Abstract
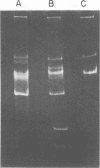
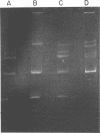
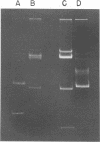
Procedures for effective cellular lysis and plasmid deoxyribonucleic acid (DNA) isolation from group N streptococci were developed. Cells were grown at 32 degrees C for 4 h in a modified Elliker broth containing 20 mM DL-threonine. After cellular digestion with 2 mg of lysozyme per ml for 7 min at 37 degrees C, 1% sodium dodecyl sulfate exposure resulted in complete and immediate lysis. Lactose (Lac) plasmid species in Streptococcus lactis C2 and S. cremoris B1 (30 and 37 megadaltons, respectively) were demonstrated upon examination of DNA from the cleared lysates by agarose gel electrophoresis. Increasing the lysozyme treatment to 20 min or more resulted in loss of the Lac plasmid, whereas other resident plasmids were unaffected and demonstrable in agarose gels. Diethylpyrocarbonate added before lysis prevented Lac plasmid loss in 20-min lysozyme-treated cells, but was not effective after 40 min of lysozyme treatment. The results suggested that endogenous nuclease activity during the lysozyme treatment period initiated Lac plasmid DNA loss. The development of an efficient lysis procedure for the group N streptococci allowed rapid identification and characterization of plasmid DNA by agarose gel electrophoresis. The plasmid composition of S. lactis C2 and S. cremoris B1, as determined by agarose gel electrophoresis, compared favorably to previous electron microscopic observations.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Plasmids, loss of lactose metabolism, and appearance of partial and full lactose-fermenting revertants in Streptococcus cremoris B1. J Bacteriol. 1977 Jan;129(1):367–377. doi: 10.1128/jb.129.1.367-377.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassy B. M. A gentle method for the lysis of oral streptococci. Biochem Biophys Res Commun. 1976 Jan 26;68(2):603–608. doi: 10.1016/0006-291x(76)91188-8. [DOI] [PubMed] [Google Scholar]
- Chassy B. M., Gibson E., Giuffrida A. Evidence for extrachromosomal elements in Lactobacillus. J Bacteriol. 1976 Sep;127(3):1576–1578. doi: 10.1128/jb.127.3.1576-1578.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Cords B. R., McKay L. L., Guerry P. Extrachromosomal elements in group N streptococci. J Bacteriol. 1974 Mar;117(3):1149–1152. doi: 10.1128/jb.117.3.1149-1152.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou J. D., McKay L. L. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J Bacteriol. 1977 Apr;130(1):257–265. doi: 10.1128/jb.130.1.257-265.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou J. D., McKay L. L. Plasmids in Streptococcus lactis: evidence that lactose metabolism and proteinase activity are plasmid linked. Appl Environ Microbiol. 1976 Jul;32(1):38–44. doi: 10.1128/aem.32.1.38-44.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg L., Fedorcsak I., Solymosy F. Diethyl pyrocarbonate in nucleic acid research. Prog Nucleic Acid Res Mol Biol. 1976;16:189–262. doi: 10.1016/s0079-6603(08)60758-8. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Hassell F. P. Transformation of Streptococcus sanguis Challis by plasmid deoxyribonucleic acid from Streptococcus faecalis. J Bacteriol. 1976 Oct;128(1):347–355. doi: 10.1128/jb.128.1.347-355.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledeboer A. M., Krol A. J., Dons J. J., Spier F., Schilperoort R. A., Zaenen I., van Larebeke N., Schell J. On the isolation of TI-plasmid from Agrobacterium tumefaciens. Nucleic Acids Res. 1976 Feb;3(2):449–463. doi: 10.1093/nar/3.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Efstathiou J. D. Transductional evidence for plasmid linkage of lactose metabolism in streptococcus lactis C2. Appl Environ Microbiol. 1976 Jul;32(1):45–52. doi: 10.1128/aem.32.1.45-52.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Plasmid distribution and evidence for a proteinase plasmid in Streptococcus lactis C2-1. Appl Microbiol. 1975 Apr;29(4):546–548. doi: 10.1128/am.29.4.546-548.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Simultaneous loss of proteinase- and lactose-utilizing enzyme activities in Streptococcus lactis and reversal of loss by transduction. Appl Microbiol. 1974 Sep;28(3):342–346. doi: 10.1128/am.28.3.342-346.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers J. A., Sanchez D., Elwell L. P., Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976 Sep;127(3):1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. Plasmid-protein relaxation complexes in Staphylococcus aureus. J Bacteriol. 1976 Sep;127(3):1177–1187. doi: 10.1128/jb.127.3.1177-1187.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubens C., Heffron F., Falkow S. Transposition of a plasmid deoxyribonucleic acid sequence that mediates ampicillin resistance: independence from host rec functions and orientation of insertion. J Bacteriol. 1976 Oct;128(1):425–434. doi: 10.1128/jb.128.1.425-434.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]