Abstract
Polymorphonuclear leukocytes are essential for host defense to infectious diseases. CCAAT/enhancer binding protein ɛ (C/EBPɛ) is preferentially expressed in granulocytes and lymphoid cells. Mice with a null mutation in C/EBPɛ develop normally and are fertile but fail to generate functional neutrophils and eosinophils. Opportunistic infections and tissue destruction lead to death by 3–5 months of age. Furthermore, end-stage mice develop myelodysplasia, characterized by proliferation of atypical granulocytes that efface the bone marrow and result in severe tissue destruction. Thus, C/EBPɛ is essential for terminal differentiation and functional maturation of committed granulocyte progenitor cells.
Cellular proliferation, differentiation, and survival during hematopoiesis depend, partly, on the action of regulatory transcription factors (1, 2). Several of these transcription molecules have been shown to be critical for proper development of cells of erythroid and lymphoid lineages (2, 3). However, the molecular aspects of myeloid stem cell development and the interplay of transcription factors involved in this process have not been fully elucidated. Regulation of essential gene expression during normal myeloid differentiation is achieved by the combinatorial action of regulatory molecules that include transcription factors. Among the relatively few transcription factors that have been implicated in myelopoiesis are several members of the CCAAT/enhancer binding protein (C/EBP) family. C/EBP members belong to the basic leucine zipper class (bZIP) of transcription factors. At present, there are six members of this family (C/EBPα to ζ) that include both transcriptional activators and repressors. C/EBPs play important roles in the control of cellular proliferation and differentiation and as essential transducers of intracellular responses to extracellular control signals.
C/EBPs homodimerize and heterodimerize with each other, which is a prerequisite to binding the same C/EBP consensus DNA site. They display a high degree of homology in the carboxyl-terminal portion that includes the bZIP domain. The prototype member of this family, C/EBPα is expressed in many tissues including the liver, blood, and adipose tissue. C/EBPα −/− mice show multiple abnormalities including impaired glycogenesis, lung abnormalities (4), and impaired neutrophil development due to the absence of granulocyte colony-stimulating factor (G-CSF) signaling (5). A number of studies indicate that C/EBPβ also plays a role in myeloid cells (6, 7). Furthermore, targeted inactivation of C/EBPβ in mice results in macrophage dysfunction, impaired tumor killing and lymphoproliferative disorders (8, 9). C/EBPγ −/− and C/EBPδ −/− mice have been generated, but do not appear to have any obvious abnormalities in hematopoiesis (D. Ron, personal communication).
C/EBPα, C/EBPβ, and C/EBPδ are all expressed during hematopoiesis, suggesting that the combinatorial action of these proteins may regulate a variety of genes in myelopoiesis (10). C/EBPɛ is a recently cloned member of this family. The human C/EBPɛ gene locus can generate four transcripts by alternative use of promoters and differential splicing that encode three proteins of different transcriptional activation potentials (11). However, all C/EBPɛ RNA transcript isoforms share the same bZIP domain. In mice, only one promoter is used (see Fig. 1A), but two proteins are generated by alternative use of translational start sites. Unlike the other C/EBP proteins, expression of C/EBPɛ in both human and mice is limited to cells of hematopoietic origin (refs. 11 and 12; R.Y., unpublished data). C/EBPɛ is preferentially expressed during granulocytic differentiation, suggesting a possible role in myeloid development (11). Ectopic expression of a cDNA clone of the human C/EBPɛ results in significantly increased proliferation of NB-4 myeloid cells (13). To ascertain the role of the C/EBPɛ protein in vivo, we have generated C/EBPɛ-deficient mice by homologous recombination in embryonic stem (ES) cells. Here we report that C/EBPɛ is critical for proper granulocytic development and function, despite overlapping expression of C/EBPα, another critical gene for myeloid differentiation.
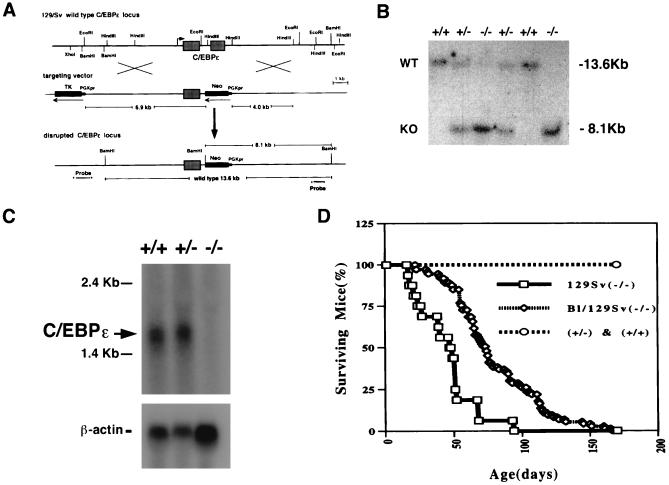
Figure 1.
(A) Targeted inactivation of the C/EBPɛ gene. Restriction map (Top) of the murine locus containing the C/EBPɛ gene. The 6.9-kb EcoRI 5′ fragment and the 4.0-kb HindIII 3′ fragment used for targeting vector construction are shown (Middle). (B) Southern blot analysis of BamHI-digested genomic DNA extracted from a litter of a heterozygote cross. The 13.6-kb wild-type (WT) fragment from the nontargeted allele and the 8.1-kb targeted (KO) fragment from the mutated allele are shown. The genotypes are indicated above the lanes, +/+ represents wild-type, +/− heterozygous, and −/− mutant animals. The blot was probed with the HindIII–EcoRI flanking probe indicated in A. (C) Northern blot analysis of RNA extracted from bone marrow. The presence of the C/EBPɛ transcripts in +/+ and +/− animals is shown. The blot was probed with part of exon 1 of C/EBPɛ and reprobed with β-actin. (D) Early lethality in C/EBPɛ −/− mice. Mortality of C/EBPɛ −/−, 129/SvEv pure background (□), 129Sv/NIH Black-Swiss mixed background (⋄), C/EBPɛ +/−, and +/+ (○) are shown.
MATERIALS AND METHODS
Generation of C/EBPɛ-Deficient Mice.
A C/EBPɛ gene targeting construct (Fig. 1A) was generated by subcloning a 6.9-kb EcoRI genomic fragment containing the 5′ end of the locus and a 4.0-kb HindIII fragment containing 3′ end in a pPNT backbone (14). The resulting vector (Fig. 1A) replaces the 1.2-kb HindIII fragment containing the 334 bp of bZIP region with a PGKneo gene. The targeting vector was linearized and electroporated into TC1 ES cells as described (14). BamHI-digested DNA from ES cells was hybridized to a 1.4-kb EcoRI/HindIII fragment representing the region 3′ of the genomic sequence (Fig. 1A). Seven (6.6%) of 106 individual targeted ES clones were selected and verified by a extensive restriction analysis using flanking and internal probes to demonstrate that the locus was targeted correctly (data not shown). Two TC1 ES cells with one targeted C/EBPɛ allele were injected into C57BL/6 blastocysts and implanted into (CBA × C57BL/6)F1 foster mothers. Resulting 14 male chimeras were mated to C57BL/6 females to verify germ-line transmission by coat color. Agouti offspring was screened for presence of the targeted C/EBPɛ allele according to Laird et al. (15). Heterozygous offspring were then intercrossed to obtain homozygous mice with both C/EBPɛ alleles targeted. Several of the chimeras transmitted the targeted allele at high frequency, allowing the mutation to be established in a mixed (129/SvEv × NIH Black Swiss) and a complete inbred (129/SvEv) background. All mouse experiments complied with the National Human Genome Research Institute Animal Care and Use Committee and American Association Laboratory Animal Care regulations.
Hematological and Histopathological Analysis.
Blood was collected into heparinized tubes and bone marrow was collected from femoral bone. For in vitro differentiation assays, a total of 2 × 105 bone marrow cells of 2-week-old C/EBPɛ +/+ and −/− mice were incubated at 37°C in methylcellulose MethoCult M3435 (StemCell Technologies, Vancouver) which includes 0.9% methylcellulose in Iscove’s modified Dulbecco’s media, 1% BSA, 10−4 M 2-mercaptoethanol, 2 mM l-glutamine, 15% fetal bovine serum, 10 μg/ml bovine pancreatic insulin, 200 μg/ml human transferrin, 10 ng/ml recombinant murine interleukin 3 (IL-3), 10 ng/ml recombinant human IL-6, and 50 ng/ml recombinant murine stem cell factor for 7–10 days. Individual colonies and bone marrow cells were placed onto glass slides by cytospin centrifugation, stained with Wright–Giemsa dye, and differential cell counts were performed. Thymus, spleen, lymph node, bone marrow, and peripheral blood were analyzed by flow cytometry. Cells extracted from each of these tissues were mixed with antibodies conjugated to either fluorescein isothiocyanate, R-phycoerythrin, or Cy-Chrome (CY). The antibodies include CD3e, CD4, CD5, CD8a, CD11b, CD38, CD43, CD45, CD45.2, CD45R/B220, CD62L, Thy-1.1, Thy-1.2, Gr-1, and c-Kit (PharMigen). Tissues were collected and fixed in 10% neutral buffered formalin. Hematoxylin/eosin staining was performed according to standard procedures.
Thioglycollate (TGC) Challenge and Dihydrorhodamine 123 (DHR) Assay.
One-month-old −/− mice and age-matched littermates received 2 ml 4% TGC broth by i.p. injection. Mice were bled at 4.5- and 24-h time points, killed by CO2 inhalation, and peritoneal exudate cells harvested by lavage with 8 ml Hanks’ balanced salt solution. Peripheral blood smears and bone marrow and lavage fluid cytospins were stained with Wright–Giemsa and leukocyte alkaline phosphatase stains at each time point. DHR assays were performed as described (16, 17) with flow cytometric analysis of the granulocytes collected from peripheral blood and peritoneal lavage prior to, and subsequent to, receiving i.p. TGC.
Northern Blot Analysis and Reverse Transcription–PCR Assay.
Total and messenger RNA were isolated from whole bone marrow and spleen cells in mice 2–4 weeks old using the RNA purification system (Pharmacia). Cellular content of bone marrow and spleen was the same in C/EBPɛ +/+ and −/− mice. Northern blot analysis was carried out using 20 μg of total RNA as described (11, 12). The RT-PCR condition used were 94°C for 1 min, 68°C for 30 s, and 72°C for 2 min followed by an extension of 5 min at 72°C, with 20 to 30 cycles as described (11). Plasmid standard curves using actin and tublin DNA (0.1, 1, 10, and 100 ng) were co-amplified with the samples to ascertain the linearity of the amplification. All PCR products were examined for specificity by Southern blot hybridization using internal probes. Primers and internal probes for each of the cytokine gene tested and β-actin were from CLONTECH, and primers used for C/EBPɛ gene were RY48 (AGCCCCCGACACCCTTGATGA), RY49 (TGGCACACTGCGGGCAGACAG), RY50 (GTCCCCTTCTCAAGGCACCCT), and internal probe RY52, RY62 (11). Gene expression from both exon 1 and 2 was examined by combination of primers RY49/RY48 and RY50/RY48, respectively. PCR-assisted mRNA analysis was repeated at least twice and performed on six to seven separate cDNA samples, yielding similar results.
RESULTS
C/EBPɛ-Deficient Mice.
The same phenotypic effects were observed in all C/EBPɛ −/− mice derived from all chimeras. Southern blot analysis of tail DNA showed successful transmission of the targeted-allele (Fig. 1B). Northern blot analysis (Fig. 1C) and RT-PCR (data not shown) with RNA extracted from bone marrow showed complete absence of C/EBPɛ RNA in C/EBPɛ −/− mice. Mice heterozygous for the C/EBPɛ mutation appeared normal. Homozygous mice were born approximately at the expected 1:2:1 Mendelian ratio (24.1% +/+, 54.1% +/−, 21.6% −/−, n = 393; χ2 test, P > 0.10), and appeared healthy at birth. Young C/EBPɛ −/− mice showed no difference in behavior and reproductive ability as compared with wild-type and heterozygous littermates. However, C/EBPɛ −/− animals raised under specific pathogen-free conditions became moribund and died within 3 to 5 months (Fig. 1D). The majority of 129/SvEv pure background −/− mice died by 2 months, 129Sv/NIH Black-Swiss mixed background −/− mice survived longer but died by 5 months of age (Fig. 1D).
Hematological Analysis.
We evaluated the hematological parameters of peripheral blood and bone marrow from C/EBPɛ +/+ and −/− mice at different ages (Table 1). A marked increase in granulocytic progenitors was evident in C/EBPɛ −/− bone marrow. In addition, C/EBPɛ −/− mice had increased numbers of hyposegmented morphologically atypical neutrophils in peripheral blood, and decreased numbers of eosinophils in both peripheral blood and bone marrow. In vitro differentiation analysis of bone marrow cells from C/EBPɛ +/+ and −/− mice at 2 weeks of age showed no quantitative differences in the number of colonies formed. Cytospin analysis of the cell morphology, however, indicated an impairment of neutrophil differentiation in colony-forming unit granulocyte/macrophage colonies from C/EBPɛ −/− mice (Fig. 2 G and H). Although wild-type colonies showed cells in various stages of differentiation from promyelocytes to neutrophils, colonies from C/EBPɛ −/− mice contained mostly (>90%) promyelocytes and myelocytes. Few metamyelocytes were observed. Further observations were done up to 15 days of culture; however, no significant differences were seen compared with observations done at day 10. We also used flow cytometry to analyze the expression of cell lineage markers. The most striking finding was the decreased frequencies of the Gr-1-positive mature granulocyte population in bone marrow and spleen in 3–4 months age of C/EBPɛ −/− mice, consistent with the result of bone marrow differential cell counts. Although the relative frequencies of B and T cells subsets were decreased in bone marrow, spleen, and peripheral blood, the total numbers of lymphocyte subsets were normal owing to the proportionate increases in cellularity. In C/EBPɛ −/− mice, the cellular composition of the thymus and lymph node were essentially normal (data not shown). Therefore, based on both cell morphology and cell surface differentiation marker analysis, C/EBPɛ deficiency blocks the maturation of granulocytic lineages, with no effect on either erythropoiesis and lymphopoiesis.
Table 1.
Hematological parameters of bone marrow and peripheral blood from C/EBPɛ +/+ and −/− mice
| C/EBPɛ | Peripheral blood
|
Bone marrow
|
||||
|---|---|---|---|---|---|---|
| (+/+) (n = 3) 2 weeks | (−/−) (n = 4) 2 weeks | (+/+) (n = 6) 3–4 months | (−/−) (n = 4) 3–4 months | (+/+) (n = 4) 3–4 months | (−/−) (n = 6) 3–4 months | |
| Differential cell counts, % | ||||||
| Segmented neutrophils | 15.3 ± 4.7 | 7.8 ± 4.2 | 4.0 ± 7.3 | 10.1 ± 2.2 | 4.1 ± 3.3 | |
| Hyposegmented/atypical neutrophils | 0 | 10.2 ± 7.4 | 0 | 30.5 ± 21.0 | ||
| Band neutrophils | 3.0 ± 0.8 | 5.5 ± 2.3 | 1.3 ± 1.5 | 12.5 ± 0.5 | 15.8 ± 1.1 | 19.0 ± 10.7 |
| Metamyelocytes | 0 | 1.5 ± 3.0 | 0 | 20.0 ± 4.5 | 8.5 ± 0.9 | 40.1 ± 7.0 |
| Myelocytes | 0 | 0.5 ± 1.0 | 0 | 2.5 ± 3.0 | 7.7 ± 2.2 | 19.7 ± 6.6 |
| Promyelocytes | 0 | 0 | 0 | 0 | 0.4 ± 0.2 | 2.0 ± 1.2 |
| Myeloblasts | 0 | 0 | 0 | 0 | 0.5 ± 0.2 | 2.5 ± 1.5 |
| Eosinophils | 2.6 ± 3.7 | 0.2 ± 0.5 | 0.8 ± 0.7 | 0 | 4.4 ± 1.4 | 0.1 ± 0.1 |
| Lymphocytes | 76.6 ± 6.6 | 80.0 ± 11.1 | 89.1 ± 5.5 | 30.0 ± 15.8 | 23.0 ± 4.6 | 5.3 ± 3.1 |
| Monocytes | 2.3 ± 0.5 | 2.0 ± 1.4 | 0.8 ± 1.3 | 0.5 ± 1.0 | 0.1 ± 0.1 | 0.1 ± 0.1 |
| Nuclear erythroid cells | 0 | 0 | 0 | 0 | 28.8 ± 6.8 | 6.4 ± 5.8 |
| Megakaryocytes | 0 | 0 | 0 | 0 | 0.1 ± 0.2 | 0.1 ± 0.1 |
| White blood cell, 103/μl | 9.6 ± 2.3 | 15.9 ± 6.8 | ||||
| Red blood cell, 106/μl | 8.8 ± 1.0 | 8.3 ± 1.2 | ||||
| Platelet, 103/μl | 753.0 ± 201.0 | 884.0 ± 285.9 | ||||
Note that hematocrit and hemoglobin values were similar between the different genotypes. Differential cell counts were performed by identifying 300 cells per peripheral blood smear and 500 cells per marrow smear. Numbers are given as means ± SE.
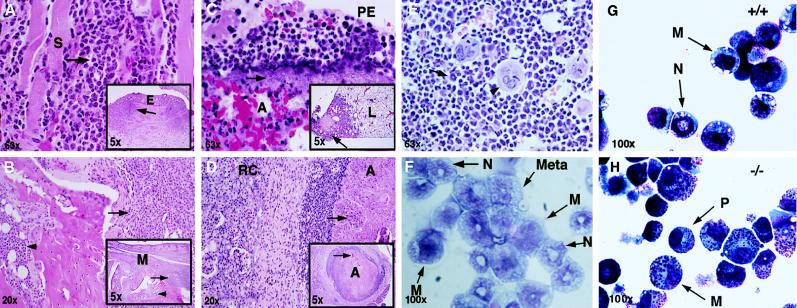
Figure 2.
Histopathology of C/EBPɛ −/− mice. In A–E, histological sections of various organs are shown, and in F bone marrow cytospin from 4 month-old −/− mice and cytospin preparations of colony-forming unit granulocyte/macrophage colonies from +/+ (G) and −/− (H) animals are shown. (A) Eyelid (E); intense inflammatory infiltrate consisting of predominantly atypical neutrophils. The infiltrate extends into the palpebral skeletal muscle (S) with associated myofiber degeneration. This conjunctivitis is likely of bacterial etiology. (B) Metatarsus (M); deep cellulitis extending to involve the cortex of the metatarsal bones. The inflammatory cell infiltrate (arrows) is composed predominantly of atypical neutrophils. Myeloid hyperplasia is noted within the marrow cavity (arrowheads). (C) Right caudal lung lobe (L); focally extensive pleuritis and necrotizing pneumonia (Inset). Bacterial rods (arrow) are prominent in the subpleural space. A mixed cell infiltrate consisting of atypical neutrophils and macrophages are present in the pleural exudate (PE) as well as within the affected alveoli (A). (D) Kidney; abscess (A) within renal medulla. Central zone of necrotic cell debris (arrows) admixed with bacterial rods surrounded by a densely cellular infiltrate comprised of atypical neutrophils, macrophages and lymphocytes. A band of fibrous connective tissue separates the abscess from the adjacent renal cortex (RC). (E) Bone marrow; the marrow space is packed with immature myeloid cells (arrow). A number of mitotic figures are present. Some megakaryocytes (arrowhead) are also present. (F) Bone marrow cytospin; immature myeloid cells are noted. Myelocyte (M), metamyelocyte (Meta), and band neutrophil (N) are seen. (G and H) In vitro differentiation. Representative examples of cell morphology by cytospin centrifugation. (G) Cells from wild-type colonies; M, myelocytes; N, neutrophils. (H) Cells from C/EBPɛ −/− colonies. P, promyelocytes; M, myelocytes.
Histopathological Examination.
Necropsy examination of 90 C/EBPɛ −/− mice ranging in age from 8 to 20 weeks revealed a variety of lesions, most of which appeared to be associated with opportunistic bacterial infections. Many of these mice had necrotizing enterocolitis, associated peritonitis, and abdominal and renal abscesses because of infection with Pseudomonas aeruginosa. Representative histopathological sections are shown in Fig. 2. In addition, some of the mice developed pulmonary abscesses, pneumonia, pleuritis, and atrial thrombosis because of Pseudomonas infection. Some of these lesions were because of septic embolization. Subcutaneous abscesses, superficial and deep cellulitis, and osteomyelitis were present in some mice, with involvement of the maxilla and distal hindlimbs. Inflammatory infiltrates in these affected tissues were characterized histologically by large numbers of atypical hyposegmented granulocytes, admixed with variable numbers of macrophages and lymphocytes. Bacterial rods consistent with P. aeruginosa were evident in some lesions (Fig. 2C). Furthermore, many mice developed conjunctivitis and rhinitis due to infection with Pasteurella pneumotropica (data not shown). Lesions were characterized by a cellular infiltrate of atypical granulocytes, extending from the submucosa into the palpebral skeletal muscle. Reactive regional lymphadenopathy, with plasma cell hyperplasia, and splenic and hepatic extramedullary hematopoiesis, which was predominantly myeloid was apparent in many cases. In the bone marrow of C/EBPɛ −/− mice, variable numbers of distorted macrophages containing intracytoplasmic eosinophilic acicular crystals were seen. Bone marrow hyperplasia was present with proliferation predominantly of myeloid precursor cells. Myeloid cell differentiation appeared to progress only to the level of hyposegmented neutrophils, lacking the condensed chromatin and low nuclear to cytoplasmic ratio seen in the peripheral blood of wild-type littermates (Fig. 3A, third panel). These data demonstrate that all C/EBPɛ −/− mice develop signs of myelodysplasia and abnormal granulopoiesis.
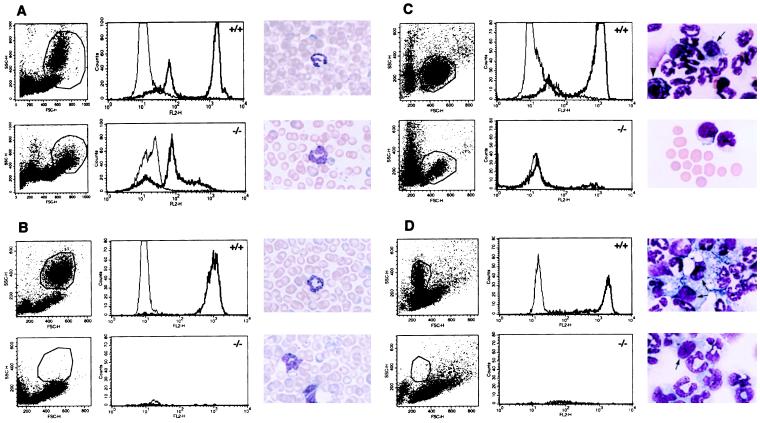
Figure 3.
Granulocytes in C/EBPɛ −/− mice are not functional. Oxidative burst was measured in granulocytes by the conversion of DHR to fluorescent rhodamine-123 (which occurs with PMA stimulation in the presence of reactive oxygen intermediates) and assayed by flow cytometry. White cells from C/EBPɛ +/+ and −/− mice were analyzed, with a total of 10,000 cells gated in both samples (A). PMA stimulation of cells demonstrated no oxidative burst in the C/EBPɛ −/− cells, as seen in the +/+ cells (thick line, PMA-stimulated cells; thin line, unstimulated). Wright–Giemsa stain shows atypical hyposegmented neutrophils are present in the peripheral blood of −/− mice (A). Following TGC-induced peritonitis, DHR assays were performed on the peripheral blood (B) and peritoneal exudate of +/+ and −/− littermates (C and D). Myelocytes, metamyelocytes, and a significantly reduced number of band neutrophils in the −/− mice are noted, compared with 85% segmented neutrophils seen in the +/+ littermates (smears, B–D) in both the peritoneal fluid and peripheral blood. Arrows show peritoneal macrophages. Arrowhead shows basophil.
Granulocyte Function.
To further understand the role of opportunistic infections in leading to the death of the C/EBPɛ −/− animals, we examined granulocyte function. The DHR assay (16, 17) measures the oxidative burst in phorbol 12-myristate 13-acetate (PMA)-stimulated neutrophils. By using this assay it was demonstrated that the peripheral blood of C/EBPɛ −/− animals was clearly abnormal compared with wild-type littermates (Fig. 3A). The amount of fluorescence can be expressed as a normal oxidative index (NOI), which is calculated by dividing the mean fluorescence channel of stimulated cells by the mean fluorescence channel of unstimulated cells. NOI in wild-type animals averaged 74.1 ± 23.6, comparable to normal humans (>30); however NOI in C/EBPɛ −/− animals averaged 6.2 ± 2.1, similar to levels seen in chronic granulomatous disease patients (<10) (17). Furthermore, the pattern of DHR excitation in the C/EBPɛ −/− gated cells closely resembled that of wild-type monocytes, suggesting that the fluorescence seen in the C/EBPɛ −/− sample was because of monocytes typically found in the granulocyte gated pool. We also examined the ability of polymorphonuclear leukocytes to migrate and function in response to chemically induced peritonitis following intraperitoneal TGC challenge (Fig. 3 B–D). TGC (4%), when injected i.p. into normal mice, results in a well characterized migration of polymorphonuclear leukocytes into the peritoneal space during the first 24 h. Morphologic examination of peritoneal and peripheral blood cells at 4.5 and 24 h following i.p. TGC demonstrated a increased number of myelocytes, metamyelocytes, and a greatly reduced number of band neutrophils in the C/EBPɛ −/− mice, compared with 85% segmented neutrophils seen in the wild-type littermates. DHR assay performed both 4.5 and 24 h following TGC challenge showed a large granulocyte population seen by forward and side scatter on flow cytometry in the peripheral blood and peritoneal exudate of wild-type animals (Fig. 3 B–D Left). This population was greatly reduced in C/EBPɛ −/− mice. PMA stimulation of both blood and peritoneal exudate showed no NADPH oxidase activity in C/EBPɛ −/− mice. Thus, both the morphology and the functional ability of C/EBPɛ-deficient neutrophils is abnormal.
Growth Factor and Cytokine Gene Expression.
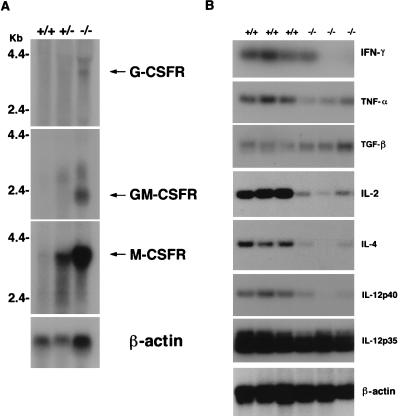
Members of the C/EBP family of transcription factors participate in the regulation of a variety of hematologically and immunologically relevant genes, including the receptors of hematopoietic growth factors (11, 18). Northern blot analysis of mRNA derived from bone marrow cells revealed an increase in mRNAs for all tested growth factor receptors (Fig. 4A). Possible explanations for this finding might be that the C/EBPɛ repressor isoform suppresses these genes in wild-type animals. Alternatively, the increased number of myeloid precursor cells in the bone marrow of C/EBPɛ-deficient mice may account for the elevated levels of receptor mRNAs. It is highly unlikely that the block observed in granulocytic differentiation is because of reduced production of G- or granulocyte/macrophage-CSF in C/EBPɛ −/− mice. Transplantation of bone marrow from C/EBPɛ −/− into lethally irradiated, C/EBPɛ +/+ recipients failed to produce normal granulocytes, only generated nonfunctional and hyposegmented granulocytes (data not shown). These results suggest a cell-autonomous block in granulocytic maturation and place it downstream of the receptors of growth factors involved in regulating neutrophil growth and differentiation. Many important aspects of the immune system are controlled by cytokine-induced signaling (19, 20). Our previous results have shown that C/EBPɛ is expressed in myeloid as well as lymphoid cells. Furthermore, some cytokine genes (e.g., IL-4, IL-6, IL-12, tumor necrosis factor α) are known to contain functional C/EBP binding sites within their promoters. To assess the effect of C/EBPɛ deficiency on splenic T cells, we determined the constitutive resident levels of several cytokine mRNAs. Several cytokine genes were indeed found to be affected by the absence of C/EBPɛ (Fig. 4B). The mRNAs encoding interferon γ, IL-2, IL-4, IL-12p40, and to a lesser extend tumor necrosis factor α were expressed at lower levels in C/EBPɛ-deficient mice (Fig. 4B). In contrast, the mRNA levels of transforming growth factor β and IL-12p35 were not altered. Taken together these results indicate an important role of C/EBPɛ in the regulation of cytokine genes. However, it is possible that the changes in cytokine mRNA levels contribute to the increased susceptibility of C/EBPɛ-deficient mice to opportunistic infections.
Figure 4.
Growth factor receptor and constitutive cytokine expression levels in C/EBPɛ −/− mice. (A) Northern blot analysis of mRNA for several growth factor receptors. Total bone marrow RNA (20 μg) from C/EBPɛ +/+ (lane 1), +/− (lane 2), and −/− (lane 3) mice were analyzed with murine cDNA probes for each of the receptor indicated. (B) Semiquantitive RT-PCR analysis of constitutive levels of mRNAs from a variety of cytokines in spleens of three +/+ and −/− littermates.
DISCUSSION
Our analysis of the effect of the targeted disruption of the C/EBPɛ gene has demonstrated a block in the development of mature granulocytes and myelodysplasia. These findings suggest that C/EBPɛ plays an essential role as a regulator of myelopoiesis. In addition, C/EBPɛ is critical for the maintenance of the constitutive levels of some cytokines.
C/EBPɛ −/− mice raised under specific pathogen free conditions invariably become moribund by 2–5 months of age, depending on the genetic background. Serological and bacteriological analyses in a number of clinically end-stage animals showed that in more than 60% of the cases P. aeroginosa was isolated from multiple organs. Thus, it appears that C/EBPɛ −/− animals die either from opportunistic infections, mainly by P. aeroginosa, or from complications of myeloproliferation. The myeloproliferation may also be explained by the lack of proper neutrophil function which may stimulate migration of myeloid cells to the site of primary stimulus leading to nonspecific tissue destruction. Mice engineered to lack G-CSF receptor have impaired production and increased apoptosis of neutrophils (21), whereas G-CSF-deficient mice have chronic neutropenia and myeloid progenitor cell deficiency (22). However, these mice display no signs of myelodysplasia and do not die from opportunistic infections. It is likely that the myelodysplasia displayed in these animals is intrinsic to C/EBPɛ-deficient myeloblasts and is augmented by a secondary myeloreactive response to inflammation. Interestingly, allelic loss of the C/EBPɛ gene was detected in 4 of 20 human cases evolving myelodysplastic syndrome (H. P. Koeffler, personal communication). There are several other mouse models of myelodysplasia. Targeted inactivation of the interferon consensus sequence binding protein results in a myeloproliferative disease with similarities to human chronic myelogenous leukemia (23). Interestingly, our mice also have decreased expression of interferon γ. Expression of BCL/ABL P210 (24), or promyelocytic leukemia–retinoic acid receptor (25) in transgenic mice develop myelolymphoproliferative disease or acute myeloid leukemia, respectively. On the other hand, several lymphoproliferative disorders have been described as a result of targeted inactivation of Ctl-4 (26), Fas (27), transforming growth factor β (28), and C/EBPβ (9), suggesting multiple mechanisms to regulate lymphocyte expansion, proliferation and survival. Analysis of the steady state mRNA expression levels of several critical hematopoietic growth factor receptor and cytokine genes demonstrates that C/EBPɛ deficiency alters the levels of constitutively expressed cytokines. The levels of resident mRNAs for interferon γ, tumor necrosis factor α, IL-4, and IL-12p40 are significantly reduced in spleens of C/EBPɛ-deficient mice. At present we cannot distinguish whether this imbalance is because of an intrinsic abnormality of hematopoietic cells in these mice or that the production of some or all of these cytokines may be a direct target of C/EBPɛ transcriptional regulation.
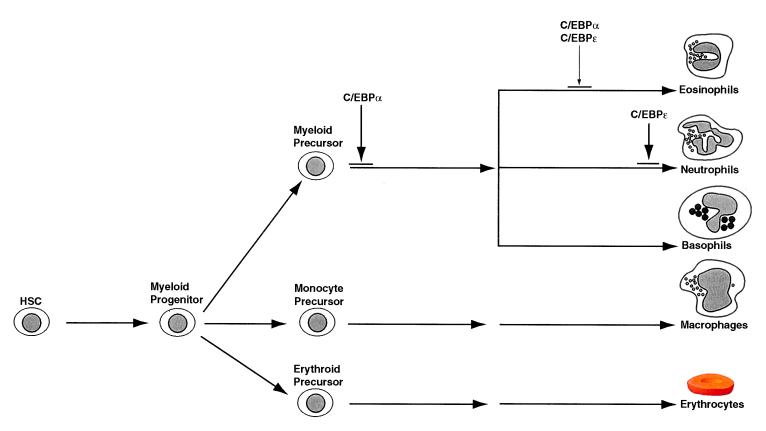
In myeloid cells, C/EBPα and C/EBPɛ are expressed in a remarkably overlapping manner (ref. 11; D. G. Tenen, personal communication). C/EBPɛ displays both activating and repressing functions, in vitro (G. D. Kim, personal communication). Consequently, the ratio of the specific C/EBPɛ isoform to C/EBPα may have opposing effects in the regulation of genes involved in myelopoiesis. Our data place C/EBPɛ downstream of C/EBPα and demonstrate its role as a critical and indispensable component of granulocytic differentiation. A model for the action of both C/EBPα and C/EBPɛ during granulopoiesis is shown in Fig. 5. Overall, our results imply that C/EBPɛ is a key regulator of myeloid homeostasis. C/EBPɛ-deficient mice may also be useful in understanding the mechanisms involved in cytokine signaling. Finally, these mice may provide a useful animal model to dissect molecular events underlying the progression to myelodysplasia.
Figure 5.
A model for the role of C/EBPɛ in granulopoiesis. C/EBPɛ acts downstream of C/EBPα and is essential for the generation of eosinophils and mature neutrophils. HSC, hematopoietic stem cell.
Acknowledgments
We are grateful to L. Garrett and T. Hernandez for excellent technical assistance, Drs. T. Fredrickson and D. G. Tenen for expert advice and reagents, and D. Bodine, S. Holland, T. Lawrence, A. Merriweather, P. Murphy, D. Horne, K. Ozato, and L. Taddesse-Hedth for critical input. We also thank Dr. R. M. Blaese for creating an inspiring environment and providing constant support. Mouse granulocyte-, granulocyte/macrophage-, and macrophage-CSF receptor probes were kindly provided by Dr. D. G. Tenen.
ABBREVIATIONS
- C/EBP
CCAAT/enhancer binding protein, RT, reverse transcription
- bZIP
basic leucine zipper
- G-CSF
granulocyte colony-stimulating factor
- ES cell
embryonic stem cell
- TGC
thioglycollate
- DHR
dihydrorhodamine 123
- PMA
phorbol 12-myristate 13-acetate
- IL
interleukin
References
- 1.Ness S A, Engel J D. Curr Opin Gen Dev. 1994;4:718–724. doi: 10.1016/0959-437x(94)90139-t. [DOI] [PubMed] [Google Scholar]
- 2.Shivdasani R, Orkin S. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 3.Tenen D G, Hromas R, Licht J, Zhang D-E. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 4.Wang N, Finegold M J, Bradley A, Ou C N, Abdelsayed S V, Wilde M D, Taylor L R, Wilson D R, Darlington G J. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D-E, Zhang P, Wang N D, Hetherington C J, Darlington G J, Tenen D G. Proc Natl Acad Sci USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz S, Kowenzleutz E, Muller C, Meese K, Ness S A, Leutz A. EMBO J. 1993;12:1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 9.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, Bistoni F, Frati L, Cortese R, Gulino A, Ciliberto G, Costantini F, Poli V. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott L M, Civin C I, Rorth P, Friedman A D. Blood. 1992;180:1725–1735. [PubMed] [Google Scholar]
- 11.Yamanaka R, Kim G D, Radomska H S, Lekstrom-Himes J, Smith L T, Antonson P, Tenen D G, Xanthopoulos K G. Proc Natl Acad Sci USA. 1997;94:6462–6467. doi: 10.1073/pnas.94.12.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonson P, Stellan B, Yamanaka R, Xanthopoulos K G. Genomics. 1996;35:30–38. doi: 10.1006/geno.1996.0319. [DOI] [PubMed] [Google Scholar]
- 13.Chumakov A M, Grillier I, Chumakova E, Chih D, Slater J, Koeffler H P. Mol Cell Biol. 1997;17:1375–1386. doi: 10.1128/mcb.17.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng C, Wynshaw-Boris A, Kuo A, Zhou F, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 15.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vowells S J, Sekharia S, Malech H L, Shalit M, Flesher T A. J Immunol Methods. 1995;178:89–97. doi: 10.1016/0022-1759(94)00247-t. [DOI] [PubMed] [Google Scholar]
- 17.Rose N R. Manual of Clinical Laboratory Immunology. 5th Ed. Washington, DC: Am. Soc. Microbiol.; 1997. [Google Scholar]
- 18.Cappelletti M, Alonzi T, Fattori E, Libert C, Poli V. Cell Growth Differ. 1996;3:29–35. [PubMed] [Google Scholar]
- 19.Ihle J N. Nature (London) 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 20.Trinchieri G, Perrit D, Gerosa F. Cytokine Growth Factor Rev. 1996;2:123–132. doi: 10.1016/1359-6101(96)00018-4. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Wu H Y, Wesselschmidt R, Kornaga T, Link D C. Immunity. 1996;5:491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 22.Lieschke G J, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, Fowler K J, Basu S, Zhan Y F, Dunn A R. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 23.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch K, Gabriele L, Waring J F, Bachmann M F, Zinkernagel R M, Morse H C, III, Ozato K, Horak I. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 24.Voncken J W, Kaartinen V, Pattengale P K, Germeraad W T V, Groffen J, Heistererkamp N. Blood. 1995;86:4603–4611. [PubMed] [Google Scholar]
- 25.Early E, Moore M A S, Kakizuka A, Nason-Burchenal K, Martin P, Evans R M, Dmitrovsky E. Proc Natl Acad Sci USA. 1996;93:7900–7904. doi: 10.1073/pnas.93.15.7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterhouse P, Penninger J M, Timms E, Wakenham A, Shahinian A, Lee K P, Thompson C B, Griesser H, Mak T W. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe-Fukunaga R, Brannan C I, Copeland N G, Jenkins N A, Nagata S. Nature (London) 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 28.Shull M M, Ormsby I, Kier A B, Pawlowski S, Diebold R J, Yin M, Allen R, Sidman C, Proetzel G, Calvin D. Nature (London) 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]