, Wu I-H, Han J, Ulevitch R J, Davis R J. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 34.Lin A, Minden A, Martinetto H, Claret F X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Nature (London) 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 36.Moriguchi T, Kawasaki H, Matsuda S, Gotoh Y, Nishida E. J Biol Chem. 1995;270:12969–12972. doi: 10.1074/jbc.270.22.12969. [DOI] [PubMed] [Google Scholar]
- 37.Gardner A M, Johnson G L. J Biol Chem. 1996;271:14560–14566. doi: 10.1074/jbc.271.24.14560. [DOI] [PubMed] [Google Scholar]
- 38.Verheij M, Bose R, Lin X H, Yao B, Jarvis W D, Grant S, Birrer M J, Szabo E, Zon L I, Kyriakis J M, Haimovitz-Friedman A, Fuks Z, Kolesnick R N. Nature (London) 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 39.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 40.Shim J, Lee H, Park J, Kim H, Choi E J. Nature (London) 1996;381:804–807. doi: 10.1038/381804a0. [DOI] [PubMed] [Google Scholar]
- 41.Acs P, Szallasi Z, Kazanietz M G, Blumberg P M. Biochem Biophys Res Commun. 1995;216:103–109. doi: 10.1006/bbrc.1995.2597. [DOI] [PubMed] [Google Scholar]
- 42.Hambleton J, Weinstein S L, Lem L, DeFranco A L. Proc Natl Acad Sci USA. 1996;93:2774–2778. doi: 10.1073/pnas.93.7.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franklin C C, Adler V, Kraft A S. Methods Enzymol. 1995;254:550–564. doi: 10.1016/0076-6879(95)54039-3. [DOI] [PubMed] [Google Scholar]