Abstract
Asthma is a complex heritable inflammatory disorder of the airways associated with clinical signs of atopy and bronchial hyperresponsiveness. Recent studies localized a major gene for asthma to chromosome 5q31-q33 in humans. Thus, this segment of the genome represents a candidate region for genes that determine susceptibility to bronchial hyperresponsiveness and atopy in animal models. Homologs of candidate genes on human chromosome 5q31-q33 are found in four regions in the mouse genome, two on chromosome 18, and one each on chromosomes 11 and 13. We assessed bronchial responsiveness as a quantitative trait in mice and found it linked to chromosome 13. Interleukin 9 (IL-9) is located in the linked region and was analyzed as a gene candidate. The expression of IL-9 was markedly reduced in bronchial hyporesponsive mice, and the level of expression was determined by sequences within the qualitative trait locus (QTL). These data suggest a role for IL-9 in the complex pathogenesis of bronchial hyperresponsiveness as a risk factor for asthma.
Atopic asthma is a serious chronic illness characterized by one or more symptoms, including episodic shortness of breath, wheezing, coughing, and chest tightness (1). These nonspecific symptoms are generally characterized as intermittent and are associated with reversible airway obstruction and bronchial hyperresponsiveness. The recognition that specific genetic and environmental factors appear to be important in the etiology of atopic asthma has led to an intensive search for those factors that significantly influence susceptibility to this disorder (2).
Clinical and physiologic studies of asthma reveal an association with widespread narrowing of the airways caused by edema and the infiltration of numerous inflammatory cells into the wall of the airway lumen. These cells include eosinophils, mast cells, IgE-producing plasma cells, and a specific subset of activated T cells referred to as Th2 lymphocytes (1, 2). Although the precise biologic mechanism underlying these cellular responses is unknown, it is clear that complex interactions between these cells in the airway in response to antigen lead to the elaboration of cytokines and other inflammatory proteins that produce chemical mediators that are critical in this disorder (2). Despite these insights, a clear understanding of the pathogenesis of atopic asthma is lacking.
Genetic susceptibility to atopic asthma is likely to be multigenic (2–4). Biologic variability in the allergic and inflammatory response, and a clear recognition of the importance of IgE and various cytokines in this response, has stimulated analyses of candidate genes for genetic variability in structure and function. Recently, a major gene for asthma was localized to chromosome 5q31-q33 in humans (3, 4). Although the gene(s) predisposing to asthma, bronchial hyperresponsiveness, and atopy have not yet been identified from this chromosomal region, 5q31-q33 is known to be syntenic, or to share chromosomal architecture with segments of mouse chromosomes 11, 13, and 18 (5). Chromosome 5q31-q33 contains numerous gene candidates that may potentially play a role in the airway inflammation associated with atopic asthma, such as cytokines, growth factors, and growth factor receptors. In particular, interleukin 9 (IL-9) was suggested as a likely candidate on the basis of linkage disequilibrium between log serum total IgE levels and a marker within this gene (6).
We show here that significant differences in bronchial responsiveness between hyporesponsive C57BL/6J (B6) and hyperresponsive DBA/2J (D2) mice is determined in part by a qualitative trait locus (QTL) which maps to the syntenic region of chromosome 13. Based on these findings, we evaluated IL-9 as a gene candidate. We found that bronchial hyporesponsiveness is associated with a greatly reduced steady-state of IL-9 levels which is related to a genetic alteration at the B6 locus. This finding was reflected in reduced levels of this cytokine in the lungs of B6 mice as compared to the bronchial hyperresponsive D2 mice. These data identify IL-9 as one gene candidate in the complex pathogenesis of bronchial hyperresponsiveness, a critical factor in the asthmatic response.
MATERIALS AND METHODS
Mice.
The following studies conformed to the principles for laboratory animal research outlined by the Animal Welfare Act and the Department of Health, Education, and Welfare (National Institutes of Health) guidelines for the experimental use of animals and were approved by the Institutional Animal Care and Use Committee. Male and female DBA/2 (D2), C57BL/6 (B6), and BXD recombinant inbred (RI) mice 5 to 6 weeks of age were purchased from The Jackson Laboratory. The animals were not maintained in specific pathogen free conditions. The animal housing facilities were maintained at 22°C with a light cycle of 10/14 h light/dark. Food and water were present ad libitum.
Phenotyping.
To determine the bronchoconstrictor response, respiratory system pressure was measured at the trachea and recorded before and during exposure to drug. Mice were anesthetized and instrumented as described (7). The bronchoconstrictor response to atracurium was assessed by the change in peak inspiratory pressure (Ppi) integrated over time. This parameter termed the airway pressure time index (APTI) is a simple and repeatable measure of the change in Ppi highly correlated with and respiratory system resistance and elastance following a bronchoconstrictor challenge (8).
Linkage Analysis.
Genetic mapping methods were used to test for evidence of linkage between airway responsiveness and markers in the mouse genome in the regions syntenic to human chromosome 5q31-q33. Four intervals on three mouse chromosomes were considered, 11, 13, and 18 (see Fig. 1). Linkage analyses were performed by using publicly available genotype data for the 26 BXD RI compiled by R. W. Elliot and electronically distributed at http://mcbio.med.buffalo.edu/rweRIdata5.97.seq.hqx. The QTL used was the log10 transformed mean value for the APTI phenotype measured for each line.
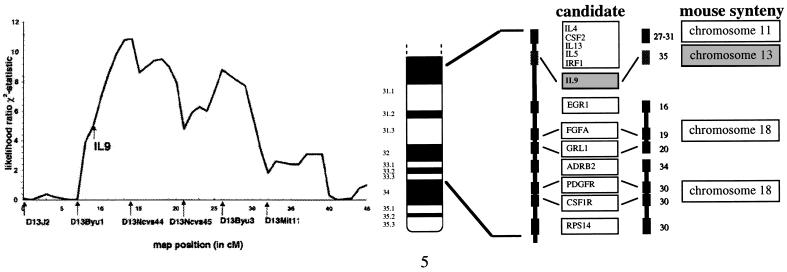
Figure 1.
Synteny and linkage homology for bronchial responsiveness implicate IL-9 candidate. (A) The likelihood ratio χ2 statistic curve on mouse chromosome 13 for bronchial responsiveness in 24 BXD RI strains that are derived from the hyporesponsive C57BL/6J and the hyperresponsive DBA/2J progenitor strains. The log10 transformed mean value for the APTI phenotype measured for each RI line was used to generate the likelihood ratio χ2 statistic curve. The permutation test of Churchill and Doerge (10) was used to assess significance after adjusting on a comparison basis for the multiple comparisons performed on chromosome 13. The threshold for significant linkage was 9.1 for the χ2 statistic after this multiple comparisons adjustment. (B) Illustration of the genetic map of human chromosome 5q31-q33 and syntenic regions in the mouse. The region of human chromosome 5q31-q33 demonstrating significant evidence for linkage with BHR is homologous to portions of mouse chromosomes 11, 13, and 18 which contain numerous candidate genes including chromosome 11: IL-4 (IL4), IL-5 (IL5), granulocyte macrophage stimulating factor (CSF2); IL-13 (IL13), immune regulatory factor 1 (IRF1); γ amino butyric acid receptor A1 (GABRA1); chromosome 13: IL-9 (IL9), and chromosome 18: adrenergic receptor β 2 (ADRB2); glucocorticoid receptor 1 (GRL1); fibroblast growth factor acidic (FGFA); platelet-derived growth factor receptor (PDGFR); colony stimulating factor receptor 1 (CSF1R); early growth response element 1 (EGR1). Relative map positions in cM are illustrated to the right of each candidate in the mouse (centromere representing 0 cM).
Single locus association and interval mapping results were calculated by using a likelihood ratio statistic (9), as implemented in mapmanager/qt 9 (version 1.5b). The APTI QTL was walked through the chromosome maps of interest in 1-cM steps. Multiple comparison adjustments for the chromosome-wide linkage tests performed were made by using the mapmanager/qt implementation of the permutation method of Churchill and Doerge (10). In this analysis, the QTL phenotypes were randomly permuted while holding the marker genotyping constant. A total of 10,000 permutations were performed. This test provides an empiric multiple comparisons adjusted assessment of the observed linkage to a given location.
Genotyping.
Exons and portions of the surrounding introns of the murine IL-9 gene were amplified from genomic DNA from the B6 and D2 mouse by PCR with the following primers: exon 1 sense (5′-AGC TAG ACT GGA AGA TGC TG-3′) and antisense (5′-CCT ACA GTA GGC AAA GAG-3′) (product size, 250 bp); exons 2 and 3 sense (5′-AAG GGG GCT CAA CTC ACT G-3′) and antisense (5′-CGT GAT TTC TCT TCA AAG CG-3′) (product size 254 bp); exon 4 sense (5′-TGA AAG CAG TCC TAG GCA G-3′) and antisense (5′-TGG GCC TAC CTG CTC TAA C-3′) (product size 330 bp); exon 5 sense (5′-CTG CTA GGA GTG TAT TGC TG-3′) and antisense (5′-CAC AAA AGG GCC TTG GTC-3′) (product size 295 bp); 5′ flanking region sense (5′-CAA GGC CAA TGC TAG CAA G-3′) and antisense (5′-AGG ATG TAT GTC ACC AAC ATG-3′) (product size 657 bp); 3′ flanking region sense primer (5′-TGC AAA GGC AGA AAA GCC G-3′) and antisense (5′-CCA AGT CTT TGC TCC ATC C-3′) (product size 317 bp).
Primary Cell Cultures and Cell Lines.
Murine splenocytes were isolated from age matched mice and prepared as described (11). One hundred thousand cells were plated per well in 96-well plates and stimulated with Con A (Sigma) and harvested as described above (11).
Reverse Transcriptase–PCR (RT-PCR).
Total RNA were isolated from cells and lung tissues by using the Trizol method as described by the manufacturer (GIBCO/BRL). Lung tissues were carefully dissected from hilar lymph nodes prior to use. Equal amounts of total RNA were reverse transcribed and amplified by using appropriate primers by PCR as described (12). Products were electrophoresed on 2% agarose gels and analyzed by ethidium bromide staining and/or Southern blot by using internal oligonucleotides as probes as described (13). Primer sets used were as follows: murine (m)IL-4 (GenBank database accession no. M25892): 5′-ACC ACA GAG AGT GAG CTC G-3′ (centered at nt 280 of the cDNA) and 5′-ATG GTG GCT CAG TAC TAC G-3′ (centered at position nt 505 of the cDNA) which produce a 254-bp product; mIL-5 (GenBank database accession no. X06270): 5′-TTG AGT GTT CTG ACT CTC AG-3′ (centered at nt 74 of the cDNA) and 5′-GAA CTG CCT CGT CCT CCG-3′ (centered at nt 375 of the cDNA), which produce a 321-bp product; mIL-9: 5′-AGC TAG ACT GGA AGA TGC TG-3′ [centered at nt 603 of the gene (17)] and 5′-AAA TAG CAT CTG TCT TCA TGG-3′ [centered at nt 3512 of the gene (17)] which produces a 497-bp product; mγ-interferon (IFN) (GenBank database accession no. K00083): 5′-GCT CTG AGA CAA TGA ACG-3′ (centered at nt 69 of the cDNA) and 5′-CAG GTG TGA TTC AAT GAC GC-3′ (centered at nt 354 of the cDNA), which produce a 304-bp product; β-actin was amplified as previously described (12); Dhfr (GenBank database accession no. L26316): 5′-TCG ACC ATT GAA CTG CAT CG-3′ (centered at position nt 70 of the cDNA) and 5′-CTT TAC TTG CCA ATT CCG G-3′ (centered at position nt 372 of the cDNA), which produce a 323-bp product; Gli-3 (GenBank database accession no. X95255): 5′-ACG AGA ACA GAT GTC AGC G-3′ (centered at position nt 253 of the cDNA) and 5′-AAG GCA GGG AAA AGA TGA GG-3′ (centered at position nt 554 of the cDNA), which produce a 320-bp product; Caml (GenBank database accession no. U21960): 5′-CGA GAA GAA GGT GAA GAC G-3′ (centered at position nt 739 of the cDNA) and 5′-CCC CTT TTA GAG ACT CTG C-3′ (centered at position nt 980 of the cDNA), which produces a 260-bp product. The oligonucleotide probes used for Southern blots were as follows: mIL-4 (5′-CTT CCA AGG TGC TTC GC-3′) (centered at position nt 305 of the cDNA); mIL-5 (5′-TGA GCA CAG TGG TGA AAG AG-3′) (centered at position nt 126 of the cDNA); mIL-9 (5′-AAT TAC CTT ATT GAA AAT CTG AAG-3′) (centered at position nt 643 of the gene); mγ-IFN (5′-CTA CAC ACT GCA TCT TGG C-3′) (centered at position nt 84 of the cDNA).
Heteronuclear RNA Transcript Analyses.
Total RNA were prepared from 24-h Con A-stimulated (B6D2)F1 splenocytes as described above. RNA samples were treated twice with RNase-free DNase (Promega) following the manufacturer’s suggestion. RNA were reverse transcribed and PCR amplified for the intronic segment containing the polymorphic CA repeat marker in intron 3 of the murine IL-9 gene (14) by using the sense primer 5′-CTG TGG TAA GTC CAG ATT TG-3′ (centered at nt 1295 of the published gene) and the antisense primer 5′-GGA AAG AGT AGG AAG ATG CC-3′ centered at nt 1425 of the published gene) resulting in a product of approximately 150 bp. The PCR fragment was then cloned into the T-tailed TA cloning vector (Invitrogen) following the manufacturer’s protocol. Recombinant clones were analyzed via direct PCR by using an internal sense primer 5′-GGC AGG GAT CAT CTA GAT ATC-3′ (centered at nt 1341) and the same antisense primer containing a 5′ FAM-6 label (Applied Biosystems). Products were electrophoresed on an Applied Biosystems model 373 automated sequencer, and allele sizes were determined by using genescan/genotyper software programs. To further confirm the identity of the resultant products, 10 recombinant clones were sequenced by using M13 forward and reverse primers contained within the cloning vector.
Western Blot Analyses.
Lung tissues were derived from various mice, dissected free of hilar lymph nodes, and snap frozen in liquid nitrogen. Sections (3 mm3) were resuspended in 500 μl of 2× SDS lysis buffer (60 mM Tris, pH 6.8/2% SDS/0.1 M 2-mercaptoethanol/0.1% bromo-phenol blue) and boiled for 5 min. Twenty microliters of each lysate was electrophoresed on 18% Tris-glycine SDS/PAGE gels and electroblotted onto Immobilon-P (Millipore) membrane in transfer buffer (48 mM Tris/40 mM glycine/0.0375% SDS/20% methanol). Filters were blocked overnight in blocking buffer (TBS, 0.05% Tween-20/5% powdered milk). Filters were probed with polyclonal goat anti-murine IL-4, IL-5, and IL-9 (R & D Systems) and a secondary horseradish peroxidase conjugated anti-goat (Pierce) and prepared for chemiluminescence.
RESULTS
Synteny and Linkage Homology Implicates the IL-9 Candidate Locus.
Evidence for linkage was found for only one of the syntenic regions tested. Single locus linkage results determined in the BXD RI lines showed a significant relationship between marker D13Ncvs44 and the APTI QTL (nominal P = 0.0019) (Fig. 1). No significant evidence for QTL linkage was observed for chromosomes 11 or 18. The permutation test of Churchill and Doerge (10) was used to assess significance after adjusting on a comparison basis for the multiple comparisons performed on chromosome 13. Even after this multiple comparisons adjustment, the APTI QTL was still significantly linked to this region (P = 0.0177).
Four of the RI lines (BXD-2, -6, -18, and -32) demonstrated an APTI intermediate in comparison to either parental strain (Table 1). To test whether these strains were potentially misclassified with respect to phenotype, and whether this might impact linkage, analyses were conducted without BXD strains 2, 6, 18, and 32. These strains display a mean response greater than 1 standard deviation below the D2 and above the B6 mean responses. When these four lines were excluded from the analysis, the significance of the putative QTL association increased to P = 0.00094 (multiple comparisons adjusted P = 0.0226). The inclusion of these intermediate strains, therefore, does not alter the localization of this QTL.
Table 1.
BXD RI strain distribution pattern for atracurium-induced APTI
| Strain | n | APTI, cmH2O⋅s |
|---|---|---|
| DBA/2 | 13 | 404 ± 40 |
| C57BL/6 | 8 | 86 ± 18 |
| BXD-1 | 7 | 109 ± 15 |
| BXD-2 | 7 | 142 ± 15 |
| BXD-5 | 5 | 65 ± 11 |
| BXD-6 | 5 | 169 ± 8 |
| BXD-8 | 4 | 247 ± 26 |
| BXD-9 | 5 | 273 ± 22 |
| BXD-11 | 4 | 107 ± 10 |
| BXD-12 | 7 | 408 ± 33 |
| BXD-13 | 6 | 281 ± 40 |
| BXD-14 | 8 | 53 ± 13 |
| BXD-15 | 2 | 462 ± 203 |
| BXD-16 | 7 | 521 ± 115 |
| BXD-18 | 4 | 164 ± 13 |
| BXD-19 | 6 | 84 ± 21 |
| BXD-21 | 7 | 317 ± 47 |
| BXD-22 | 5 | 54 ± 24 |
| BXD-24 | 6 | 83 ± 13 |
| BXD-25 | 6 | 984 ± 125 |
| BXD-27 | 5 | 570 ± 120 |
| BXD-28 | 5 | 291 ± 14 |
| BXD-29 | 4 | 102 ± 23 |
| BXD-30 | 4 | 346 ± 48 |
| BXD-31 | 6 | 304 ± 21 |
| BXD-32 | 4 | 142 ± 15 |
The mean APTI responses (±SEM) to 20 mg/kg atracurium given i.v. to 24 BXD RI strains are shown. n represents the number of animals phenotyped per strain. The data represent the untransformed values of the mean APTI phenotype data measured for each strain. A log10 transformed mean value for these APTI phenotype data was used for the associated linkage analyses.
This QTL is located on the midportion of chromosome 13 (Fig. 1). The D13Ncvs locus was observed to explain 44% of the APTI phenotype variance with an additive component of 0.45. The known positional candidates in the linked region of chromosome 13 include calcium modulating cyclophilin ligand (Caml) (Mouse Genome Database WWW site) and IL-9 (Il9) (15, 16).
IL-9 mRNA and Protein Levels Differ Significantly Between B6 and D2 Mice in Splenocytes and Lungs.
Homology mapping of bronchial responsiveness in mice to the syntenic region including IL-9 prompted us to search for functional or structural changes at this candidate locus. We first sequenced each exon and the surrounding RNA spice site consensus sequence of the coding regions of the IL-9 gene from the B6 and D2 strains and found no sequence variation. We next isolated RNA from mitogen-stimulated splenocytes to see whether alternative splice variants could be detected that would suggest a lesion within an intron. Although no splice variants were identified, a distinct difference in IL-9 expression was observed between the two strains (Fig. 2). Induction of IL-9 mRNA occurs by 12 h in D2-stimulated splenocytes and returns to baseline after approximately 48 h. In contrast, the B6 splenocytes did not produce detectable levels of IL-9 message (Fig. 2A). These data suggest a genetic alteration at the B6 locus that effects steady-state levels of mRNA either through reduced production or more rapid elimination. In an attempt to identify an alteration in potential regulatory or stability elements, we analyzed the published 531-bp 5′ and the 285-bp 3′ flanking regions of the IL-9 gene (17). Analyses of these regions of the IL-9 gene did not identify any variants between the B6 and D2 mice that could have explained the variability in message levels.
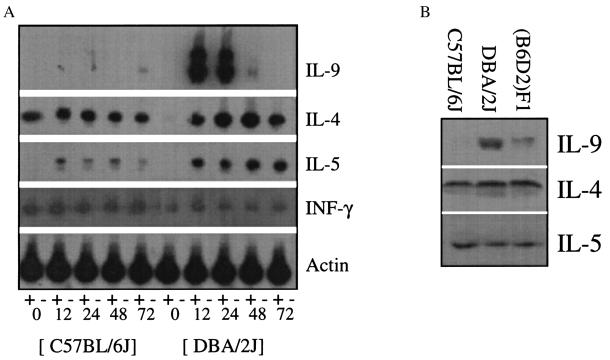
Figure 2.
Reduced abundance of the IL-9 candidate in hyporesponsive B6 mice. (A) RNA were isolated from B6 and D2 splenocyte in vitro after 0, 12, 24, 48, and 72 h of Con A stimulation. Southern blots of RT-PCR for IL-9, IL-4, IL-5, and interferon-γ are shown. Actin RNA were used as an internal control. The symbols (+) and (−) designate with and without reverse transcriptase, respectively. (B) Western blot analysis of cytokines from the lungs of hyporesponsive B6, intermediate (B6D2)F1, and hyperresponsive D2 mice demonstrating reduced IL-9 levels in B6 mice.
Western blot analyses were performed on the lungs of D2 and B6 mice to determine whether IL-9 levels differed in the lungs of these mice. As shown in Fig. 2B, significant differences in IL-9 levels were detected between these strains whereas the TH2 cytokines IL-4 and IL-5 appear to have similar steady-state levels. Interestingly, IL-9 levels of (B6D2)F1 mouse lungs appeared to be intermediate to B6 and D2 levels, perhaps reflecting expression of a single allele, presumably from the D2 strain. These protein levels were corroborated by RT-PCR data, which also detected similar steady-state levels of interferon- γ (data not shown).
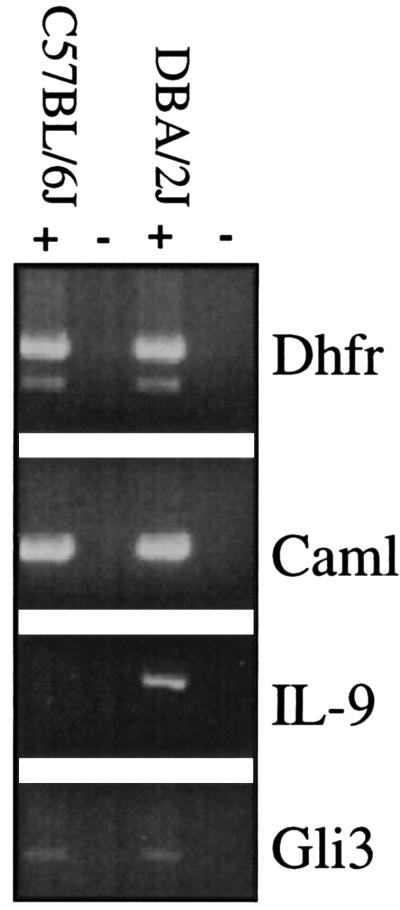
To examine whether a generalized reduction in B6 transcripts occurs relative to the D2 mouse, RT-PCR was used to compare the expression of surrounding loci on chromosome 13 in B6 and D2 splenocytes 24 h after Con A stimulation. The glioblastoma oncogene homolog 3(Gli3) is 8 cM from the centromere, dihydrofolate reductase (Dhfr) is located at 51 cM, and calcium modulating ligand is located within the linked QTL at 35 cM (Caml). No significant differences were noted in transcript abundance or size between these strains of mice for these other loci (Fig. 3).
Figure 3.
Comparison of expression for surrounding loci in B6 and D2 mice. Analyses of B6 and D2 splenocytes 24 h after Con A stimulation by RT-PCR for expression of surrounding loci on chromosome 13. The glioblastoma oncogene homolog 3(Gli3) is 8 cM from the centromere, dihydrofolate reductase (Dhfr) is located at 51 cM, and calcium modulating ligand is located within the linked QTL at 35 cM (Caml).
IL-9 Gene Expression in the (B6D2)F1 Cellular Environment.
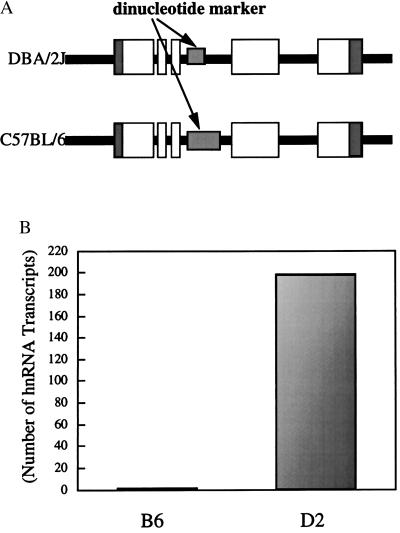
We reasoned that if our genetic mapping of biologic variability in bronchial responsiveness to the IL-9 locus was correct in B6 and D2 mice then the (B6D2)F1 cell would demonstrate a reduced abundance of B6 transcript related to an unidentified alteration at the B6 locus. A polymorphic dinucleotide CA repeat marker that is contained within intron 3 of the IL-9 gene was used to identify which parental allele each transcript was generated from (Fig. 4A). The B6 allele has three more dinucleotides than the D2 allele (14). To assess the steady-state levels of IL-9 message produced by the B6 and D2 loci in the same cell, heteronuclear RNA were isolated, RT-PCR amplified, and cloned from splenocytes of (B6D2)F1 mice stimulated for 24 h with Con A. Total RNA were amplified by using RT-PCR with intronic primers surrounding the CA repeat marker (see Materials and Methods), and cloned into T-tailed vectors. Two hundred recombinant clones were analyzed. The B6 message was significantly less abundant in comparison to the D2 allele (1:100 respectively), suggesting that a cis-acting element at the B6 locus results in the lack of induction to Con A and the reduced abundance of IL-9 mRNA and protein in this animal (Fig. 4B). Genomic DNA were PCR amplified for the CA repeat, cloned into T-tailed vector, and analyzed. Twenty recombinant clones found equal amounts of both alleles thus ruling out a cloning bias for the D2 allele.
Figure 4.
Characterization of the molecular defect at the B6 IL-9 locus. (A) Illustration of the IL-9 locus with an intragenic polymorphic dinucleotide repeat (see arrows) within intron 3 which is 3 dinucleotides larger in the B6 gene than in the D2 gene. The black line represents the chromosome, the dark grey represents the untranslated regions, and the open boxes are the exons. (B) Quantification of cloned transcripts obtained from hnRNA in (B6D2)F1 splenocyte.
DISCUSSION
In this study we identified the IL-9 gene as a positional gene candidate in allergic asthma based on linkage homology between humans and mice for bronchial responsiveness. We have utilized the BXD RI strains to identify a QTL which influences the expression of bronchial responsiveness that maps to a small region of chromosome 13 in the mouse that is syntenic with human chromosome 5q31-q33. By using this strategy, we were able to significantly reduce the effort required in the positional cloning of this candidate. The results described here suggest that allelic variation in IL-9 is responsible, in part, for biologic variability in baseline bronchial responsiveness between these inbred strains of mice. The bronchial hyporesponsive B6 strain was found to express markedly lower levels of IL-9 in lung than the hyperresponsive D2 strain, whereas (B6D2)F1 mice appear to express intermediate levels. Interestingly, these F1 mice have an intermediate bronchial response (18), demonstrating a tight genotype–phenotype correlation. The marked reduction of IL-9 expression between hyporesponsive B6 mice compared with hyperresponsive D2 mice was not observed for surrounding genes. Caml, which maps to the linked QTL and several other genes that span chromosome 13, failed to demonstrate any strain difference in expression. Thus, the genetic alteration associated with the differential expression of IL-9 in these mice is specific to this locus.
Significant biologic variability in airway responsiveness occurs in humans and baseline bronchial hyperresponsiveness is recognized as a risk factor for asthma (19–21). Baseline airway responsiveness also differs between inbred strains of mice and rats, which is strongly controlled by genetic factors (7, 18, 22–24). Recently, T cells were shown to regulate baseline murine airway responsiveness in the absence of an antigen challenge or gross airway inflammation (25). Airway hyperresponsiveness was transferred to the hyporesponsive B6 mouse by administering T cell-enriched preparations from hyperresponsive mice. Thus, although factors affecting T cell growth and differentiation are well known to affect antigen response (26, 27), they also are likely to be essential in determining the baseline airway response phenotype. IL-9 is a T cell growth factor, and experimental data have shown a correlation between IL-9 production and TH2-responses in vitro and in vivo (11, 28, 29). TH2 cells are well documented to have a crucial role in the development of the allergic response and are presumed to be associated with the pathogenesis of asthma (26, 27).
A pleiotropic role for IL-9 in allergic asthma is suggested by several independent studies describing its functions in vitro and in vivo (30). Mediator release from mast cells by allergen has long been considered a critical initiating event in allergy. IL-9 was originally identified as a mast cell growth factor, and appears to up-regulate the expression of mast cell proteases including mMCP-1, mMCP-2, mMCP-4 (31), and granzyme B (32). Thus, IL-9 may serve a role in the proliferation and differentiation of mast cells. In addition, elevated IgE levels are considered to be a hallmark of atopic allergy and a risk factor for asthma, and in vitro and in vivo studies have shown IL-9 to potentiate the release of IgE from primed B cells (3, 33, 34).
Based on the data presented, there is substantial support for the IL-9 gene candidate in asthma. First, we demonstrate linkage homology between humans and mice suggesting the same gene may be affected in both species. Second, differences in expression of the murine IL-9 candidate gene were associated with biologic variability in bronchial responsiveness. In particular, a loss of function is associated with a lower baseline bronchial response in B6 mice. Third, consistent with a role for this gene in both species, recent linkage disequilibrium data in humans suggest IL-9 may be associated with atopy and bronchial hyperresponsiveness (6). Finally, the pleiotropic functions of this cytokine in the allergic immune response strongly support a role for IL-9 in the complex pathogenesis of asthma. Carefully executed epidemiologic studies will be required to describe the full extent to which this gene may play a role in the natural history of allergy and asthma. Nevertheless, our data suggest that further study of this gene, and perhaps associated genes in a related biologic pathway, may provide potentially useful insights into the pathogenesis of asthma.
Acknowledgments
We thank Dr. Benjamin Taylor for his advice and assistance, and Drs. Jacques Van Snick and Jean Christophe Renauld for their intellectual discussions. We are indebted to Bert Vogelstein for his critical review of this manuscript.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: IL, interleukin; RI, recombinant inbred; APTI, airway pressure time index; RT-PCR, reverse transcriptase–PCR; QTL, qualitative trait locus; hnRNA, heteronuclear RNA.
References
- 1.McFadden E R, Jr, Gilbert I A. N Engl J Med. 1992;327:1928–1937. doi: 10.1056/NEJM199212313272708. [DOI] [PubMed] [Google Scholar]
- 2.Holgate S T, Church M K, Howarth P H, Morton N E, Frew A J, Ratko D. Int Arch Allergy Immunol. 1995;107:29–33. doi: 10.1159/000236921. [DOI] [PubMed] [Google Scholar]
- 3.Postma D S, Beecker E R, Amelung P J, Holroyd K J, Xu J, Panhuysen C I M, Meyers D A, Levitt R C. N Engl J Med. 1995;333:894–900. doi: 10.1056/NEJM199510053331402. [DOI] [PubMed] [Google Scholar]
- 4.The Collaborative Study on the Genetics of Asthma (CSGA) Nat Genet. 1997;15:389–392. [Google Scholar]
- 5.DeBry R W, Seldin M F. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- 6.Doull I, Lawrence S, Watson M, Begishvili T, Beasley R, Lampe F, Holgate S T, Morton N E. Am J Respir Crit Care Med. 1996;153:1280–1284. doi: 10.1164/ajrccm.153.4.8616554. [DOI] [PubMed] [Google Scholar]
- 7.Levitt R C, Mitzner W. FASEB J. 1988;2:2605–2608. doi: 10.1096/fasebj.2.10.3384240. [DOI] [PubMed] [Google Scholar]
- 8.Ewart S L, Levitt R C, Mitzner W. J Appl Physiol. 1995;79:560–566. doi: 10.1152/jappl.1995.79.2.560. [DOI] [PubMed] [Google Scholar]
- 9.Haley C S, Knott S A. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- 10.Churchill G A, Doerge R W. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gessner A, Blum H, Rollinghoff M. Immunobiology. 1993;189:419–435. doi: 10.1016/S0171-2985(11)80414-6. [DOI] [PubMed] [Google Scholar]
- 12.Nicolaides N C, Carter K C, Shell B K, Papadopoulos N, Vogelstein B, Kinzler K W. Genomics. 1995;30:195–206. doi: 10.1006/geno.1995.9885. [DOI] [PubMed] [Google Scholar]
- 13.Nicolaides N C, Gualdi R, Casadevall C, Manzella L, Calabretta B. Mol Cell Biol. 1991;11:6166–6176. doi: 10.1128/mcb.11.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich W, Katz H, Lincoln S E, Shin H S, Freidman J, Dracopli N C, Lander E S. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mock B A, Krall M, Kozak C A, Nesbitt M N, McBride O W, Renauld J-C, Van Snick J. Immunogenetics. 1990;31:265–270. doi: 10.1007/BF00204898. [DOI] [PubMed] [Google Scholar]
- 16.DeBry R W, Seldin M F. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- 17.Renauld J-C, Goethals A, Houssiau F, Merz H, Van Roost E, Van Snick J. J Immunol. 1990;144:4235–4241. [PubMed] [Google Scholar]
- 18.Levitt R C, Ewart S L. Am J Respir Crit Care Med. 1995;151:1537–1542. doi: 10.1164/ajrccm.151.5.7735612. [DOI] [PubMed] [Google Scholar]
- 19.Townley R G, Bewtra A K, Wilson A F, Hopp R J, Elston R C, Nair N, Watt G D. J Allergy Clin Immunol. 1986;77:101–107. doi: 10.1016/0091-6749(86)90330-1. [DOI] [PubMed] [Google Scholar]
- 20.Hopp R J, Bewtra A K, Biven R, Nair N M, Townley R G. Ann Allergy. 1988;61:184–186. [PubMed] [Google Scholar]
- 21.Hopp R J, Townley R G, Biven R E, Bewtra A K, Nair N M. Am Rev Respir Dis. 1990;141:2–8. doi: 10.1164/ajrccm/141.1.2. [DOI] [PubMed] [Google Scholar]
- 22.Levitt R C, Mitzner W. J Appl Physiol. 1989;67:1125–1132. doi: 10.1152/jappl.1989.67.3.1125. [DOI] [PubMed] [Google Scholar]
- 23.Levitt R C. Pharmacogenetics. 1991;1:94–97. doi: 10.1097/00008571-199111000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Pauwels R, Van Der Straeten M, Weyne J, Bazin H. Eur J Respir Dis. 1985;66:98–104. [PubMed] [Google Scholar]
- 25.De Sanctis G, Itoh A, Green F H Y, Qin S, Kimura T, Grobholz J K, Martin T R, Maki T, Drazen J M. Nat Med. 1997;3:460–462. doi: 10.1038/nm0497-460. [DOI] [PubMed] [Google Scholar]
- 26.Robinson D S, Hamid Q, Ying S, Tsicopoulos A, Barkans J, Bentley A M, Corrigan C, Durham S R, Kay A B. N Engl J Med. 1992;326:289–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- 27.Del Prete G F, De Carly M, D’Elios M M, Maestrelli P, Ricci M, Sabbri L, Romagnini S. Eur J Immunol. 1993;23:1445–1449. doi: 10.1002/eji.1830230707. [DOI] [PubMed] [Google Scholar]
- 28.Grencis R K, Hultner L, Else K J. Immunology. 1991;74:329–332. [PMC free article] [PubMed] [Google Scholar]
- 29.Svetic A, Madden K B, Zhou X, Lu P, Katora I M, Finkelman F D, Urban J F, Gause W C. J Immunol. 1993;150:3434–3441. [PubMed] [Google Scholar]
- 30.Renauld J-C, Kermouni A, Vink A, Louahed J, Van Snick J. J Leukocyte Biol. 1995;57:353–360. doi: 10.1002/jlb.57.3.353. [DOI] [PubMed] [Google Scholar]
- 31.Eklund K K, Ghildyal N, Austen K F, Stevens R L. J Immunol. 1993;151:4266–4273. [PubMed] [Google Scholar]
- 32.Louahed J, Kermouni A, Van Snick J, Renauld J-C. J Immumol. 1995;154:5061–5070. [PubMed] [Google Scholar]
- 33.Dugas B, Renauld J-C, Pene J, Bonnefoy J Y, Petit-Fere C, Braquet P, Bousquet J, Van Snick J, Mencia-Huerta J M. Eur J Immunol. 1993;23:1134–1138. doi: 10.1002/eji.1830230743. [DOI] [PubMed] [Google Scholar]
- 34.Petit-Frerer C, Dugas B, Braquet P, Mencia-Huerta J M. Immunology. 1993;79:146–151. [PMC free article] [PubMed] [Google Scholar]