Abstract
A major problem facing the effective treatment of patients with cancer is how to get the specific antitumor agent into every tumor cell. In this report we describe the use of a strategy that, by using retroviral vectors encoding a truncated human CD5 cDNA, allows the selection of only the infected cells, and we show the ability to obtain, before bone marrow transplantation, a population of 5-fluouraci-treated murine bone marrow cells that are 100% marked. This marked population of bone marrow cells is able to reconstitute the hematopoietic system in lethally irradiated mice, indicating that the surface marker lacks deleterious effects on the functionality of bone marrow cells. No gross abnormalities in hematopoiesis were detected in mice repopulated with CD5-expressing cells. Nevertheless, a significant proportion of the hematopoietic cells no longer expresses the surface marker CD5 in the 9-month-old recipient mice. This transcriptional inactivity of the proviral long terminal repeat (LTR) was accompanied by de novo methylation of the proviral sequences. Our results show that the use of the CD5 as a retrovirally encoded marker enables the rapid, efficient, and nontoxic selection in vitro of infected primary cells, which can entirely reconstitute the hematopoietic system in mice. These results should now greatly enhance the power of studies aimed at addressing questions such as generation of cancer-negative hematopoiesis.
Keywords: cancer treatment, gene transfer, bone marrow transplant, gene therapy, leukemia
Recombinant retroviruses provide an attractive vehicle for gene transfer based on their capacity for highly efficient infection and nontoxic integration into the genome of a wide range of cell types. Although the potential for retrovirus-mediated gene transfer efficiency approaches 100%, this may not be realized because of low viral titers or the failure to stimulate cells to divide, which is required for successful integration. Moreover, expression of the transferred gene(s) may not reach desired levels or be sustained. Such problems are evident in current efforts to apply retroviral gene transfer to the gene therapy of cancer, because if we cannot deliver the antitumor specific drug (1–3) into every tumor cell, then any malignant cell that remains unaffected will emerge as a resistant clone. Recently we have tested the utility of coexpressing the specific antitumor agent and a cDNA encoding a truncated human cell surface antigen CD5 for the immediate postinfection selection of the hematopoietic cells transduced with the retrovirus containing the specific antitumor drug (4). Fluorescence-activated cell sorter (FACS) analysis in combination with functional studies showed that under the conditions used, all cells selected expressed CD5 within 48 hr of termination of the infection procedure. Moreover, the fraction of CD5-positive cells did not decrease with extended propagation of selected cells, indicating that the surface marker lacks deleterious effects on cell growth effect and viability. The use of the truncated CD5 cell surface antigen as a selectable marker of gene transfer offers significant advantages over those components that confer resistance to toxic compounds, including the rapid and quantitative detection of transferred gene expression in the desired target cell population by flow cytometry and the efficient and nontoxic selection of transduced target cells by FACS (4, 5).
These results suggest an approach to in vitro gene therapy in hematopoietic cancer. Cancer-negative hematopoiesis might be restored by autografting patients with bone marrow cells infected with the virus expressing the specific antitumor agent against the tumor-associated fusion junctions (which only exist in the tumor cells but not in the normal cells of the patient) and the selectable marker, followed by selecting to ensure that all reinfused stem cells had integrated functional retroviral sequences.
In this study we show the ability to obtain, before bone marrow (BM) transplant, a population of BM cells that are 100% marked. This marked population of BM cells is able to reconstitute the hematopoietic system in lethally irradiated mice, indicating that the surface marker lacks deleterious effects on functionality of BM cells. To our knowledge, these data provide the first direct evidence of generation of a functional hematopoietic system in which 100% of the cells have been marked. This type of strategy should be useful as a preclinical model for the development of effective gene transfer strategies for long-term human repopulating stem cells and should now greatly enhance the power of studies aimed at addressing questions such as generation of cancer-negative hematopoiesis.
MATERIALS AND METHODS
Retroviral Vector.
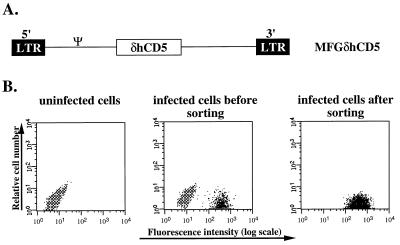
The MFGδCD5 retroviral vector is based on the simplified MFG retroviral vector, which does not contain a dominant selectable marker gene (6). The precise sequences subcloned into the MFG vector were the minimal 1,218-bp portion of the human CD5 cDNA encompassing the complete extracytoplasmic coding region (4). The specific CD5 DNA fragment to be inserted into the retroviral vector was generated by reverse transcriptase–PCR with primers that included initiation and termination codons. The authenticity of this construct was confirmed by DNA sequencing and revealed no PCR errors or cloning artifacts. The proviral structure of this vector is shown in Fig. 1A.
Figure 1.
The structure and characterization of the retroviral vector used to study the hematopoietic reconstitution after tagging and selection of BM cells. (A) Proviral structure of the MFGδhCD5 vector. The MFG retroviral vector (6) incorporates a 1,218-bp portion of the human CD5 cDNA encompassing the complete extracytoplasmic coding region (4). No selectable marker exists in the vector. (B) Selection of CD5 virus-infected Ba/F3 cells by FACS. The CD5 expression profiles of uninfected Ba/F3 cells (Left), Ba/F3 cells cocultured with CD5 viral producer cells 48 hr postinfection (Center), and sorted Ba/F3 cells (Right) are shown. Cells were stained with anti-human CD5 antibody and analyzed by flow cytometry.
Cell Lines.
The ecotropic retrovirus packaging cell line ψ-cre was used to generate helper-free recombinant retrovirus. The cell line was maintained in DMEM supplemented with 10% calf serum. Viral packaging cell lines were maintained in the same medium supplemented with 1 mg/ml of the neomycin analog G418 (GIBCO/BRL). The mouse hematopoietic Ba/F3 cell line was maintained in DMEM with 10% fetal calf serum (FCS) and 5% of WEHI-3B conditioned medium as a source of interleukin 3 (IL-3). All cells were cultured at 37°C in a humidified atmosphere of 5% CO2 in air.
Retrovirus Production.
ψ-cre cells were first transfected with 20 μg of the MFGδCD5 retroviral vector along with 1 μg of the pMC1neo vector using the calcium phosphate precipitation method. After G418 selection (1 mg/ml), the best clones were selected on the basis of their CD5 expression. Ba/F3 cells were infected by 2 days of coculture with the MFGδCD5 viral producer cell lines in DMEM supplemented with 10% calf serum, polybrene (8 μg/ml), and IL-3. After coculture Ba/F3 cells infected by the virus were monitored by CD5 expression.
Transduction of Murine Bone Marrow Cells.
Donor bone marrow cells were harvested from the limbs of C57 BL/6J male mice 5 days after injection with 5-fluorouracil (5-FU) (150 mg/kg of body weight) (7). BM cells were prestimulated in the presence of growth factors and cocultivated for 2 days over vector-producing fibroblasts in the presence of polybrene (8 μg/ml) according to the methods described by Weinthal et al. (8). The growth factors used for the prestimulation were 200 units of murine IL-3 per ml (Amgen), 200 units of human IL-6 per ml (Amgen), 200 units of human IL-1α per ml (Immunex), and 100 ng of mast cell growth factor per ml (Immunex). Transduced bone marrow cells (20%, approximately) were monitored by CD5 expression.
Bone Marrow Transplant (BMT) and Sample Collection.
Recipient female C57BL/6J mice (8–12 weeks old) were irradiated with two split doses of 600 cGy 2 hr apart. Sorted transduced BM cells were injected into the tail vein of the irradiated mice at 2–4 × 106 cells per mouse for long-term reconstitution. Animals were sacrificed and hematopoietic tissues were collected for FACS analysis.
Labeling of Cells.
To sort CD5-positive cells, MFGδCD5-infected cells were incubated on ice for 40 min with the fluorescein isothiocyanate (FITC)-directed, conjugated anti-human CD5 monoclonal antibody (PharMingen), washed twice with PBS + 1% FCS, and resuspended in PBS + 1% FCS. Propidium iodide (Sigma) was included in the last wash to distinguish dead cells before analysis by flow cytometry using a FACScan cell analyzer (Beckton Dickinson). Cells were sorted on a FACStar+ sorter (Beckton Dickinson) equipped with a 5-W argon and a 30-mW helium neon laser. Cells were collected in sterile Eppendorf vials in medium with 50% FCS. CD5 expression among specific hematopoietic cell types was analyzed by staining hematopoietic tissues (BM, spleen and thymus) with anti-human CD5-PE in combination with FITC-labeled Sca-1 to identify hematopoietic stem cells (HSC); Gr-1 for granulocytes; CD11b (Mac-1) for macrophages; TER-119 for erythroid cells; CD3-ɛ, CD4 (L3T4), and CD8 (Ly-2) for T cells; and CD45R (B220) for B cells.
Methylation Analysis.
The methylation status of the proviral 5′ long terminal repeat (LTR) was determined by digestion of genomic DNA (15–25 μg) with BamHI to reduce the size of the DNA fragments, followed by NheI digestion. The DNA was then precipitated with ethanol, redissolved in TE buffer (10 mM Tris/0.1 mM EDTA), and divided into two equal portions, one of which was subjected to digestion with the methylation-sensitive enzyme SmaI. Completeness of the genomic DNA digestions was monitored by mixing a sample of the digestion mixture with λ DNA (BRL), which was subsequently run on a 1% gel. Digested DNA was electrophoresed and blotted to nylon membranes. The blots were probed with a 32P-labeled fragment of the MFGδCD5 vector from the SmaI site in the untranslated leader region to a NcoI site near the 5′ end of the CD5 gene (see Fig. 4B).
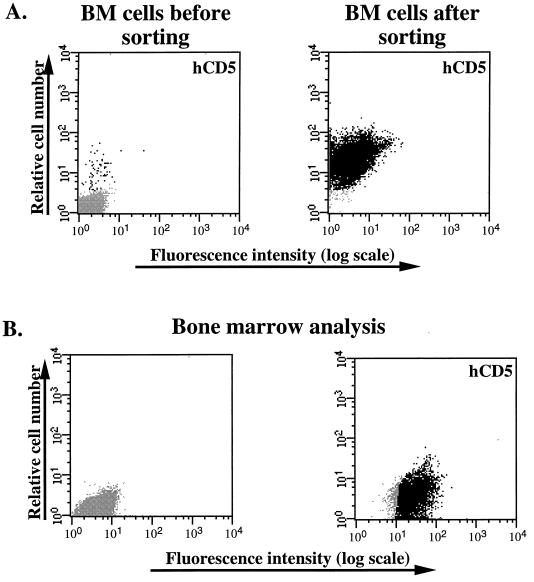
Figure 4.
(A) Selection of CD5 virus-infected BM cells by FACS in cases where the efficiency of gene transfer obtained was poor. The CD5 expression profiles of BM cells cocultured with CD5 viral producer cells 48 hr postinfection (Left) and sorted BM cells (Right) are shown. Cells were stained with anti-human CD5 antibody and analyzed by flow cytometry. (B) Analysis of CD5 expression in BM cells. The CD5 expression profile in cells derived from a representative BMT recipient engrafted with preselected cells (Right) is shown.
RESULTS
The MFGδCD5 Retroviral Vector.
To explore the possible use of the CD5 as cell surface marker for tagging and selecting functional retrovirally transduced bone marrow cells, the retroviral vector depicted in Fig. 1A was constructed. This vector incorporates a 1,218-bp portion of the human CD5 cDNA encompassing the complete extracytoplasmic coding region (4). The inserted truncated human CD5 cDNA will be transcribed from promoter/enhancer sequences in the retroviral LTR. MFGδCD5 viral producers were generated using the ecotropic ψ-cre-packaging cell line. The best clones were selected on the basis of their CD5 expression, and they had a titer of 0.5 (copy number), as assessed by titering in NIH 3T3 cells (data not shown).
Mouse hematopoietic Ba/F3 cells were infected with the CD5-retrovirus. Approximately 10% of Ba/F3 cells were found to express high levels of surface CD5 antigen after 2 days of cocultivation with MFGδCD5 viral producers, thereby tagging them (Fig. 1B Center). The Ba/F3 cells infected with MFGδCD5 virus expressed sufficient truncated human CD5 (which is not expressed in the parental mouse cell line), encoded by the retrovirus, to allow selection of infected cells by FACS sorting and regrowth of viable CD5-positive cells (Fig. 1B Left). As shown in a representative FACS profile for one experiment, approximately 10% of the Ba/F3 cells recovered after cocultivation infection was positive for the CD5 cell surface antigen. In three, independent experiments 100% of the Ba/F3 cells recovered in the CD5+ fraction were CD5+ compared with 10% in the unsorted population. Thus, retrovirally transduced Ba/F3 cells can be successfully enriched based on their immediate expression of a transduced CD5 gene in vivo. Moreover, because the fraction of CD5+ cells did not decrease with extended propagation, the surface marker can be assumed to lack deleterious effects on cell growth and viability.
FACS Selection of CD5-Transduced Bone Marrow Cells.
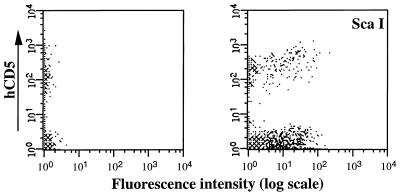
Day 5 5-FU BM cells were cocultivated with MFGδCD5 viral producers for 48 hr and recovered nonadherent cells were cultured for a further 24 hr to allow expression of the transferred CD5 gene before flow cytometric analysis and cell sorting. As shown in a representative FACS profile for one experiment, approximately 20% of the BM cells recovered after cocultivation infection were positive for the human CD5 cell surface antigen (see Fig. 2 Left). A significant proportion of the CD5+-BM cells were HSC as defined for the coexpression of the stem cell marker ScaI (Fig. 2 Right). These retrovirally transduced BM cells can be successfully purified based on their immediate expression of a transduced CD5 gene in vivo, as is illustrated in Fig. 3 Top.
Figure 2.
Transduction of murine HSC with the MFGδCD5 retroviral vector. The CD5 expression profile of BM cells cocultured with CD5 viral producer cells 48 hr postinfection is shown (Left). Cells were stained with anti-human CD5 antibody and analyzed by flow cytometry. Detailed analysis with HSC specific marker (ScaI) is shown at Right.
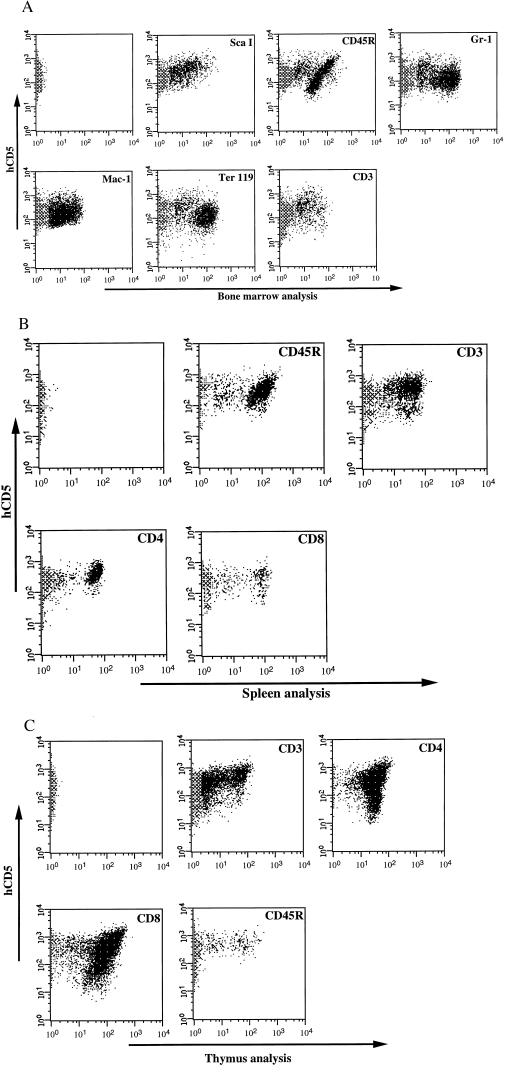
Figure 3.
Analysis of hematopoietic system in mice reconstituted with CD5+ HSC by FACS. Expression of the transferred hCD5 gene on cells of multiple hematopoietic lineages from a mouse that had been repopulated with MFGδCD5 retrovirus-infected BM cells 3 months previously. The first panel in each part of the figure shows the flow cytometric analysis of hCD5 expression in the hematopoietic tissues stained with anti-hCD5. Bone marrow (A), spleen (B), and thymus (C) samples were subjected to double-antibody labeling with anti-hCD5 in combination with ScaI to identify HSC; Ter-119 for erythroid cells; Gr-1 for granulocytes; Mac-1 for macrophages; CD3, CD4, and CD8 for T lymphocytes; and CD45R for B lymphocytes, and the samples were then analyzed by flow cytometry.
Long-Term Reconstituted Mice.
Sorted transduced BM cells were injected into the tail vein of lethally irradiated mice at 2–4 × 106 cells per mouse for long-term reconstitution. Animals were sacrificed between 3 and 9 months after BMT. No gross abnormalities in hematopoiesis were observed in mice expressing CD5 after transplantation with CD5+ BM cells compared with normal control animals. The hematopoietic cells in the organs 3 months after BMT from mice reconstituted with sorted transduced BM were analyzed for CD5 expression (Fig. 3). The CD5 cell surface antigen can be used not only as a marker for the selection of retrovirally infected target cells but also as a reporter molecule for the phenotyping of target and progeny cells. As shown in a representative FACS profile for one experiment, all of the hematopoietic cells in the BM (Fig. 3A), spleen (Fig. 3B), and thymus (Fig. 3C) from mice reconstituted with sorted transduced BM cells were found to express high levels of surface CD5 antigen 3 months after BMT. Double-staining procedure performed on different hematopoietic tissues of irradiated recipients repopulated with CD5+ BM cells showed that all hematopoietic lineages were able to express the transferred CD5 gene, including HSC, granulocytes, macrophages, erythrocytes, and B and T lymphocytes (Fig. 3). The successful outcome of our experiments was directly related to the level of transduction and selection of CD5+ BM cells. The preselection of BM transduced cells was successful even in cases in which the efficiency of gene transfer obtained was poor (less than 2%) based on the cells’ immediate expression of the transduced CD5 gene, as illustrated in Fig. 4A. All of the hematopoietic cells derived from six mice reconstituted with these cells (2–3 × 105 cells per mouse) were found to express high levels of surface CD5 antigen 3 months after BMT (Fig. 4B).
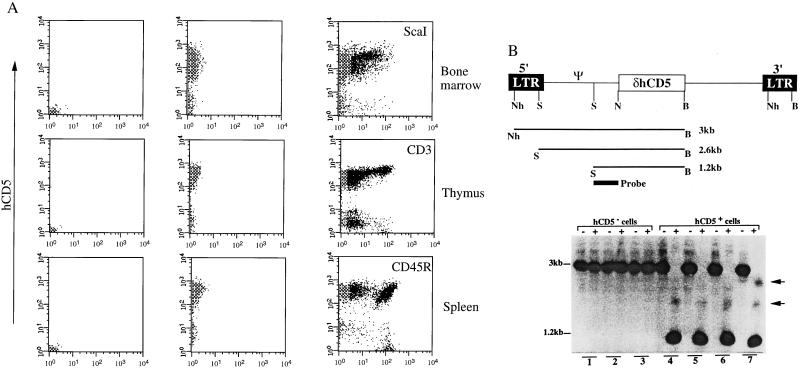
Although these results indicate that the surface marker in the sorted BM cells lacks deleterious effects (as judged by the ability to reconstitute a functional hematopoietic system), even the hematopoietic cells in the organs 3 months after BMT may be derived from progenitor cells capable of short-term but not long-term engraftment (9). Therefore, we analyzed the phenotyping of target and progeny cells in the hematopoietic organs 9 months after BMT (Fig. 5A). The majority of the hematopoietic cells in the BM, spleen, and thymus from mice reconstituted with sorted transduced BM cells were found to express high levels of surface CD5 antigen 9 months after BMT. This sustained multilineage expression in recipient mice at least 9 months posttransplantation shows that the use of a foreign antigen such as a retroviral marker is compatible with long-term expression in hematopoietically reconstituted, lethally irradiated recipients with sorted BM cells. Nevertheless, a significant proportion of the hematopoietic cells no longer express the surface marker CD5 in the 9-month-old recipient mice (Fig. 5A). This transcriptional inactivity of the proviral LTR has been observed in several systems and is accompanied by de novo methylation of the proviral sequences (10, 11)
Figure 5.
Lack of expression from the MFGδCD5 retroviral vector is associated with methylation in vivo. (A) FACS analysis of cells in bone marrow, spleen, and thymus obtained from 9-month-old mice after BMT. A significant percentage of cells does not express the hCD5 marker. Analysis with lineage markers is shown at Right. (B Upper) The map of the MFGδCD5 retroviral vector showing restriction sites and probe used for DNA methylation analysis. (Lower) A Southern blot analysis of methylation of the MFGδCD5 provirus in thymus from three different mice (lanes 1 and 4; 2 and 5; 3 and 6). DNA was subjected to NheI and BamHI with (+) or without (−) SmaI digestions. Lanes 1, 2, and 3 represent DNA from hCD5− cells, and lanes 4–6 correspond to DNA obtained from hCD5+ cells. Lane 7 represents DNA from hCD5+ cells coming from a thymus before hCD5− cells were detected. Nh, NheI; S, SmaI; N, NcoI; B, BamHI.
Methylation Analysis of the CD5− Hematopoietic Cells.
Genomic DNA from CD5− and CD5+ cells sorted from thymus of recipient mice at 9 months posttransplantation were compared for SmaI resistance according to the protocol described in Material and Methods and illustrated in Fig. 5B. SmaI is a methylation-sensitive enzyme that can cleave DNA at the CCCGGG site only if the CpG sequence is not methylated; therefore, SmaI resistance is used as a measurement of DNA methylation. Complete digestion by SmaI shows the absence of methylation of the 5′ LTR. All SmaI digestions of DNA from CD5+ cells show reduction in size of the vector-specific band to 1.2 kb, which corresponds to lack of methylation of the SmaI sites of the provirus 5′ LTR. In contrast, the provirus in all CD5− cells shows SmaI resistance, reflecting various degrees of methylation of the 5′ LTR in these tissues. We conclude that the transcriptional inactivity observed in CD5− cells is associated with methylation of the 5′ provirus LTR at the SmaI sites.
Our analysis has indicated striking differences in the methylation patterns of this sequence. The SmaI sites described are extensively methylated in the CD5− cells, which do not show LTR proviral transcription, but is not methylated in the CD5+ cells, which do express vector transcripts.
Although this failure of transcription from the LTR in hematopoietic tissues is in accord with prior observations (12), the observed association between proviral methylation and expression inactivity does not show whether methylation plays a causal role in suppressing expression or is merely a secondary event after failure of expression has occurred. Nevertheless, the studies presented here suggest that the wild-type LTR may not be the ideal transcriptional unit for long-term expression in pluripotent HSCs and their progeny cells. Characterization of a transcriptionally active retroviral vector in HSCs may provide a better understanding of the regulation of gene expression occurring in these cells.
DISCUSSION
Murine Hematopoietic Reconstitution After Tagging and Selection of Retrovirally Transduced Bone Marrow Cells.
In this report we describe the use of a strategy to generate BMT recipients in which 100% of hematopoietic cells carry and express retroviral vector sequences. The strategy uses retroviral vectors encoding a truncated human CD5 cDNA. The use of a cell surface antigen as a selectable marker of gene transfer (4, 5, 13–16) offers significant advantages over those components that confer resistance to toxic compounds, including the rapid and quantitative detection of transferred gene expression in the desired target cell population by flow cytometry and the efficient and nontoxic selection of transduced target cells by FACS. Our truncated human CD5 cDNA allows us to select only the infected cells, and we show the ability to obtain, before bone marrow transplant, a population of 5-fluouraci-treated murine bone marrow cells that are 100% marked. Moreover, the preselection of BM transduced cells was successful even in cases where the efficiency of gene transfer obtained was poor. The marked population of BM cells is able to reconstitute the hematopoietic system in lethally irradiated mice, indicating that the surface marker lacks deleterious effects on functionality of bone marrow cells. No gross abnormalities in hematopoiesis were detected in mice repopulated with CD5-expressing cells, although a more careful evaluation of hematological parameters will be necessary before making a definitive statement about the lack of effects of marker expression on hematopoietic cell function. The CD5 cell surface antigen can be used not only as a marker for the selection of retrovirally infected target cells but also as a reporter molecule for the phenotyping of target and progeny cells. In this way, all of the hematopoietic cells in the hematopoietic organs from mice reconstituted with sorted transduced BM cells were found to express high levels of surface CD5 antigen 3 months after BMT. However, a significant proportion of the hematopoietic cells no longer express the surface marker CD5 in the 9-month-old recipient mice. Overall, our results show that the use of the CD5 as a retrovirally encoded marker enables the rapid, efficient, and nontoxic selection in vitro of infected primary cells, which can entirely reconstitute the hematopoietic system in mice. These data provide evidence of generation of a functional hematopoieitc system using only the retrovirally transduced CD5 cells.
Implications of the Generation of a Functional Hematopoietic System Using only Retrovirally Transduced Cells for Treatment of Leukemia.
A key problem in the effective treatment of patients with cancer is how to distinguish between tumor and normal cells. This largely explains why current cancer treatments are often ineffective. There have been remarkable advances in our understanding of the molecular biology of cancer, providing new mechanisms for selective tumor destruction. The molecular characterization of tumor-specific chromosomal abnormalities has revealed that fusion proteins are the consequence in the majority of cancers. These fusion proteins are encoded by chimeric genes generated by the chromosomal rearrangement. These chimeric molecules represent ideal therapeutic targets because they are unique to the disease state (they exist only in the tumor cells, not in the normal cells of the patient). Inhibition of the chimeric gene expression by antitumor agents specifically kills the leukemic cells without affecting the normal cells (1–4). A major challenge is to deliver these specific antitumor agents into every tumor cell. If this is not achieved, any malignant cell that remains unaffected will emerge as a resistant clone. Although the efficiency for retrovirus-mediated gene transfer approaches 100%, this may not be realized either because of low viral titers or because of the failure to stimulate cells to divide, which is required for successful integration. Such problems are especially evident in current efforts to apply the technique to gene therapy of cancer; if the specific antitumor agent is not delivered to every tumor cell, any malignant cell that is not affected will emerge as a resistant clone. The possibility of generating a functional hematopoietic system using only retrovirally transduced cells should now greatly enhance the power of studies aimed at addressing questions such as generation of cancer-negative hematopoiesis. Our results suggest an approach to in vitro gene therapy in hematopoietic cancer. Cancer-negative hematopoiesis might be restored by autografting patients with bone marrow cells infected with the virus expressing the specific antitumor agent (1–4) against the tumor-associated fusion junctions (which only exist in the tumor cells but not in the normal cells of the patient) and the selectable marker, followed by selecting to ensure that all reinfused stem cells had integrated functional retroviral sequences. The selected normal stem cells will reconstitute the hematopoietic system, and the specific antitumor agent will eradicate the tumor stem cells.
Moreover, our results show that the expression of the transferred gene is not sustained in cells, because a significant proportion of the hematopoietic cells no longer express the surface marker in the 9-month-old recipient mice. This fact has to be taken into account when using treatments that depend on the generation of drug-resistant hematopoietic cells and strategies in which the inhibition of the leukemic phenotype relies on the sustained expression of the transferred gene.
Acknowledgments
We thank Dr. T. H. Rabbitts and the Laboratory of Molecular Biology–Medical Research Council in Cambridge (U.K.) for support. We also thank Dr. D. Martín-Zanca for helpful advice and support and Dr. R. Mulligan for the MFG vector. This work has been supported by Fundación Internacional José Carreras (FIJC-94/INT), the European Commission (BMH4-CT96-0375), Dirección General de Investigación Cientifica y Técnica (UE96-0041), and Fundación Científica of the Associación Española Contra Cáncer.
ABBREVIATIONS
- FACS
fluorescence-activated cell sorter
- LTR
long terminal repeat
- HSC
hematopoietic stem cells
- FCS
fetal calf serum
- 5-FU
5-fluorouracil
- BM
bone marrow
- BMT
bone marrow transplant
- IL
interleukin
References
- 1.Szczylik C, Skorski T, Nicolaides N C, Manzella L, Malaguarnera L, Venturelli D, Gewirtz A M, Calabretta B. Science. 1991;253:562–565. doi: 10.1126/science.1857987. [DOI] [PubMed] [Google Scholar]
- 2.Skorski T, Nieborowska-Skorska M, Nicolaides N C, Szczylik C, Iversen P, Iozzo R V, Zon G, Calabretta B. Proc Natl Acad Sci USA. 1994;91:4504–4508. doi: 10.1073/pnas.91.10.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo Y, Sánchez-García I, Klug A. Nature (London) 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 4.García Hernández B, Sánchez-García I. Mol Med. 1996;2:125–133. [PMC free article] [PubMed] [Google Scholar]
- 5.Pawliuk R, Kay R, Lansdorp P, Humphries K. Blood. 1994;84:2868–2877. [PubMed] [Google Scholar]
- 6.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, Jackson V, Hamada H, Pardoll D, Mulligan R C. Proc Natl Acad Sci USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison D E, Lerner C P. Blood. 1991;78:1237–1240. [PubMed] [Google Scholar]
- 8.Weinthal J, Nolta J A, Yu X J, Lilley J, Uribe L, Kohn D B. Bone Marrow Transplant. 1991;8:403–412. [PubMed] [Google Scholar]
- 9.Lemischka I R, Raulet D H, Mulligan R C. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- 10.Habers K, Schnieke A, Stuhlman H, Jahne D, Jaenisch R. Proc Natl Acad Sci USA. 1981;78:7609–7613. doi: 10.1073/pnas.78.12.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaehner D, Jaenisch R. Nature (London) 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 12.Challita P-M, Kohn D B. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson C, Bank A. Blood. 1995;86:2579–2589. [PubMed] [Google Scholar]
- 14.Mavilio F, Ferrari G, Rossini S, Nobili N, Bonini C, Casorati G, Traversari C, Bordignon D. Blood. 1994;83:1988–1997. [PubMed] [Google Scholar]
- 15.Pawlivk R, Kay R, Lansdorp P, Humphries R K. Blood. 1994;84:2868–2877. [PubMed] [Google Scholar]
- 16.Conneally E, Bardy P, Eaves C J, Thomas T, Chappel S, Shpall E J, Humphries R K. Blood. 1996;87:456–464. [PubMed] [Google Scholar]