Abstract
Myotonic dystrophy (DM) is associated with expansion of CTG repeats in the 3′-untranslated region of the myotonin protein kinase (DMPK) gene. The molecular mechanism whereby expansion of the (CUG)n repeats in the 3′-untranslated region of DMPK gene induces DM is unknown. We previously isolated a protein with specific binding to CUG repeat sequences (CUG-BP/hNab50) that possibly plays a role in mRNA processing and/or transport. Here we present evidence that the phosphorylation status and intracellular distribution of the RNA CUG-binding protein, identical to hNab50 protein (CUG-BP/hNab50), are altered in homozygous DM patient and that CUG-BP/hNab50 is a substrate for DMPK both in vivo and in vitro. Data from two biological systems with reduced levels of DMPK, homozygous DM patient and DMPK knockout mice, show that DMPK regulates both phosphorylation and intracellular localization of the CUG-BP/hNab50 protein. Decreased levels of DMPK observed in DM patients and DMPK knockout mice led to the elevation of the hypophosphorylated form of CUG-BP/hNab50. Nuclear concentration of the hypophosphorylated CUG-BP/hNab50 isoform is increased in DMPK knockout mice and in homozygous DM patient. DMPK also interacts with and phosphorylates CUG-BP/hNab50 protein in vitro. DMPK-mediated phosphorylation of CUG-BP/hNab50 results in dramatic reduction of the CUG-BP2, hypophosphorylated isoform, accumulation of which was observed in the nuclei of DMPK knockout mice. These data suggest a feedback mechanism whereby decreased levels of DMPK could alter phosphorylation status of CUG-BP/hNab50, thus facilitating nuclear localization of CUG-BP/hNab50. Our results suggest that DM pathophysiology could be, in part, a result of sequestration of CUG-BP/hNab50 and, in part, of lowered DMPK levels, which, in turn, affect processing and transport of specific subclass of mRNAs.
Myotonic dystrophy (DM), the most common form of adult muscular dystrophy, although primarily a muscle disorder, also affects other organs including the heart, eye, testes, lungs, and brain (1). The primary genetic defect in DM is a (CTG)n trinucleotide repeat expansion in the 3′-untranslated region of the myotonin-protein kinase (DMPK) gene (2–7). The length of this expansion correlates with the severity of the clinical symptoms (8–10). The mechanism by which the CTG repeat expansion induces the DM phenotype remains puzzling, particularly because the CTG repeat is located in the 3′-untranslated region. In homozygous knockout mice, where DMPK is absent, only muscle weakness and myopathy are observed (11, 12). Cardiomyopathy develops in mice overexpressing the DMPK gene (12), but disease symptoms in other organs, including skeletal muscle myotonia are not observed. These data show that alteration of DMPK expression is only a part of DM mechanism suggesting that other genes are involved.
In pursuit of alternative mechanisms for the pathogenesis of DM, we identified novel proteins that bind specifically to triplet repeats (13) in DNA and others that bind to triplet repeats in RNA (13, 14). One of these RNA CUG-binding proteins, CUG-BP/hNab50, is identical to hNab50 protein and binds specifically to RNA (CUG)8 repeats (14), the triplet repeat found in the DMPK mRNA, responsible for DM. Two hybrid interaction studies suggest that CUG-BP/hNab50 protein is involved in mRNA processing and/or nucleoplasmic transport (14, 15). We hypothesize that CUG-BP/hNab50 is specific not only for DMPK mRNA, but it also facilitates nucleocytoplasmic transport/processing of many mRNAs with CUG repeats, and therefore, is essential for the expression of multiple genes. Expanded regions of CUG repeats as in DM (up to several thousand) could sequester the CUG-BP/hNab50 and disrupt this protein’s normal function.
We have recently shown that CUG-BP/hNab50 exists in two forms CUG-BP1 and CUG-BP2 (14). In this study we demonstrate that CUG-BP1 and CUG-BP2 are hyper- and hypophosphorylated forms of a (CUG)n triplet repeat binding protein. The existence of phosphorylated and hypophosphorylated forms of this protein suggests that its function might be controlled by phosphorylation. Because CUG expansion is located in DMPK gene, we suggest that DMPK kinase in some way might participate in phosphorylation of CUG-BP/hNab50. By using two biological systems where DMPK levels are reduced, homozygous DM patient and DMPK knockout mice, we have demonstrated that the intracellular distribution of CUG-BP isoforms is altered in the absence of DMPK. Our data show that DMPK phosphorylates CUG-BP protein in vitro, suggesting the regulation of nuclear CUG-BP localization via phosphorylation.
METHODS
Preparation of Whole Cell Extracts (WCE), Cytoplasm, and Nuclear Protein Extracts.
The procedures for isolation of WCEs from cultured cells and for fractionation of cytoplasm and nuclear extracts have been described by us (13, 14). The quality of the separation of cytoplasm and nuclear proteins was examined by determination of binding activity of the nuclear protein upstream stimulatory factor (13). Isolation of WCE from tissues was carried out by using homogenization of the tissues in high salt buffer (50 mM Tris⋅HCl, pH 7.5/0.42 M NaCl/25% sucrose/5 mM EDTA/1 mM phenylmethylsulfonyl fluoride). After incubation on ice for 30 min samples were centrifuged and supernatans (WCE) were frozen and kept at −80°C.
Electrophoretic Mobility-Shift Assay.
Conditions for electrophoretic mobility-shift assay are described (13, 14). (CUG)8 RNA oligonucleotide nucleotide was labeled by [γ-32P]ATP with T4 kinase and incubated with 5 μg of nuclear extracts or with 30 μg of WCEs.
Western Blot Analysis.
One hundred micrograms of proteins (for cytoplasm and nuclear extract) and 50 μg (for WCE) were loaded on 8–10% polyacrylamide/0.1% SDS gels, transferred on the nitrocellulose filter (Nitro Bind) and incubated with monoclonal 3B1 (14) (1:2,000) or polyclonal anti-CUG-BP (1:1,000). Polyclonal antibodies against human CUG-BP protein were generated by immunization of the rabbits in Research Genetics Co. with a recombinant CUG-BP fused with a maltose-binding protein (14). These antibodies interact with both human and mouse CUG-BP proteins (data not shown). The conditions for Western assay are described in details in our previously published paper (14). To verify the measured protein concentration, a preliminary filter was stained with Coomassie blue. After detection of specific proteins, each filter was reprobed with antibodies to β-actin or with antibodies to cdk4. Immunodetection was repeated 3–10 times with different preparations of protein extracts.
UV-Cross-Link Assay.
Nuclear and cytoplasmic protein extracts were incubated with RNA (CUG)8 probe as described (14) for 30 min at room temperature and subjected to UV treatment for 30 min by using UV-light box (Stratagene). The samples were loaded on 10% PAGE/0.1% SDS gel, transferred on the membrane and autoradiographed. The same membrane was stained with Coomassie blue to verify protein loading.
Coimmunoprecipitation and Kinase Assay.
Coimmunoprecipitation of DMPK and CUG-BP/hNab50 was performed as follows. Total proteins from cardiac tissue of patients with dilated cardiomyopathy (DCM) and DM were prepared as described above and incubated with rabbit antibodies to DMPK (16) for 4 hr and overnight with protein A agarose (Santa Cruz Biotechnology). Samples were washed four to five times with PBS (1 ml) and resuspended in sample buffer for SDS electrophoresis or in 40 μl of PBS for kinase assay. Four μl of proteins suspended in PBS were incubated in kinase buffer in the presence of [γ-32P]ATP as described (16). Phosphorylated proteins were analyzed by electrophoresis. For the second immunoprecipitation, the labeled proteins were stripped by 1M NaCl and separated from protein A agarose by centrifugation. Supernatant was diluted with PBS and incubated with monoclonal antibodies to CUG-BP for 4 hr. Protein A agarose (30 μl) were then added and incubated overnight at 4°C. Protein A agarose was washed four times with PBS and loaded on 10% PAGE/0.1% SDS gel. Proteins were transferred onto membrane and autoradiographed.
RESULTS
CUG-BP1 and CUG-BP2 Are Hyper- and Hypophosphorylated Forms of Protein.
Sequence analysis of the CUG-BP/hNab50 predicts that this protein contains multiple phosphorylation sites (14) suggesting the regulation of CUG-BP/hNab50 RNA binding activity by phosphorylation. To examine whether phosphorylation influences CUG-BP RNA binding activity, HeLa cell extracts were treated with phosphatase and subsequently used for (CUG)8 bandshift analysis (Fig. 1). Phosphatase treatment led to the formation of a single (CUG)8 complex with lower relative mobility than either CUG-BP1 or CUG-BP2 (Fig. 1). Addition of the 3B1 mAb to the binding reaction, which contained the dephosphorylated form of CUG-BP, led to complete neutralization of the RNA-protein complex (data not shown). Immunoblot analysis of HeLa protein extracts with antibodies specific for CUG-BP/hNab50 showed two proteins with molecular weights 49 and 51 kDa (14). To determine the level of phosphorylation of these proteins, total protein extracts from HeLa were treated with increased amounts of phosphatase, followed by Western assay with mAbs to CUG-BP/hNab50. Dephosphorylation changed the electrophoretic mobility of the immunoreactive proteins (Fig. 1) eliminating fast migrating CUG-BP1 band (49 kDa) and leaving only the slower migrating band. The slower migrating band has a molecular mass of 51 kDa. These data show that the 51-kDa protein is a hypophosphorylated form of the CUG-BP/hNab50 and that the 49-kDa protein is a hyperphosphorylated protein.
Figure 1.
(Left) Bandshift. Phosphorylation generates the different CUG-BP1 and CUG-BP2 isoforms. Bandshift assay of the CUG-BP1 and CUG-BP2 purified from HeLa cells in the presence of alkaline phosphatase. (Right) Western. CUG-BP/hNab50 Western blot analysis of the protein extracts prepared from HeLa cells treated with alkaline phosphatase. Whole cell protein extract (20 or 100 μg; lanes 1 and 2, respectively) was analyzed with mAb 3B1. Two isoforms, CUG-BP1 and CUG-BP2, are indicated. In phosphatase assay 100 μg of whole cell protein extract were used because affinity of mAb 3B1 is much higher to the hyperphosphorylated CUG-BP.
Hypophosphorylated CUG-BP Isoform Accumulates in Nuclei in DM Disease.
We have shown (14) that, in cultured cells from DM patients, the CUG-BP/hNab50 binding activity was altered and translocated to nuclei. To determine whether CUG-BP activity is altered in vivo, in tissues from the patient with DM, (CUG)8 binding activity was determined in protein extracts from several tissues including skeletal muscle, heart, brain, spleen, and liver from a severely affected patient homozygous for the DM mutation. In this patient, the enlarged (CTG)n expansion on both alleles of the DMPK gene is associated with a severe clinical phenotype and a significant reduction in DMPK expression (15). Extracts were analyzed by electrophoretic mobility shift assay by using the RNA (CUG)8 probe (Fig. 2A). This analysis showed that the hyperphosphorylated CUG-BP1 was present in all tissues with the highest level of expression in the brain, but the hypophosphorylated CUG-BP2 was observed only in skeletal muscle and heart of the patient. This is in contrast to the normal heart, where CUG-BP1 is abundant, but CUG-BP2 is undetectable (Fig. 2B). CUG-BP2 binding activity is also observed in heart from DCM patient (Fig. 2B) suggesting possible involvement of CUG-BP in DCM. Single-stranded CRRP protein (13) that has dual binding affinity to single-stranded CTG repeats and RNA CUG repeats was found almost in all tissues (except of spleen) with the highest level of expression in heart (Fig. 2A).
Figure 2.
Expression of CUG-BP isoforms in tissues from unique patient homozygous for DM mutation. Whole cell protein extracts were prepared from tissues of patient with DM (A) or cardiac tissue from normal control and DCM patient (B) and analyzed by bandshift assay with RNA (CUG)8 probe. Positions of CUG-BP1, CUG-BP2, and ssCRRP (13) are indicated. Specificity of binding was examined by addition of 100 ng of unlabeled (CUG)8 RNA oligonucleotide. (C) CUG-BP is accumulating in nuclei in cultured cells from DM patients. Cytoplasmic and nuclear protein extracts were subjected to SDS/PAGE and immunoblotted with antibody 3B1. Reprobing of the same membrane with antibodies to cdk4 or anti-β-actin (Lower) shows equal loading of proteins.
CUG-BP2 binding activity has been shown (14) to be induced and translocated to nuclei in cultured cells from DM. To determine the cellular localization of the CUG-BP/hNab50 protein in vivo, immunoblot analysis was performed by using cytoplasmic and nuclear extracts isolated from cultured normal myoblasts and myoblasts from a homozygous DM patient. The CUG-BP/hNab50 was located in both the cytoplasm and nuclei (Fig. 2C) of normal myoblasts. In contrast, myoblasts from DM homozygote showed the CUG-BP/hNab50 levels to be very low in the cytoplasm (in some experiments undetectable), but was significantly increased in the nuclear extract. Reprobing of the same membrane with antibodies to the nuclear protein cdk4 indicated good separation of cytoplasm and nuclei with equal loading of nuclear proteins. The separation of nuclei and cytoplasm was also verified by detection of the nuclear protein, upstream stimulatory factor, as described (ref. 13 and data not shown). In addition, reprobing of the membrane containing cytoplasmic proteins with anti-β-actin indicated equal loading of cytoplasmic proteins. Thus, we conclude that CUG-BP is accumulating in nuclei in DM disease and such accumulation could be explained by increased amount of the hypophosphorylated CUG-BP2 that was predominantly nuclear (14).
CUG-BP2 Isoform Is Translocated to the Nuclei in DMPK Knockout Mice.
The observation that the hypophosphorylated CUG-BP/hNab50 form (CUG-BP2) was accumulated in the nuclei of patients with DM (containing low levels of DMPK) suggested that DMPK activity may be required for CUG-BP/hNab50 phosphorylation. Knockout mice in which the DMPK gene was eliminated from both alleles have been earlier described (11, 12). These homozygous mice exhibited muscle weakness and myopathy (11, 12). Results of immunoblot analysis of CUG-BP/hNab50 expression in skeletal muscles and heart from the DMPK knockout mice, as well as wild-type littermates, are shown in Fig. 3A. In normal littermates, CUG-BP1 is observed primarily in the cytoplasm with little amount of CUG-BP2 in the nuclear extracts. On the contrary, in DMPK knockout mice, the nuclear concentration of CUG-BP2 is considerably increased in both skeletal muscle and heart. The nuclear accumulation of the CUG-BP2 was observed in the experiments with both monoclonal and polyclonal antibodies to CUG-BP/hNab50 (see Materials and Methods). To determine whether the CUG-BP/hNab50 present in the heart exhibits binding to RNA CUG repeats, UV photo-cross-linking was performed on the DMPK null mutants and wild-type littermates (Fig. 3B). The predominant CUG-BP1 RNA binding activity is detected in the cytoplasm in wild-type mice (Fig. 3B), in contrast, in the DMPK knockout mice, the predominant CUG-BP2 binding activity was observed in the nuclear extract. Bandshift analysis of the nuclear extract with the (CUG)8 probe confirmed this result (data not shown). Coomassie staining of the membrane after UV cross linking showed equal loading of proteins in position of CUG-BP. We have shown (Fig. 1) that the hyperphosphorylated CUG-BP1 has faster electrophoretic mobility than the hypophosphorylated form and, in the DMPK knockout mice, the nuclear CUG-BP/hNab50 bands had lower electrophoretic mobility, suggesting that the nuclear form of CUG-BP/hNab50 is the hypophosphorylated form.
Figure 3.
CUG-BP/hNab50 expression in skeletal muscle and heart tissue from DMPK knockout mice and genetically normal littermates. (A) Western blot analysis of cytoplasmic and nuclear protein extracts with antibody 3B1. CUG-BP/hNab50 is translocated to nuclei in DMPK null mutants. (B) CUG-BP2 binding activity is increased in nuclear extracts from cardiac tissue of DMPK null animals. Cytoplasmic and nuclear proteins from cardiac tissue of DMPK knockout and genetically normal littermates were analyzed by UV-cross link with RNA (CUG)8 probe as described in Materials and Methods. The same membrane was stained with Coomassie Blue showing equal protein loading. The positions of molecular mass markers are shown (Left).
Results of the preceding experiments show that CUG-BP1 is localized primarily in the cytoplasm of normal tissues. In contrast, CUG-BP2 was detected primarily in the nucleus of cells from homozygous or heterozygous patients (14) and in homozygous DMPK knockout mice. One of the features in common with nuclear accumulation of the hypophosphorylated CUG-BP/hNab50 is the marked reduction of DMPK in the homozygous patient (15) and its complete absence in DMPK knockout mice (11). This suggests that DMPK may interact with CUG-BP/hNab50 and that DMPK is required for phosphorylation of CUG-BP/hNab50.
CUG-BP Interacts with DMPK In Vivo.
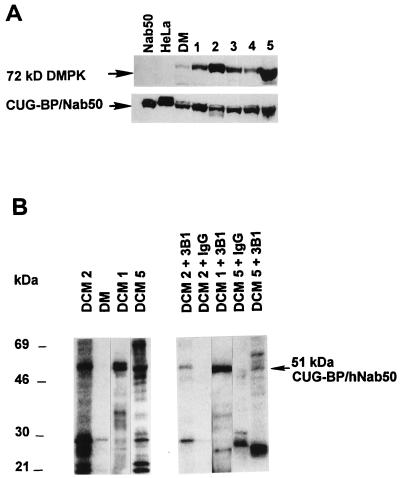
To determine whether DMPK and the CUG-BP/hNab50 interact with each other, coimmunoprecipitation analysis was carried out. Initially we have identified tissues that contained high levels of both DMPK and CUG-BP/hNab50. Tissue extracts were prepared from patients with cardiac disease unrelated to DM and analyzed by immunoblotting with antibodies to DMPK and CUG-BP/hNab50. High levels of DMPK protein were detected in two patients (patients 2 and 5) and moderate levels in the other (Fig. 4A). Reprobing of the same filter with antibodies to CUG-BP/hNab50 showed that the levels of CUG-BP/hNab50 were approximately equal in all patients (Fig. 4A). A bandshift assay with the (CUG)8 probe showed that the tissue samples derived from patients 1, 2, and 5 contained high levels of CUG-BP2 binding activity (data not shown). To examine whether CUG-BP interacts with DMPK, DMPK was immunoprecipitated from whole cell protein extracts obtained from patients 1, 2, and 5 by using a specific antibody (16). Cardiac extracts from patients homozygous for DM, which we have previously shown to have very low levels of DMPK (15), were used as an internal control for specificity. Immunoprecipitates were incubated with [γ-32P]ATP under kinase assay conditions and analyzed by denaturing electrophoresis. Electrophoresis of the cardiac immunoprecipitates showed several bands suggesting that DMPK interacts in vivo with several proteins that can be potential substrates. Immunoprecipitates (IPs) from patients 1, 2, and 5 indicate a major protein in the position of 51 kDa. In contrast, the immunoprecipitate from cardiac tissues of the homozygous patient showed one faint band (Fig. 4B). To determine whether the 51-kDa protein observed in the immunoprecipitate of cardiac patients is CUG-BP/hNab50, a second immunoprecipitation of the phosphorylated proteins was performed by using antibody to CUG-BP/hNab50 as described in Materials and Methods. A major 49–51-kDa protein was immunoprecipitated under these conditions (Fig. 4B). The levels of [32P]CUG-BP/hNab50 present in the cardiac tissue samples from different patients correlated with the levels of DMPK. In the immunoprecipitate from patient 5, two proteins with molecular masses of 51 and 72 kDa were detected (Fig. 4B). The 72-kDa protein is likely to be DMPK that underwent autophosphorylation (16) and reassociated with CUG-BP/hNab50 during incubation with protein A agarose or an unknown protein immunocrossreactive with CUG-BP. The detection of 72-kDa labeled by 32P only in IP from the patient 5 is consistent with high levels of DMPK in this patient (Fig. 4A) supporting the suggestion that the 72-kDa protein is DMPK. In the second IP, we have also observed a protein with lower molecular weight. This protein could be a degradation product of CUG-BP or a protein bound to IP nonspecifically. The presence of CUG-BP in IPs with anti-DMPK indicates that DMPK and CUG-BP/hNab50 interact with each other in vivo and that DMPK can phosphorylate CUG-BP/hNab50 in vitro.
Figure 4.
Interaction of DMPK and CUG-BP/Nab50. (A) Western analysis of proteins isolated from patients with cardiac disease unrelated to DM was carried out as described above. (B) Coimmunoprecipitation of DMPK and CUG-BP/hNab50. Left part. Total proteins from cardiac tissue of patients with DCM (patients 1, 2, and 5) and from a patient with DM initially were incubated with rabbit antibodies to DMPK (16) and protein A agarose and subjected to kinase assay as described in Materials and Methods. 32P-labeled proteins were analyzed by SDS/PAGE. (Right) Second immunoprecipitation of labeled proteins with CUG-BP antibody was carried out as described above and analyzed by SDS/PAGE.
DMPK Directly Phosphorylates CUG-BP and Affects the Mobility of CUG-BP/RNA Complex.
Because the DMPK immunoprecipitates from cardiac tissue show multiple proteins, it is possible that DMPK interacts with CUG-BP/hNab50 indirectly. To determine whether the interaction of DMPK with CUG-BP/hNab50 is direct, recombinant DMPK and CUG-BP/hNab50 were produced as described (14, 16). Recombinant DMPK was attached to protein A agarose by using specific antibodies to DMPK (Pr-A-IgG-DMPK column) and incubated with the recombinant CUG-BP/hNab50 fusion protein. For controls, CUG-BP/hNab50 was incubated either with protein A agarose treated with BSA or with protein A agarose containing anti-DMPK IgGs but lacking DMPK protein. Immunoblotting with an anti-CUG-BP/hNab50 mAb has shown that the fusion CUG-BP/hNab50 protein is detectable only in immunoprecipitates with protein A-anti-DMPK/DMPK column (Fig. 5A). No interaction was observed in control experiments with protein A agarose or protein A-anti-DMPK column indicating that CUG-BP/hNab50 interacts specifically with DMPK. To examine whether this interaction affects the phosphorylation and the binding activity of CUG-BP/hNab50, recombinant CUG-BP/hNab50 was incubated with recombinant DMPK in the presence of [γ-32P]ATP. CUG-BP/hNab50 binding activity was measured after this incubation. DMPK phosphorylates CUG-BP/hNab50 in a dose-dependent manner (Fig. 5B). Measurement of (CUG)8 binding activity after this phosphorylation has shown that intensity of the CUG-BP2/RNA complex is dramatically reduced (Fig. 5B). Slight reduction of CUG-BP1 binding activity was also detected in some experiments. These results confirm that DMPK directly phosphorylates CUG-BP/hNab50 in vitro.
Figure 5.
Recombinant DMPK and CUG-BP/hNab50 interact with each other (A) in vitro and DMPK phosphorylates CUG-BP/hNab50 (B). (A) Protein A agarose containing anti-DMPK/DMPK and two controls (protein A agarose/anti-DMPK and protein A agarose only) were incubated with bacterially expressed homogeneous CUG-BP/hNab50 and analyzed by Western blot with anti-CUG-BP/hNab50. CUG-BP/hNab50 does not bind to protein A agarose lacking DMPK protein. (B Upper) Kinase assay. Purified CUG-BP/hNab50 was incubated with electrophoretically homogeneous DMPK in the presence of [γ-32P]ATP and subjected to SDS/PAGE and exposed to x-ray film. (Lower) Gel shift analysis of CUG-BP binding activity after phosphorylation by DMPK. Increased amounts of DMPK result in reduction of binding activity of the hypophosphorylated CUG-BP2.
DISCUSSION
The severity of DM disease strongly correlates with amount CTG repeats located in the 3′ UTR of DMPK gene. It was originally proposed that the expanded CTG repeat results in the reduction of DMPK expression (17). However, DMPK knockout mice or mice with overexpressed DMPK have shown only a partial phenotype (11, 12). These data suggest that the molecular mechanisms for DM are much more complex than expected and involve other genes. We have proposed an alternative hypothesis for DM pathogenesis. According to this hypothesis, expanded CTG repeats lead to the expression of the mutant DMPK mRNA with long CUG RNA repeats that might be a binding sites for specific RNA binding proteins (13–15). As a result, the CUG RNA-binding proteins are titrated by increased (CUG)n repeats leading to reduction of unbound CUG-binding proteins which, in turn, affects the expression of CUG repeat containing mRNAs including DMPK mRNA. In agreement with this hypothesis, we have identified several CTG and CUG-binding proteins (13). One of these proteins, CUG-BP/hNab50, has been purified and cloned (14).
We have described (14) the changes of CUG-BP binding activity in cultured cells from DM patients. In this paper we present evidence that concentration of hypophosphorylated CUG-BP isoform is increased in nuclei in DM disease and that DMPK, at least in part, is involved in phosphorylation of CUG-BP. Two biological models were used in our investigations: (1) a homozygous patient that contained expansion of CTG repeats in both alleles (15) and (2) DMPK knockout mice (11). In the homozygous patient, DMPK expression has been shown to be dramatically reduced (15). Analysis of (CUG)n RNA binding activity of the CUG-BP/Nab50 in different tissues shows that this protein is affected in heart and skeletal muscle, where, in normal controls, the expression of DMPK is abundant. Thus, alteration of the CUG-BP binding activity in the heart and skeletal muscle are the strong evidence that this protein is involved in DM pathogenesis.
Accumulation of hypophosphorylated CUG-BP in the patient with reduced levels of DMPK prompted us to investigate the phosphorylation of CUG-BP by DMPK and to determine intracellular localization of hypo- and hyperphosphorylated CUG-BPs. We suggested that direct or indirect interaction between DMPK and CUG-BP/hNab50, and the resulting phosphorylation of CUG-BP/hNab50 by DMPK, regulate CUG-BP phosphorylation status and its intracellular localization (15). Mice lacking DMPK have been studied and shown that hypophosphorylated CUG-BP/hNab50 was accumulated in the nucleus. This is a strong genetic evidence that DMPK regulates CUG-BP intracellular localization. We suggest that the increased nuclear concentration of CUG-BP2 in DM is the major change that affects the expression of other mRNAs that might be under CUG-BP control. It is interesting to note that DM patients have the abnormal processing of DMPK mRNAs (18) and that DMPK mRNAs are accumulating in nuclei (19). Consistent with our hypothesis and experimental data, two recent independent studies have shown nuclear retention of DMPK transcripts in DM cells (20, 21). Because CUG-BP interacts with DMPK mRNA (14) and hypophosphorylated form is accumulating in nuclei, it is reasonable to speculate that nuclear retention of DMPK transcripts, at least in part, is mediated by CUG-BP protein. We, therefore, propose an autoregulatory loop whereby DMPK phosphorylates CUG-BP/hNab50, which, in turn, facilitates the transport or processing of the mRNA for DMPK and other CUG-BP/hNab50 dependent mRNAs (Fig. 6). According to this autoregulatory loop, the sequestration of CUG-BP by increased CUGn repeats leads to abnormal processing and to reduced levels of DMPK mRNA and protein. Direct interaction between DMPK and CUG-BP and phosphorylation of CUG-BP by DMPK suggest that the reduction in DMPK levels, in turn, result in failure to phosphorylate CUG-BP protein and in accumulation of the hypophosphorylated CUG-BP2 isoform in nuclei. This hypothetical loop is consistent with the observation that DMPK phosphorylates CUG-BP in vitro and this phosphorylation leads to reduction CUG-BP2-RNA complex that was observed in DMPK knockout mice. Thus, our observations support hypothetical autoregulatory loop and show that both DMPK and CUG-BP are involved in DM pathogenesis. It is likely that the expansion of the triplet repeats in DM cells impairs the ability of DMPK to phosphorylate CUG-BP/hNab50. Decreased phosphorylation of CUG-BP/hNab50 results in decrease of DMPK mRNA expression and possibly other mRNAs resulting in the multiple organ involvement that is observed in DM. We have previously proposed (15) a model of DMPK regulation by CUG-BP. The results presented in this paper confirm this model and show that DM pathogenesis involves alteration of at least two genes, DMPK and CUG-BP. We have shown previously (14) that CUG-BP/hNab50 contains three RNA binding domains (14) and that the CUG-BP/hNab50 binds to a synthetic (CUG)8 probe and to the DMPK 3′-UTR containing the (CUG)n repeats in vitro (14). Our recent observations indicate that, although the binding site for CUG-BP/Nab50 is (CUG)n where n ≥ 8, CUG-BP can also bind to various RNA probes containing only two CUG repeats surrounded by G+C-rich sequence (L.T.T., unpublished observation). These data suggest that CUG-BP can bind to a broad set of RNAs and regulate their expression. We are now looking for the RNAs that are under control of CUG-BP to examine the regulation of these RNAs by CUG-BP.
Figure 6.
A hypothetical model showing autoregulatory loop. Expanded CUG repeats sequester CUG-BP protein and cause the reduction the DMPK expression. The DMPK reduction leads to increase of hypophosphorylated CUG-BP that is accumulating in nuclei of DM patients.
Acknowledgments
We are very grateful to Dr. E. Roeder for providing blood and tissue samples from a patient with DM, Dr. P. Clemens for help and advice with establishing of primary muscle culture, Dr. A. J. Marian for providing samples of cardiac tissues from patients with DCM, and to Drs. H. Bellen, H. Zoghbi, T. Cooper, and D. Nelson for critical reading of the manuscript. This work was supported by National Institutes of Health Grants HL543-13 and 1 R01 AR10D44387-01; Grant 97G-256 from the American Heart Association, Texas Affiliate, Inc.; and an Established Investigator Award from the American Heart Association (to M.S.S.).
ABBREVIATIONS
- DCM
dilated cardiomyopathy
- DM
myotonic dystrophy
- CUG-BP/hNab50
RNA CUG-binding protein, identical to hNab50 protein
- DMPK
myotonin-protein kinase
- WCE
whole cell protein extract
References
- 1.Harper P S. Myotonic Dystrophy. 2nd Ed. London: Saunders; 1989. [Google Scholar]
- 2.Buxton J, Shelbourne P, Davies J, Jones C, Van Tongeren T, Aslanidis C, de Jong P, Jansen G, Anvret M, Riley B, Williamson B, Johnson K. Nature (London) 1992;355:547–548. doi: 10.1038/355547a0. [DOI] [PubMed] [Google Scholar]
- 3.Harley H G, Brook J D, Rundle S A, Crow S, Reardon W, Buckler A J, Harper P S, Housman D E, Shaw D J. Nature (London) 1992;355:545–546. doi: 10.1038/355545a0. [DOI] [PubMed] [Google Scholar]
- 4.Aslanidis C, Jansen J, Amemiya C, Shutler G, Mahadevan M, Tsilfidis C, Chen C, Alleman J, Wormskamp N G, Vooijs M, Buxton J, Johnson K, Sweets H J M, Lennon G G, Carrano A V, Korneluk R G, Wieringa B, de Jong P J. Nature (London) 1992;355:548–551. doi: 10.1038/355548a0. [DOI] [PubMed] [Google Scholar]
- 5.Brook J D, McCurrach M E, Harley H G, Buckler A J, Church D, et al. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y, Pizzuti A, Fenwick R G, Jr, King J, Rajnarayan S, Dunne P W, Dubel J, Nasser G A, Ashizawa T, de Jong P, Wieringa B, Korneluk R G, Perryman M B, Epstein H F, Caskey C T. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 7.Mahadevan M S, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C E, Narang M, Barcelo J, O’Hoy K. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 8.Redman J B, Fenwick R G, Fu Y-H, Pizzuti A, Caskey C T. J Am Med Assn. 1993;269:1960–1965. [PubMed] [Google Scholar]
- 9.Tsilfidis C, MacKenzie A E, Mettler G, Barcelo J, Korneluk R G. Nat Genet. 1992;1:192–195. doi: 10.1038/ng0692-192. [DOI] [PubMed] [Google Scholar]
- 10.Hunter A, Tsilfidis C, Mettler G, Jacob P, Mahadevan M, Surh L, Korneluk R G. J Med Genet. 1992;29:774–779. doi: 10.1136/jmg.29.11.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy S, Smith D B J, Rich M M, Leferovich J M, Reily P, Davis B M, Tran K, Rayburn H, Bronson R, Cros D, Balice-Gordon R J, Housman D. Nat Genet. 1996;13:325–335. doi: 10.1038/ng0796-325. [DOI] [PubMed] [Google Scholar]
- 12.Jansen G, Croenen P J T A, Bachner D, Jap P H K, Coerwinkel M, Oerlemans F, van den Brock W, Gohlsch B, Pette D, Plomp J J, Molenaar P C, Nederhoff M G J, van Echteld C J A, Dekker M, Berns A, Hameister H, Wieringa B. Nat Genet. 1996;13:316–324. doi: 10.1038/ng0796-316. [DOI] [PubMed] [Google Scholar]
- 13.Timchenko L T, Timchenko N A, Caskey C T, Roberts R. Hum Mol Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- 14.Timchenko L T, Miller J, Timchenko N A, DeVore D R, Datar K V, Lin L, Roberts R, Caskey C T, Swanson M S. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caskey C T, Swanson M S, Timchenko L T. Cold Spring Harbor Symp Quant Biol. 1996;61:607–614. [PubMed] [Google Scholar]
- 16.Timchenko L T, Nastainczyk W, Schneider T, Patel B, Hofmann F, Caskey C T. Proc Natl Acad Sci USA. 1995;92:5366–5370. doi: 10.1073/pnas.92.12.5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y-H, Friedman D L, Richards S, Pearlman J A, Gibbs R A, Pizzuti A, Ashizawa T, Perryman M B, Scarlato G, Fenwick R G, Jr, Caskey C T. Science. 1993;260:235–238. doi: 10.1126/science.8469976. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Pegoraro E, Menegazzo E, Gennarelli M, Hoop R C, Angelini C, Hoffman E P. Hum Mol Genet. 1995;4:599–606. doi: 10.1093/hmg/4.4.599. [DOI] [PubMed] [Google Scholar]
- 19.Taneja K L, McCurrach M, Schalling M, Housman D, Singer R H. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis B M, McCurrach M E, Taneja K L, Singer R H, Housman D E. Proc Natl Acad Sci USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamshere M G, Newman E E, Alwazzan M, Athwal B S, Brook J D. Proc Natl Acad Sci USA. 1997;94:7394–7399. doi: 10.1073/pnas.94.14.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]