Abstract
Hereditary hemochromatosis (HH) is a common autosomal recessive disease associated with loss of regulation of dietary iron absorption and excessive iron deposition in major organs of the body. Recently, a candidate gene for HH (also called HFE) was identified that encodes a novel MHC class I-like protein. Most patients with HH are homozygous for the same mutation in the HFE gene, resulting in a C282Y change in the HFE protein. Studies in cultured cells show that the C282Y mutation abrogates the binding of the recombinant HFE protein to β2-microglobulin (β2M) and disrupts its transport to the cell surface. The HFE protein was shown by immunohistochemistry to be expressed in certain epithelial cells throughout the human alimentary tract and to have a unique localization in the cryptal cells of small intestine, where signals to regulate iron absorption are received from the body. In the studies presented here, we demonstrate by immunohistochemistry that the HFE protein is expressed in human placenta in the apical plasma membrane of the syncytiotrophoblasts, where the transferrin-bound iron is normally transported to the fetus via receptor-mediated endocytosis. Western blot analyses show that the HFE protein is associated with β2M in placental membranes. Unexpectedly, the transferrin receptor was also found to be associated with the HFE protein/β2M complex. These studies place the normal HFE protein at the site of contact with the maternal circulation where its association with transferrin receptor raises the possibility that the HFE protein plays some role in determining maternal/fetal iron homeostasis. These findings also raise the question of whether mutations in the HFE gene can disrupt this association and thereby contribute to some forms of neonatal iron overload.
Keywords: iron, major histocompatibility complex class I protein, neonatal iron overload, placenta
Feder et al. (1) reported the positional cloning of a candidate gene for hereditary hemochromatosis (HH) that is now called the HFE gene. [Although Feder et al. (see ref. 1) originally designated the HH candidate gene HLA-H, this designation had already been assigned to a pseudogene and the HH locus had already been assigned the name HFE by the nomenclature committee (27).] They found 83% of 178 HH patients to be homozygous for the same missense mutation (C282Y) in the HFE gene. Eight of nine HH patients who were heterozygous for this mutation were found to have a different missense mutation (H63D) on the other HFE allele (1). On the basis of these findings, they proposed that a mutation in the HFE gene is the molecular basis for most cases of HH. The high frequency of the C282Y mutation in HH patients has been confirmed by at least five other studies (2–6).
The human HFE protein predicted from the cDNA sequence is composed of 343 amino acids. It is most homologous to major histocompatibility complex (MHC) class I molecules that are integral membrane proteins with three extracellular loops (α1, α2, and α3), a transmembrane region, and a short cytoplasmic tail. The C282Y mutation was predicted to disrupt a critical disulfide bond in the α3 loop of the HFE protein and abrogate binding of the mutant HFE protein to β2-microglobulin (β2M) and its transport to and presentation on the cell surface. Feder et al. (7) confirmed these predictions by demonstrating a failure of the C282Y mutant HFE protein to associate with endogenous β2M in human embryonic kidney cells (293 cells) stably transfected with the mutant cDNA. A recent study by Waheed et al. (8) demonstrated that the wild-type HFE protein expressed in transfected COS-7 cells associates with coexpressed β2M and is transported to the cell surface, but these capabilities are lost by the C282Y mutant HFE protein. Much of the C282Y mutant protein remains in high Mr aggregates, and fails to undergo late Golgi processing. While these studies provided a mechanism whereby the C282Y mutation might impair the function of the mutant HFE protein, they did not disclose how HFE protein is related to the maintenance of iron homeostasis or how the C282Y mutation might produce HH.
We recently used immunohistochemistry to demonstrate the localization of the HFE protein in the gastrointestinal tract (9). The unique localization of the HFE protein in the crypts of the small intestine suggested a possible role for the HFE protein in iron sensing in the cryptal cells. In the present study, we examined human placenta because it was one of the tissues in which the HFE transcript was identified and it seemed a potentially convenient tissue source to isolate proteins that associate with the HFE protein and might be more directly involved in iron metabolism (1). We used immunohistochemistry to define the site of the HFE protein expression in placenta and used immunoaffinity chromatography and immunoprecipitation to identify some of the proteins with which it is associated. The studies reported here demonstrate that the HFE protein is expressed on the apical plasma membrane of the syncytiotrophoblasts, where iron is normally transported to the placenta via transferrin receptor-mediated endocytosis. Furthermore, our results demonstrate that the HFE protein is physically associated with the transferrin receptor that plays a central role in iron transport across the placenta. Thus, the novel HFE protein could be one link in regulating transfer of iron from maternal blood to the fetus.
MATERIALS AND METHODS
Antibodies.
The production and characterization of the antibody raised against a polypeptide of 16 C-terminal amino acids based on the cDNA of the HFE protein have been described (9). We refer to this antibody as CT16. Using a corresponding peptide-Affigel 10 affinity resin, affinity-pure, peptide-specific IgG was isolated and stored in 50% glycerol at −20°C. Rabbit anti-human β2M antibody and normal human IgG were purchased from Sigma. Monoclonal anti-human transferrin receptor antibody was a product of Dakopatts (Glostrup, Denmark).
Human Placenta Specimens and Immunohistochemistry.
The full-term placentas (n = 2) were collected immediately after vaginal delivery. There were no known pathological aspects affecting placental structure or function. The placental specimens for biochemical studies were frozen in liquid nitrogen and stored at −80°C before use. The specimens for immunohistochemistry were fixed and embedded in paraffin (10) and immunostaining was performed using an immunoperoxidase technique as described (9).
Preparation of Placental Membrane and Biotinylation of the Proteins.
The frozen placenta was thawed and homogenized in ice-cold 50 mM sodium phosphate buffer, pH 7.5, containing 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 1 mM o-phenanthroline (as protease inhibitors) using a Brinkmann Polytron homogenizer. The tissue homogenates were centrifuged at 135,000 × g for 30 min. The cytosol and total membrane pellets were recovered, and the membrane pellets were suspended in the homogenization buffer. The total membrane proteins were biotinylated as described (11).
Chemical Cross-Linking of HFE Protein-β2M-Transferrin Receptor Complex.
The membrane suspension of a human term placenta specimen was mixed with a reversible bifunctional cross-linker, 1 mM dithiobis (succinimidyl propionate) in 50 mM sodium phosphate buffer, pH 7.5, containing the protease inhibitors, and the mixture was incubated at room temperature for 10 min (12). The cross-linking was quenched with 10 mM ethanolamine. The cross-linked proteins were solubilized with 1% Nonidet P-40, and the membrane extract obtained after cross-linking was applied to a CT16 IgG Affigel-10 column. Unbound proteins were removed by washing with PBS. The bound proteins were eluted with 0.1 M glycine⋅HCl solution, pH 2.5, containing 0.1% Nonidet P-40, and the eluted material was mixed with 1 M Tris base to neutralize the pH of the solution. The eluted proteins were concentrated using Centricon P-10 tubes (Amicon) and analyzed by SDS/PAGE followed by Western blot analysis. The cross-linked placental membrane extracts were immunoprecipitated using anti-human transferrin receptor antibody and the immunocomplex was analyzed by SDS/PAGE followed by Western blot analysis.
Affinity Purification of Neonatal Fc Receptor (hFcRn) and HFE Protein from Human Placenta.
The biotinylated placental membrane proteins were extracted with 50 mM sodium phosphate buffer, pH 7.5, containing 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM o-phenanthroline, 1 mM benzamidine, and 2% Nonidet P-40. After centrifugation at 40,000 × g for 30 min, the supernatant was applied to a human IgG Affigel-10 column, and pH of the extract was adjusted to pH 6.2. Unbound proteins were washed out with 50 mM sodium phosphate buffer, pH 6.5, while bound hFcRn and associated proteins were eluted with 50 mM sodium phosphate buffer, pH 8.0, containing 0.1% Nonidet P-40 (13). The eluted proteins were concentrated using Centricon P-10 tubes (Amicon) and dialyzed against PBS.
For purification of HFE protein and associated proteins, the flow-through from the human IgG Affigel-10 column was adjusted to pH 7.5 and applied to a CT16 IgG Affigel-10 column. Unbound proteins were washed out with PBS, and bound proteins were eluted with 0.1 M glycine⋅HCl, pH 2.5. The pH of the eluted protein fraction was titrated to pH 7.0 with 1 M Tris base, and the protein fraction was concentrated using Centricon P-10 tubes (Amicon) and dialyzed against PBS. A nonspecific association of proteins with Affigel-10 resin was ruled out using a mock Affigel-10 column.
Biotinylated placental membrane proteins eluted from the CT16 IgG Affigel-10 column or from the human IgG Affigel-10 column were subjected to immunoprecipitation using a monoclonal anti-human transferrin receptor antibody. The immunocomplex was analyzed by SDS/PAGE followed by electrophoretic transfer of the polypeptides and visualization using a streptavidin-peroxidase complex (Sigma).
SDS/PAGE and Western Blot Analysis.
SDS/PAGE was performed under reducing and nonreducing conditions in a Mini-Protean electrophoresis unit (Bio-Rad) according to Laemmli (14). The polypeptides were transferred electrophoretically from the gel to a polyvinylidene difluoride membrane (Millipore). After the transblotting, the biotinylated polypeptides were visualized by a streptavidin-peroxidase complex, and HFE protein and β2M were immunostained using the anti-HFE protein (CT16) and β2M antibodies followed by incubation with peroxidase-conjugated goat anti-rabbit IgG (Sigma). The peroxidase activity was visualized using a chemiluminescent substrate.
RESULTS
Demonstration of the HFE Protein in Placenta.
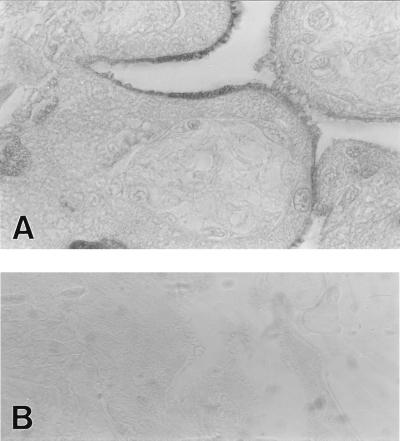
Fig. 1A shows an intense immunoreaction for the HFE protein in the syncytiotrophoblasts of human placenta, with the most prominent signal confined to the apical plasma membrane. The positive staining was inhibited by excess CT16 peptide (Fig. 1B). Additional control stainings using 1:100 diluted nonimmune rabbit serum showed no immunoreaction (data not shown). The cellular and subcellular localization of the HFE protein place it at a strategic site to participate in the regulation of iron transport across the placenta. In fact, the transferrin receptor, which is thought to mediate iron transport across the placenta, shows the same cellular and subcellular localization (15). This localization is also similar to that of hFcRn, a homologous protein that mediates maternal/fetal transport of IgG (16).
Figure 1.
Immunohistochemical localization of HFE protein in full-term human placental villi. Positive immunoreaction is confined to the apical plasma membrane of the syncytiotrophoblasts (A). The positive reaction was blocked by addition of the CT16 peptide (B). Magnifications, ×1,000 (A); ×400 (B).
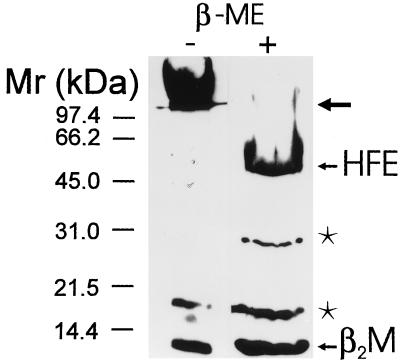
To confirm the presence of HFE protein and β2M in placenta, total placental homogenate was analyzed by Western blot using a mixture of the CT16 antibody to the C-terminal portion of HFE protein and anti-human β2M antibody. Fig. 2 shows that under nonreducing conditions, the β2M runs at the Mr expected for free β2M. However, most of the HFE protein is in high Mr complexes too large to enter the separating gel, suggesting that HFE protein exists as disulfide bridge-linked high Mr protein complexes in the placental membrane. Following reduction, the major polypeptides identified by these antibodies are the 48-kDa monomeric HFE protein and the 12-kDa β2M. Additional 30- and 18-kDa polypeptides are also seen that are thought to be proteolytic fragments of the HFE protein present in placental extracts.
Figure 2.
Western blot of HFE protein and β2M in human placenta. Total placental cell homogenate containing 25 μg equivalent protein was analyzed by SDS/PAGE in nonreducing (−) or reducing (+) conditions followed by immunostaining using the mixture of CT16 and anti-human β2M antibodies. In the first lane (under nonreducing conditions), the β2M runs at the Mr expected for free β2M while most of the HFE protein is in high molecular weight complexes too large to enter the separating gel (arrow). In the second lane (under reducing conditions), the polypeptides for monomeric HFE protein and β2M are evident in addition to polypeptides of 30 and 18 kDa (asterisks) that represent proteolytic products of HFE protein.
Immunoaffinity Chromatography of HFE and Associated Proteins.
The association of recombinant HFE protein with β2M was recently demonstrated in Western blot analysis of membrane proteins from transfected cells expressing both proteins (7, 8). The complex of HFE protein and β2M could be isolated from transfected COS-7 cell membranes by an Affigel-10 column containing immobilized CT16 antibody (8). Here we provide evidence that this column can be used to isolate the complex of HFE and associated proteins from placental membranes that was evident on the nonreducing gel in Fig. 2. To be certain that the column was isolating the HFE protein specifically, and to remove any contaminating hFcRn, which has the same molecular weight as HFE, we used two different affinity columns sequentially. The first contained immobilized human IgG, and retains the hFcRn, but should not retain HFE protein. The second column contained immobilized C-terminal peptide-specific IgG purified from the CT16 antiserum that we showed previously (8) could retain the complex of the HFE protein and β2M from COS-7 cell membranes.
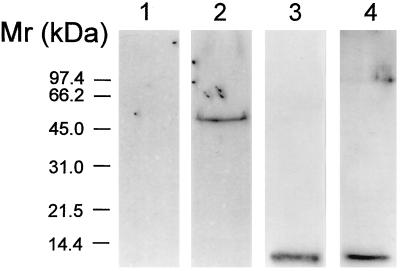
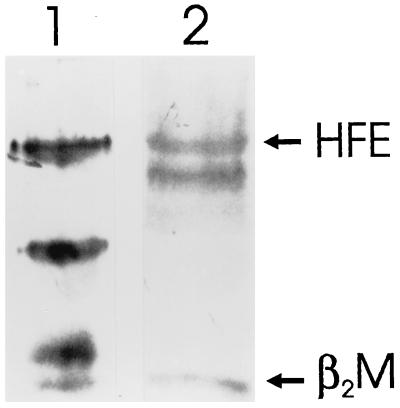
An extract of total biotinylated placental membranes was applied first to the human IgG Affigel-10 column, after which the flow-through of this column was applied to the rabbit CT16 IgG Affigel-10 column. Then, the proteins eluted from the two columns were analyzed by Western blot after SDS/PAGE. The results are shown in Fig. 3. Lanes 1 and 3 show the blots of the proteins eluted from the human IgG Affigel-10 column, while lanes 2 and 4 show blots of proteins eluted from the CT16 IgG Affigel-10 column. Lanes 1 and 2, which were probed with the CT16 antibody, indicate that the 48-kDa HFE protein was not retained on the first column (lane 1), but was retained and eluted from the CT16 IgG Affigel-10 column (lane 2). Lanes 3 and 4, which were probed with antibody to human β2M, indicate that β2M is retained and eluted from both columns.
Figure 3.
Sequential affinity purification of placental hFcRn and HFE protein complexes and characterization of HFE-associated proteins. Lanes 1 and 3 illustrate blots of biotinylated hFcRn and associated proteins eluted from a human IgG Affigel-10 column. The flow-through from the human IgG Affigel-10 column was subjected to a second immunoaffinity purification step using immobilized CT16 antibodies. Lanes 2 and 4 contain blots of biotinylated proteins that were eluted from the CT16 IgG Affigel-10 column. On Western blots, the CT16 antibody did not cross-react with any polypeptide present in the hFcRn preparation (lane 1), but did identify a 48-kDa polypeptide band corresponding to the molecular weight of monomeric HFE protein in the fraction eluted from the CT16 IgG Affigel-10 column (lane 2). Western blot analysis using anti-human β2M antibody (lanes 3 and 4) identified a 12-kDa polypeptide in both the hFcRn (lane 3) and HFE protein preparations (lane 4).
Identification of the Transferrin Receptor in the Proteins Associated with HFE Protein.
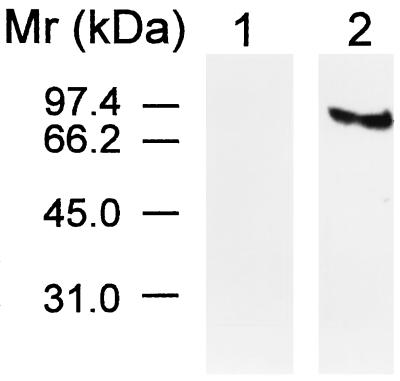
The transferrin receptor is known to be abundant in the apical plasma membrane of syncytiotrophoblasts (15) in which both the HFE protein and hFcRn are found. To determine whether the transferrin receptor copurified with either hFcRn or HFE protein, the eluates from both columns were treated with monoclonal anti-human transferrin receptor antibody and the immunoprecipitates were analyzed by SDS/PAGE. Fig. 4 shows that a polypeptide ≈93 kDa is immunoprecipitated from the proteins isolated from the CT16 IgG Affigel-10 column (lane 2) but not from the proteins isolated from the human IgG Affigel-10 column (lane 1). These data suggest that the transferrin receptor is specifically associated with the immunoaffinity-isolated complex that includes the HFE protein and β2M but is not present in the immunoaffinity-isolated complex that includes the hFcRn and β2M.
Figure 4.
Immunoprecipitation of transferrin receptor from human placental membrane proteins coisolated with the HFE protein. The biotinylated proteins eluted from the human IgG Affigel-10 (lane 1) and CT16 IgG Affigel-10 columns (lane 2) were immunoprecipitated using monoclonal anti-human transferrin receptor antibody and analyzed using a streptavidin-peroxidase complex. A polypeptide of 93 kDa was present only in the protein preparation isolated by the second column, the CT16 IgG Affigel-10 column (lane 2).
Coimmunoprecipitation of HFE and β2M by Anti-Transferrin Receptor Antibody.
Fig. 5 (lane 1) shows the association of HFE protein with β2M on a Western blot of proteins that were cross-linked chemically, isolated on the CT16 IgG Affigel-10 column, separated by SDS/PAGE under reducing conditions, and analyzed by Western blot using antibodies to HFE protein and β2M. The antibodies identified 48- and 12-kDa proteins corresponding to monomeric HFE protein and β2M, and also the 30- and 18-kDa proteolytic fragments of the HFE protein.
Figure 5.
Association of the transferrin receptor with the HFE protein and β2M. Proteins in the membrane suspension from placenta were chemically cross-linked with a reversible crosslinker. The cross-linked complexes of HFE protein and associated proteins were isolated by immunoaffinity using a CT16 IgG Affigel-10 column (lane 1). The transferrin receptor-associated proteins were isolated by immunoprecipitation using anti-human transferrin receptor antibody (lane 2). The proteins in the complexes isolated by the two methods were dissociated and separated by SDS/PAGE under reducing conditions followed by Western blot analysis. Both the complexes isolated by CT16 IgG Affigel-10 column and by immunoprecipitation with the anti-human transferrin receptor antibody contained polypeptides reacting with CT16 and anti-human β2M antibodies. Lane 1 shows a 48-kDa band of the HFE protein and the 12-kDa β2M in addition to the 30- and 18-kDa HFE-species. Lane 2 shows a 45/48-kDa doublet of HFE proteins and the 12-kDa β2M.
If the transferrin receptor is associated with the complex of HFE protein and β2M in placental membranes, as suggested by the fact that it copurifies with HFE and β2M over a CT16 IgG Affigel-10 column, we would expect the HFE protein and β2M to coimmunoprecipitate with transferrin receptor when the cross-linked proteins in placental membranes are exposed to antibody to the transferrin receptor. Fig. 5 (lane 2) shows that this expectation is realized. Both HFE protein and β2M are identified by the antibodies to the respective proteins in the immunoprecipitate derived from cross-linked placental membrane proteins that were treated with the anti-human transferrin receptor antibody. The HFE protein appears as a 48/45-kDa doublet, similar to the 48/45-kDa doublet seen in cells expressing recombinant HFE protein (7, 8), suggesting partial deglycosylation or N-terminal proteolytic degradation occurred during the immunoprecipitation with anti-transferrin receptor antibody. It is of interest that the 30- and 18-kDa proteolytic fragments of the HFE protein that are identified by the CT16 antibody on the Western blot of the material eluted from immunoaffinity column (lane 1) are not present in the complexes immunoprecipitated by the anti-transferrin receptor antibody. This suggests that the transferrin receptor does not form a stable complex with the 30- and 18-kDa proteolytic fragments of the HFE protein. The results presented in Figs. 4 and 5, taken together, clearly support the association of the transferrin receptor in a complex that includes the HFE protein and β2M in placental membranes.
DISCUSSION
The major findings presented in this study are: (i) the expression of the HFE protein in the apical plasma membrane of the syncytiotrophoblast cells, (ii) the association of the HFE protein with β2M in placental membranes, and (iii) the association of the transferrin receptor with the complex of the HFE protein and β2M. Whether the HFE protein participates in the regulation of iron transport in placenta is an interesting question raised by these provocative findings.
The rapidly growing human fetus requires a large supply of iron that is taken up from the maternal blood by the syncytiotrophoblast cells of the placenta. The rate of iron transfer to the fetus increases rapidly during the last third of pregnancy and keeps up with fetal growth (17). Iron transport across the placenta is thought to be mediated by transferrin receptors at the apical side of the syncytiotrophoblasts that face the maternal circulation. The studies presented here show that these receptors are physically associated with the HFE protein/β2M complex. It has been suggested that placental iron levels regulate the expression of the transferrin receptors on the apical plasma membrane of the syncytiotrophoblasts (18, 19) and that down-regulation of transferrin receptors provides a mechanism to protect the fetus against the entry of excess iron when maternal iron levels are high (18).
The mechanisms involved in placental iron transport are highly efficient, since fetal serum iron concentrations normally exceed those in the maternal circulation (20). Therefore, premature infants and even normal newborns have some features of iron overload in that neonates have relatively high levels of saturation of serum transferrin (21). However, only rarely do infants exhibit clinical evidence of iron toxicity in the form of neonatal iron overload (22). Although perinatal hemochromatosis resembles adult HH histopathologically, there are only a few reports suggesting that they are related genetically (23, 24). Nonetheless, it should be of interest to genotype the affected individuals and their parents to determine whether mutant HFE genotypes contribute to this extreme form of iron overload in the fetus.
Given the findings reported here that the HFE protein is expressed at the site of iron transport in the apical plasma membrane of the syncytiotrophoblast, and that the HFE protein is physically associated with the transferrin receptor that mediates that transport, it should be of great interest also to identify C282Y homozygous newborns prospectively to examine their placentas and to evaluate their iron status at birth. The availability of simple screening tests for the HFE mutations should make it possible to identify couples at risk of producing such homozygous infants. Immunocytochemistry on their placentas should disclose whether the C282Y mutation blocks transport of the HFE protein to the apical plasma membrane of syncytiotrophoblasts, as one might expect from the studies of the recombinant C282Y protein expressed in cultured cells (7, 8). Iron studies on such infants and their mothers should clarify whether homozygosity for the C282Y mutation in the fetus alters the maternal/fetal iron homeostasis in these pregnancies.
The high frequency of the HH allele in Caucasians, most of whom can now be assumed to be homozygous for the C282Y mutation (1–6) has led to the speculation that heterozygosity for the ancestral HH mutation (or another closely linked gene) must confer some positive selective advantage (25, 26). The C282Y mutation may have improved survival during infancy, childhood, and pregnancy in times past, by leading to increased iron absorption and accumulation of larger body iron stores. Although this selection could operate at the level of increased dietary iron absorption, such mutations might also lead to enhanced maternal/fetal iron transport. Such an effect might confer a selective advantage on the fetus under conditions of maternal iron deprivation, which is a much greater problem globally than maternal iron overload.
The studies here demonstrated that the HFE protein is expressed on the apical surface of the syncytiotrophoblasts and is associated in a complex with the transferrin receptor and β2M. Whether a similar interaction of the HFE protein with the transferrin receptor occurs in the small intestine, where dietary iron absorption is regulated, will be important to establish. If such an interaction is important for regulation of dietary iron absorption, it might explain how mutations in the HFE protein, which prevent this interaction, could lead to adult HH.
Acknowledgments
We acknowledge Elizabeth Torno for editorial assistance. The work was supported by grants DK40163 (W.S.S.), DK41816 (B.R.B.), and GM34182 (W.S.S.) from the National Institutes of Health, and by a grant from the Sigrid Juselius Foundation (S.P).
ABBREVIATIONS
- β2M
β2-microglobulin
- MHC
major histocompatibility complex
- hFcRn
human neonatal Fc receptor
- HH
hereditary hemochromatosis
References
- 1.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 2.Beutler E, Gelbart T, West C, Lee P, Adams M, Blackstone R, Pockros P, Kosty M, Venditti C P, Phatak P D, Seese N K, Chorney K A, TenElshof A E, Gerhard G S, Chorney M. Blood Cells Mol Dis. 1996;22:187–194. doi: 10.1006/bcmd.1996.0027. [DOI] [PubMed] [Google Scholar]
- 3.Jouanolle A M, Gandon G, Jézéquel P, Blayau M, Campion M L, Yaouanq J, Mosser J, Fergelot P, Chauvel B, Bouric P, Carn G, Andrieux N, Gicquel I, Le Gall J-Y, David V. Nat Genet. 1996;14:251–252. doi: 10.1038/ng1196-251. [DOI] [PubMed] [Google Scholar]
- 4.Jazwinska E C, Cullen L M, Busfield F, Pyper W R, Webb S I, Powell L W, Morris C P, Walsh T P. Nat Genet. 1996;14:249–251. doi: 10.1038/ng1196-249. [DOI] [PubMed] [Google Scholar]
- 5.Carella M, D’Ambrosio L, Totaro A, Grifa A, Valentino M A, Piperno A, Girelli D, Roetto A, Franco B, Gasparini P, Camaschella C. Am J Hum Genet. 1997;60:828–832. [PMC free article] [PubMed] [Google Scholar]
- 6.Borot N, Roth M-P, Malfroy L, Demangel C, Vinel J-P, Pascal J-P, Coppin H. Immunogenetics. 1997;45:320–324. doi: 10.1007/s002510050211. [DOI] [PubMed] [Google Scholar]
- 7.Feder J N, Tsuchihashi Z, Irrinki A, Lee V K, Mapa F A, Morikang E, Prass C E, Starnes S M, Wolff R K, Parkkila S, Sly W S, Schatzman R C. J Biol Chem. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- 8.Waheed, A., Parkkila, S., Zhou, X. Y., Tomatsu, S., Tsuchihashi, Z., Feder, J. N., Schatzman, R. C., Britton, R. S., Bacon, B. R. & Sly, W. S. (1997) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 9.Parkkila S, Waheed A, Britton R S, Feder J N, Tsuchihashi Z, Schatzman R C, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1997;94:2534–2539. doi: 10.1073/pnas.94.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mühlhauser J, Crescimanno C, Rajaniemi H, Parkkila S, Milovanov A P, Castellucci M, Kaufmann P. Histochemistry. 1994;101:91–98. doi: 10.1007/BF00269354. [DOI] [PubMed] [Google Scholar]
- 11.Altin J G, Pagler E B. Anal Biochem. 1995;224:382–389. doi: 10.1006/abio.1995.1054. [DOI] [PubMed] [Google Scholar]
- 12.Waheed A, Hille A, Junghans U, von Figura K. Biochemistry. 1990;29:2449–2455. doi: 10.1021/bi00462a003. [DOI] [PubMed] [Google Scholar]
- 13.Gastinel L N, Simister N E, Bjorkman P J. Proc Natl Acad Sci USA. 1992;89:638–642. doi: 10.1073/pnas.89.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Parmley R T, Barton J C, Conrad M E. Br J Haematol. 1985;60:81–89. doi: 10.1111/j.1365-2141.1985.tb07388.x. [DOI] [PubMed] [Google Scholar]
- 16.Simister N E, Story C M, Chen H-L, Hunt J S. Eur J Immunol. 1996;26:1527–1531. doi: 10.1002/eji.1830260718. [DOI] [PubMed] [Google Scholar]
- 17.McArdle H J, Morgan E H. J Reprod Fertil. 1982;66:529–536. doi: 10.1530/jrf.0.0660529. [DOI] [PubMed] [Google Scholar]
- 18.Bierings M B, Baert M R M, van Eijk H G, van Dijk J P. Mol Cell Biochem. 1991;100:31–38. doi: 10.1007/BF00230807. [DOI] [PubMed] [Google Scholar]
- 19.Bergamaschi G, Bergamaschi P, Carlevati S, Cazzola M. Haematologica. 1990;75:220–223. [PubMed] [Google Scholar]
- 20.Okuyama T, Tawada T, Furya H, Villee C A. Am J Obstet Gynecol. 1985;152:344–350. doi: 10.1016/s0002-9378(85)80225-8. [DOI] [PubMed] [Google Scholar]
- 21.Milman N, Ibsen K K, Christensen J M. Acta Obstet Gynecol Scand. 1987;66:205–211. doi: 10.3109/00016348709020748. [DOI] [PubMed] [Google Scholar]
- 22.Knisely A S. Semin Liver Dis. 1994;14:229–235. doi: 10.1055/s-2007-1007314. [DOI] [PubMed] [Google Scholar]
- 23.Glista B A, Bautista A, Prudencio M, Desposito F. Pediatr Res. 1986;20:410A. [Google Scholar]
- 24.Silver M M, Beverley D W, Valberg L S, Cutz E, Phillips M J, Shaheed W A. Am J Pathol. 1987;128:538–554. [PMC free article] [PubMed] [Google Scholar]
- 25.Motulsky A. N Engl J Med. 1979;301:1291. doi: 10.1056/NEJM197912063012319. [DOI] [PubMed] [Google Scholar]
- 26.Bothwell T H, Charlton R W, Motulsky A G. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw-Hill; 1995. pp. 2246–2247. [Google Scholar]
- 27.Bodmer J G, Parham P, Albert E D, Marsh S G E. Nat Genet. 1997;15:234–235. doi: 10.1038/ng0397-234c. [DOI] [PubMed] [Google Scholar]