Abstract
Genotype by environment interactions (GEI) play a major part in shaping the genetic architecture of quantitative traits and are confounding factors in genetic studies, for example, in attempts to associate genetic variation with disease susceptibility. It is generally not known what proportion of phenotypic variation is due to GEI and how many and which genes contribute to GEI. Behaviors are complex traits that mediate interactions with the environment and, thus, are ideally suited for studies of GEI. Olfactory behavior in Drosophila melanogaster presents an opportunity to systematically dissect GEI, since large numbers of genetically identical individuals can be reared under defined environmental conditions and the olfactory system of Drosophila and its behavioral response to odorants have been well characterized. We assessed variation in olfactory behavior in a population of 41 wild-derived inbred lines and asked to what extent different larval-rearing environments would influence adult olfactory behavior and whether GEI is a minor or major contributing source of phenotypic variation. We found that ∼50% of phenotypic variation in adult olfactory behavior is attributable to GEI. In contrast, transcriptional analysis revealed that only 20 genes show GEI at the level of gene expression [false discovery rate (FDR) < 0.05], some of which are associated with physiological responses to environmental chemicals. Quantitative complementation tests with piggyBac-tagged mutants for 2 of these genes (CG9664 and Transferrin 1) demonstrate that genes that show transcriptional GEI are candidate genes for olfactory behavior and that GEI at the level of gene expression is correlated with GEI at the level of phenotype.
PHENOTYPIC plasticity is a hallmark of many quantitative traits, as the ability to adapt to changes in the environment is essential for survival. Phenotypic plasticity itself, however, can vary depending on genotype. Genotype by environment interaction (GEI) occurs when there is variation among genotypes in the rank order or relative magnitude of effects in different environments (Falconer and Mackay 1996).
The importance of GEI has been documented in several studies on human disease susceptibility. GEI in which early experience influences adult disease susceptibility has been documented for asthma. This study showed that cytokine response profiles are influenced by early exposure to immune challenges (Hoffjan et al. 2005). Furthermore, a variable number tandem repeat in the promoter of the monoamine oxidase A gene is associated with violent behavior, but only in individuals who were abused as children (Caspi et al. 2002). Another study showed that the short allele of the serotonin transporter predisposes to depression, but that this effect is more pronounced in individuals who have experienced stressful events (Caspi et al. 2003). GEI is pervasive and remains a confounding factor in the analysis of the genetics of complex traits, including the identification of risk alleles for human diseases.
Thus far, studies on GEI have been mostly descriptive. The question whether underlying rules that govern GEI exist requires a systematic analysis. We have used the effect of early developmental exposure to different growth conditions on adult olfactory behavior in Drosophila melanogaster as a model trait for the systematic analysis of GEI. Because genetically identical individuals can be reared under defined environmental conditions, D. melanogaster presents an ideal model system in which we can use whole genome approaches to ask whether there is a core set of environmentally plastic genes that are especially sensitive to GEI or to what extent plasticity of gene expression varies under different environmental conditions and in different genetic backgrounds. Furthermore, the functional organization of the olfactory system of Drosophila (Stocker 1994; Ito et al. 1998; Laissue et al. 1999; Shandbhag et al. 1999; Gao et al. 2000; Vosshall et al. 2000; Jefferis et al. 2001; Marin et al. 2002; Wong et al. 2002) and the genetic basis of odor-guided behavior of flies have been well documented (Anholt et al. 1996; Kim et al. 1998; Fanara et al. 2002; Kulkarni et al. 2002; Ganguly et al. 2003; Stoltzfus et al. 2003; Rollmann et al. 2005), including environmental sensitivity of epistatic interactions that contribute to the manifestation of the behavioral phenotype (Sambandan et al. 2006). Thus, olfactory behavior in Drosophila presents an ideal model trait for studies on phenotypic plasticity and GEI.
We have generated 41 inbred lines of flies derived from isofemale lines collected from a natural Raleigh population and grown larvae of each of these lines on three different food sources: standard fly food, tomato medium, and ethanol-supplemented medium. After eclosion, all flies were maintained on standard medium. Adults were then tested for their ability to respond to a standard odorant, benzaldehyde. Variation in adult olfactory responsiveness as a result of prior environmental exposure during larval development was assessed for each genotype and reaction norms were constructed to estimate phenotypic plasticity and GEI. We then performed whole genome transcript analysis on a subset of 8 lines that differed in patterns of phenotypic plasticity and GEI to identify genes that showed corresponding variations in transcript abundance. We identified a surprisingly small group of 20 genes associated with the manifestation of GEI for adult olfactory behavior. Finally, we performed quantitative complementation tests to demonstrate that genes that show transcriptional GEI are candidate genes for olfactory behavior and that GEI at the level of gene expression is correlated with GEI at the level of phenotype.
MATERIALS AND METHODS
Drosophila stocks:
D. melanogaster lines were derived from 41 isofemale lines collected from a Raleigh natural population by 20 generations of full-sib mating. The lines are viable and fertile and do not contain detectable residual heterozygosity as determined by whole genome marker analysis. We reared larvae on three different media, on the basis of a previous study in which the effect of genotype by environment interactions on fitness was documented (Fry et al. 1996): standard cornmeal–molasses medium, an unfavorable growth medium consisting of standard medium supplemented with 9% ethanol (Fry et al. 1996), and a nutrient-enhanced environment consisting of standard medium enriched with tomato paste (340 g of tomato paste/liter of standard medium; Fry et al. 1996). After eclosion, we transferred all flies to standard medium at 25° under a 12:12-hr light/dark cycle prior to characterization of behavioral responses to benzaldehyde and transcription profiling.
Behavioral assay:
We measured olfactory behavior on 3- to 7-day-old flies at two concentrations of benzaldehyde (0.1 and 0.3% v/v) on the basis of previous dose-response experiments (Sambandan et al. 2006), using the “dipstick” assay, as described previously (Anholt et al. 1996). Briefly, flies are taken off their food source for a period of 45 min before testing. Five adult flies of a single sex are introduced into a plastic vial divided into two marked compartments. The standard odorant benzaldehyde is introduced via a cotton swab into the plastic vial. After a period of 15 sec, the number of flies in the compartment farthest away from the source of the odorant is scored every 5 sec for a period of 60 sec and the average of the 10 scores is calculated. This corresponds to a single replicate. Ten such replicates per sex per line are performed. The mean olfactory response score of the line is calculated as the mean of the 10 replicates and ranges between 0 (complete attraction) to 5 (complete avoidance). Behavioral assays were conducted in an environmental chamber (25°, 70% humidity) between 9 am and 12 am.
Data analysis and estimation of genetic parameters:
We used factorial analysis of variance to partition variance among the genotypes for olfactory behavior into sources attributable to line (L, random effect), sex (S, fixed effect), concentration of benzaldehyde (C, fixed effect), and food (F, fixed effect). Variance components were calculated using SAS GLM and VARCOMP programs (SAS Institute, Cary, NC), according to the model
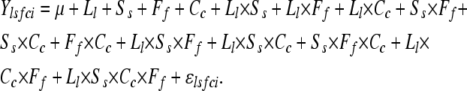 |
Ylsfci is the mean olfactory avoidance behavior of replicate i (i = 1–10) for line l (l = 1–41), s is sex s (s = 1, 2) in food medium f (f = 1, 2, 3) at benzaldehyde concentration c (c = 1, 2). μ indicates the overall mean and ɛlsfci the environmental variance between replicates. The term Ll × Ff indicates GEI. Ll × Cc indicates differences in the dose-response relationship to benzaldehyde among the lines. Ss × Ff and Ss × Cc indicate differences in olfactory behavior between the sexes, which depend on the larval-rearing environment and the concentration of benzaldehyde, respectively. Ff × Cc reflects variation in the dose-response to benzaldehyde as a result of the larval-rearing environment. Ll × Ss × Ff and Ll × Ss × Cc indicate differences in the dependence of sexual dimorphism in the behavioral response as a function of the larval food medium and the odorant concentration, respectively. Ss × Ff × Cc assesses the dependence of sexual dimorphism in the behavioral response on larval-rearing environment and benzaldehyde concentration. Ll × Ff × Cc indicates the dependence of the effect of line on benzaldehyde concentration and growth medium. Ll × Ss × Cc × Ff indicates the dependence of the effect of line on sex, benzaldehyde concentration, and growth medium.
We performed reduced ANOVAs for the two stimulus concentrations of benzaldehyde separately pooled across sexes and larval food source on the basis of the model Yilsf = μ + Ll + Ss + Ff + Ll × Ss + Ll × Ff + Ss × Ff + Ll × Ss × Ff + ɛilsf; for three larval food sources pooled across sexes and concentration of benzaldehyde on the basis of the model Yilsc = μ + Ll + Ss + Cc + Ll × Ss + Ll × Cc + Ss × Cc + Ll × Ss × Cc + ɛilsc; for two concentrations of benzaldehyde and three larval food sources pooled across sexes on the basis of the model Yils = μ + Ll + Ss + Ll × Ss + ɛils; and for two concentrations of benzaldehyde and three larval food sources by sex on the basis of the model Yil = μ + Ll + ɛil.
We estimated cross environmental genetic correlations (rGE) between adult olfactory responses and larval food media from variance components as 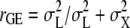 , where
, where 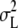 is the variance among the lines and
is the variance among the lines and 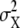 denotes the total variance of all significant line-interaction terms.
denotes the total variance of all significant line-interaction terms.
Line selection and expression analysis:
We used hierarchical clustering analysis by using the TREE procedure for centroid hierarchical cluster analysis (SAS PROC VARCLUS program) to group the 41 genotypes on the basis of their similarity in olfactory behavior and defined eight clusters. We selected 1 genotype from each cluster for whole genome expression analysis. We collected two replicates of 3- to 5-day-old flies from each genotype for each of the three rearing conditions with an equal number of males and females for each sample. We extracted total RNA from the 48 samples (eight lines × two replicates × three rearing conditions) using the Trizol reagent (GIBCO BRL, Grand Island, NY). Biotinylated cRNA probes were hybridized to high-density oligonucleotide microarrays (Affymetrix, Drosophila GeneChip 2.0) and visualized with a streptavidin–phycoerythrin conjugate, as described in the Affymetrix GeneChip Expression Analysis Technical Manual (2000), using internal references for quantification.
Microarray data analysis:
The 18,800 probe sets on the Affymetrix Drosophila GeneChip 2.0 are represented by 14 perfect-match (PM) and 14 mismatch (MM) pairs. The quantitative estimate of expression of each probe set is the Signal (Sig) metric, as described in the Affymetrix Microarray Suite, Version 5.0. The Sig metric is computed using the weighted log (PM–MM) intensity for each probe set and was scaled to a median intensity of 500. A detection call of present, absent, or marginal is also reported for each probe set. We assigned a score of “1” to present and marginal calls and “0” to absent calls. As we had 48 arrays in total, we then excluded from the analysis probe sets with detection scores <5. This filter eliminated probe sets with very low and/or insignificant expression levels. We analyzed the remaining probe sets with two-way mixed model ANOVA of the Signal metric, on the basis of the model Yilf = μ + Li + Ff + Ll × Ff + ɛilf. Yilf is the Signal for the ith replicate (i = 1, 2) of line (L, random effect) l (l = 1–8) in food (F, fixed effect) medium f (f = 1, 2, 3), μ indicates the overall mean, and ɛilf is the variance between replicate arrays. The Ll × Ff interaction is the GEI term. We corrected the P-values computed in these ANOVAs for multiple tests using a false discovery rate (FDR) of <0.05. All statistical analyses were performed using SAS procedures. Gene ontology categories were annotated with the functional annotation tool, DAVID 2.0 Bioinformatics resources 2007, National Institute of Allergy and Infectious Diseases/National Institutes of Health (http://david.abcc.ncifcrf.gov/home.jsp).
The data from this article have been deposited in NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession no. GSE10053. Link for reviewers can be found at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=bhevlkqywkgqkvy&acc=GSE10053.
Quantitative RT–PCR:
For the 48 RNA samples used for microarray analysis, cDNA was generated from 200 ng of total RNA by reverse transcription. We quantified mRNA levels of six genes, CG9664, Transferrin 1, Dopamine transporter, geko, Turandot A, and Pherokine-1, using an ABI-7900 sequence detector with a SYBR green detection method (Applied Biosystems, Foster City, CA). Transcript-specific primers were designed to amplify 50–60 bp regions of all six genes using the primer express program from Applied Biosystems. Primers were designed to encompass common regions of alternative transcripts (supplemental Table S1). We used negative controls without reverse transcriptase for all genes to exclude potential genomic DNA contamination.
Quantitative complementation tests:
We purchased P-element stocks for CG9664 and Transferrin 1 from the Bloomington Drosophila stock center. CG9664 is on the second chromosome (w[1118]; PBac{w[+mC]=PB}CG9664[c00321]) and Transferrin 1 is on the X chromosome (w[1118] PBac{w[+mC]=WH}Tsf1[f05108]). We backcrossed these stocks to the isogenic Canton-S (B) genotype for three generations and made homozygous P-element and control stocks for each of the two genes. Virgin females from each of the two mutant and two control stocks were then mated with males of each of the eight lines used for expression analysis and larvae were reared on standard, tomato, or alcohol-supplemented media. The F1 (3–7 days old) was measured for adult olfactory behavior to 0.3% (v/v) benzaldehyde.
We used factorial, fixed-effects ANOVA to analyze complementation with mutants of CG9664 on the basis of the model Yilfsg = μ + Ll + Ff + Ss + Gg + Ll × Ss + Ll × Ff + Ll × Gg + Ss × Ff + Ss × Gg + Ff × Gg + Ll × Ss × Ff + Ll × Ss × Gg + Ss × Ff × Gg + Ll × Gg × Ff + Ll × Ss × Gg × Ff + ɛilfsg. Yilfsg is the mean olfactory avoidance score of the ith replicate (i = 1–10) of line l (l = 1–8), in rearing medium f (f = 1, 2, 3), for sex s (s = 1, 2), and genotype (G, mutant or control) g (g = 1, 2). Since Transferrin 1 is on the X chromosome, only female olfactory responses were analyzed according to the reduced ANOVA model Yilfg = μ + Ll + Ff + Gg + Ll × Ff + Ll × Gg + Ff × Gg + Ll × Gg × Ff + ɛilfg. Significant Ll × Gg, Ll × Gg × Ss, or Ll × Ss × Ff × Gg interaction terms indicate quantitative failure to complement for GEI. Least square means were calculated for the Ll × Gg term for the two genes for controls and mutants using the LSMEANS program of SAS.
RESULTS
Variation in olfactory behavior:
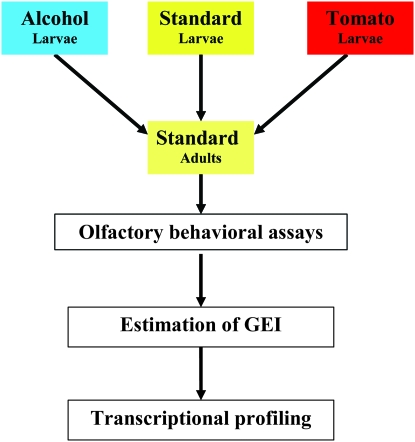
We assessed the magnitude of variation in olfactory behavior in our population of wild-derived inbred lines and asked to what extent larval exposure to different nutritional rearing environments would influence adult olfactory behavior and whether GEI is a minor or major contributing source of phenotypic variation. We reared flies on different food sources during larval development and evaluated the effects of larval exposure to standard, tomato, or alcohol-supplemented medium on adult olfactory behavior and gene expression (Figure 1). Tomato medium is an enriched food environment, whereas alcohol-supplemented medium represents an adverse growth condition.
Figure 1.—
Diagram of the experimental design.
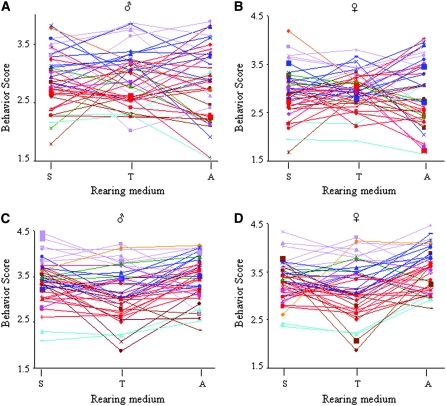
We measured olfactory responses to a standard odorant, benzaldehyde, at two submaximal concentrations (0.1% v/v and 0.3% v/v) for optimal resolution of variation in the behavioral response. Both the full-model and reduced-model ANOVA showed significant variation among the lines (Table 1; supplemental Tables S2–S5), in agreement with a previous study on the response to benzaldehyde in the same population (Wang et al. 2007). Sexual dimorphism in the response between males and females was small and did not contribute significantly to the total variance. Sex effects were mostly evident as a significant L × S interaction at lower concentrations of benzaldehyde or for flies reared as larvae under adverse nutritional conditions (alcohol-supplemented medium). To estimate the contribution of GEI to the total variance, we estimated the cross environmental genetic correlation. For the full-model ANOVA rGE = 0.47, which indicates that GEI contributes ∼50% to the observed variation in adult olfactory behavior when flies are reared on different food sources as larvae. Estimates of rGE remained high, even when calculated from reduced-model ANOVA analyses separately for odorant concentration and larval food sources (supplemental Table S4). The extent of GEI is unexpectedly high, considering that it arises entirely from differences in larval-, but not adult-rearing environments. GEI among the lines is readily visualized as extensive crossing over of reaction norms (Figure 2).
TABLE 1.
ANOVA of olfactory behavior of a population of 41 wild-derived inbred lines
| Source | d.f. | MS | F | P | σ2 |
|---|---|---|---|---|---|
| Line (L) | 40 | 14.539 | 6.55 | 0.0001 | 0.0937 |
| Sex (S) | 1 | 2.674 | 4.11 | 0.0493 | — |
| Food (F) | 2 | 4.034 | 1.28 | 0.2841 | — |
| Concentration (C) | 1 | 251.194 | 139.11 | <0.0001 | — |
| L × S | 40 | 0.650 | 0.85 | 0.6882 | 0.0000 |
| L × F | 80 | 3.155 | 1.22 | 0.2003 | 0.0207 |
| L × C | 40 | 1.806 | 0.63 | 0.9437 | 0.0000 |
| S × F | 2 | 1.784 | 3.35 | 0.0399 | — |
| S × C | 1 | 0.225 | 0.28 | 0.5974 | — |
| F × C | 2 | 13.609 | 5.19 | 0.0076 | — |
| L × S × F | 80 | 0.532 | 0.95 | 0.5866 | 0.0000 |
| L × S × C | 40 | 0.795 | 1.42 | 0.0909 | 0.0048 |
| S × F × C | 2 | 0.894 | 1.60 | 0.2082 | — |
| L × F × C | 80 | 2.623 | 4.69 | <0.0001 | 0.0887 |
| L × S × F × C | 80 | 0.559 | 1.44 | 0.0066 | 0.0166 |
| Error | 4428 | 0.388 | 0.3879 |
Significant P- and σ2-values are underlined. Correlation of line means across sexes, concentrations, and food sources is rGE = 0.4708. d.f., degrees of freedom; MS, mean squares.
Figure 2.—
Reaction norms for olfactory behavior of adult flies reared in different larval environments among wild-derived inbred lines. Crossing over of reaction norms indicates GEI. The olfactory behavior scores are plotted on the y-axis. The x-axis designates the rearing environments. S, T, and A denote standard, tomato, and alcohol larval food sources, respectively. (A and B) Behavior scores obtained at 0.1% (v/v) benzaldehyde. (C and D) Behavior scores obtained at 0.3% (v/v) benzaldehyde. A and C show reaction norms for males. B and D show reaction norms for females. Lines are color coded according to their hierarchical clustering profile shown in Figure 3.
We calculated the broad-sense heritability to be H2 = 0.37 according to the full-model ANOVA with estimates ranging from 0.20 to 0.57 for reduced-model analyses (supplemental Table S5). This value indicates both substantial genetic and environmental contributions to phenotypic variation in olfactory behavior, consolidating our notion that olfactory behavior in Drosophila is an appropriate model for studies on GEI.
Variation in transcriptional response:
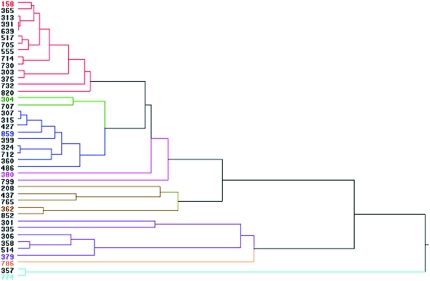
Since GEI contributes about half of the observed phenotypic variation in adult olfactory behavior (Table 1; Figure 2), we asked whether this large GEI effect at the level of phenotype would be mirrored by a large GEI effect at the level of transcription. To answer this question we compared whole-genome transcript-abundance profiles of adult flies from lines reared on different larval food sources. To reduce the scope of the experiment to within manageable limits, we selected a representative sample of lines from among the 41 wild-derived inbred strains. First, we performed a hierarchical clustering analysis for olfactory behavior measured at 0.3% (v/v) benzaldehyde (supplemental Table S6) and grouped the lines according to similarity in olfactory behavior under the different larval-rearing conditions (Figure 3). We identified eight distinct clusters. Lines within each cluster were more similar in their olfactory response to 0.3% (v/v) benzaldehyde to each other than to lines in other clusters. To capture the extent of variation in GEI among our lines, it was therefore sufficient to analyze the transcriptional profiles of eight lines, one randomly chosen from each cluster. Genetic correlations for olfactory behavior among these eight lines across sexes and odorant concentrations were rGE = 0.24 for standard medium, rGE = 0.37 for tomato medium, and rGE = 0.36 for ethanol-supplemented medium.
Figure 3.—
Hierarchical clustering for olfactory behavioral measurements at 0.3% (v/v) benzaldehyde pooled for sexes. The eight major clusters are color coded to the reaction norms in Figure 2. The numbers of the eight lines chosen from each cluster for the microarray analysis are highlighted in color.
We used ANOVA to quantify statistically significant differences in transcript levels for the probe sets on the array and calculated a false discovery rate to correct for multiple testing (Benjamini and Hochberg 1995). At FDR < 0.05, we found 6940 probe sets that varied in expression among the lines (approximately half of the genome), 329 probe sets with variation in expression attributable to the medium on which the larvae were reared (phenotypic plasticity), and 22 probe sets, representing 20 genes, in which variation in expression due to the larval growth medium was dependent on genotype. This surprisingly restricted number of genes is associated with the manifestation of GEI in adult flies (Table 2). No additional probe sets associated with GEI were evident when the FDR criterion was relaxed to 0.2.
TABLE 2.
Genes with significant GEI effects
| Gene | Biological function gene ontology |
|---|---|
| CG1486 | Carboxylic acid metabolic process |
| CG15434 | Mitochondrial electron transport, NADH to ubiquinone |
| CG17177 | Cell communication and signal transduction |
| CG17821 | Lipid metabolic process and fatty acid metabolic process |
| CG31091 | Lipid metabolic process |
| CG32238 | Protein modification process, C-terminal protein-tyrosinylation and protein metabolic process |
| CG32496/CG6788 | Defense response to bacterium, cell adhesion, signal transduction |
| CG4835 | Chitin metabolic process |
| CG9664 | Lipid metabolic process and lipid transport |
| Dopamine transporter | Neurotransmitter transport, regulates sleep and arousal in insects, target for cocaine addiction |
| geko | Implicated in olfactory behavior and response to ethanol |
| Jonah 66C | Proteolysis, intracellular signaling cascade, cyclic nucleotide biosynthetic process |
| ryan express | Transcription initiation from RNA polymerase II promoter, male meiosis, spermatogenesis |
| Transferrin1 | Iron ion transport and homeostasis, defense response |
| Ugt36Bb | Polysaccharide metabolic process, defense response, steroid metabolic process, response to toxin |
| CG13532 | Unknown function |
| CG14872 | Unknown function |
| CG15649 | Unknown function |
| CG30447 | Unknown function |
| E protein | Unknown function |
Transcripts that varied in abundance among the lines included transcripts for two odorant receptors, Or42a and the ubiquitously expressed odorant receptor Or83b, and 27 odorant-binding proteins. The latter account for about half of the family of odorant-binding proteins, but, since about half of the genome shows transcriptional variation among the lines, this multigene family is not significantly overrepresented. The paucity of odorant-receptor transcripts that show transcriptional variation is due to the low levels of transcription of these genes below or near the detection limit of our expression microarrays. In addition to odorant-binding proteins and odorant-receptor transcripts, other genes previously associated with olfactory behavior in D. melanogaster also showed significant variation in expression among the lines, including scribble (Fedorowicz et al. 1998), Semaphorin 5C (Rollmann et al. 2007), escargot, innexin 2, Merlin, CG32556, CG16708, and pipsqueak (Sambandan et al. 2006). Altered transcript abundance was also observed for several genes involved in olfactory learning and memory and genes implicated both in olfactory behavior and in response to ethanol, including the odorant-binding protein lush (Kim et al. 1998) and protein kinase A (Moore et al. 1998; supplemental Table S6).
Genes that showed phenotypic plasticity in the behavioral response to benzaldehyde, as reflected by a significant main effect of the food term in the ANOVA, included Obp49a, Pbprp1(Obp69a), and Calreticulin (Stoltzfus et al. 2003), previously identified as a candidate gene for olfactory behavior, and genes implicated in olfactory learning and memory, phototaxis, and male courtship behavior (no-on-transient A) (Greenspan and Ferveur 2000; Campesan et al. 2001), and circadian rhythm (pigment dispersing factor) (Williams et al. 2001; Mertens et al. 2005; supplemental Table S7).
Transcripts, for which the line × food interaction term in the ANOVA was significant, demonstrate GEI. This group contained only 22 probe sets representing 20 genes (Table 2). Genes that show GEI for transcript abundance in adult flies following larval growth on different food sources include CG9664, associated with lipid metabolism; Dopamine transporter, implicated in regulation of sleep and arousal in insects (Kume et al. 2005); geko, implicated in olfactory behavior and response to ethanol (Shiraiwa et al. 2000); Transferrin 1 (Nichol et al. 2002), involved in iron transport and homeostasis; ryan express, which plays a role in male meiosis and spermatogenesis (Mukai et al. 2006); Jonah 66C, involved in intracellular signaling and proteolysis (Ross et al. 2003); Ugt36Bb, involved in defense response and polysaccharide metabolism (Theopold et al. 1999); and several other genes with predicted transcripts of unknown function. Some of these gene products appear to have in common a role in responses to and metabolism of hydrophobic xenobiotics. Interestingly, no genes encoding odorant receptors or odorant-binding proteins showed GEI at the level of transcription.
Confirmation of variation in transcript abundance by quantitative RT–PCR:
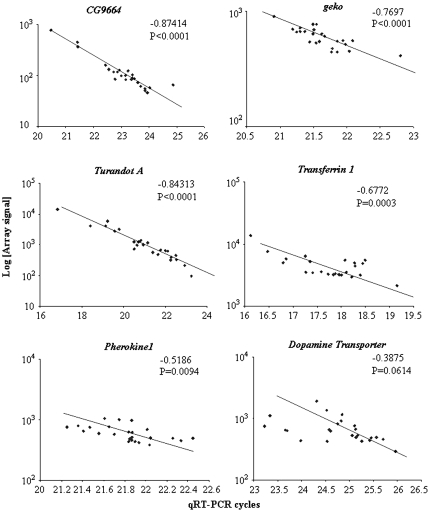
To confirm the reliability of variation in transcript abundance measured on the microarrays, we performed quantitative RT–PCR analysis for a sample of six genes that includes CG9664, Transferrin 1, Dopamine transporter, geko, and Pherokine 1. All of these genes showed significant variation among lines. Five of them also showed GEI, i.e., they were significant for the line × food term in the ANOVA, CG9664, Transferrin 1, Dopamine transporter, and geko at FDR < 0.05, and Pherokine1 at FDR < 0.1.
Quantitative RT–PCR measurements were highly correlated with expression microarray signal amplitudes for CG9664 (P < 0.0001), Transferrin 1 (P = 0.0003), Turandot A (P < 0.0001), Pherokine 1 (P = 0.0094), and geko (P < 0.0001) (Figure 4). The correlation between the microarray signal and quantitative RT–PCR signal was marginal for Dopamine transporter (P = 0.0614), most likely due to low expression levels (Figure 4).
Figure 4.—
Correlations between qRT–PCR and expression microarray signals. The y-axis shows the log transformed raw microarray signal values for the six genes. The x-axis shows the number of PCR cycles needed to reach the SYBR green detection threshold.
Quantitative tests for complementation of GEI:
We hypothesized that genes that show GEI at the level of transcription would also be implicated in GEI at the level of olfactory behavior. To determine to what extent GEI at the level of gene expression correlates with GEI at the level of phenotype, we performed quantitative complementation tests for GEI, using available mutant stocks from the Bloomington Drosophila stock center. Based on the availability of suitable stocks, we selected two genes, CG9664 and Transferrin 1, tagged by a piggyBac transposon. For each of these genes, we generated control and mutant stocks and crossed them to each of the eight wild-derived inbred lines on which expression microarrays had been performed and reared the larvae on the three different food media. We then measured olfactory behavior at 0.3% (v/v) benzaldehyde of adults that were maintained on standard medium (supplemental Tables S8 and S9). Since Transferrin 1 is located on the X chromosome, quantitative complementation tests for this gene were limited to females.
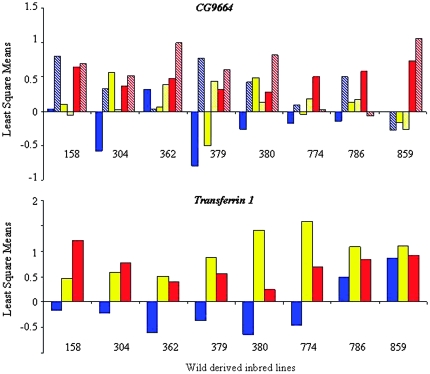
GEI can be inferred when the F × G interaction term in the ANOVA is significant, as this indicates that the effect of the rearing environment on the behavioral response to benzaldehyde is significantly different when the wild-derived inbred line is crossed to the mutant compared to its control, i.e., failure to complement. ANOVA of the behavioral responses showed a significant F × G interaction term for all but two (774 and 786) of the eight lines crossed to the CG9664 mutant, and for all but two (786 and 859) of the eight lines crossed to the Transferrin 1 mutant (Tables 3 and 4).
TABLE 3.
ANOVA for olfactory behavior to test for failure to complement pooled across three larval food sources for CG9664 (sexes pooled, A) and Transferrin 1 (females only, B)
| Source | d.f. | MS | F | P |
|---|---|---|---|---|
| A. CG9664 | ||||
| Line (L) | 7 | 3.531 | 1.15 | 0.422 |
| Sex (S) | 1 | 5.177 | 2.08 | 0.1921 |
| Food (F) | 2 | 1.8 | 1.18 | 0.3349 |
| Genotype (G) | 1 | 13.419 | 39.69 | 0.0004 |
| L × S | 7 | 2.484 | 2.9 | 0.127 |
| L × F | 14 | 1.52 | 2.71 | 0.183 |
| L × G | 7 | 0.338 | 0.61 | 0.7086 |
| S × F | 2 | 2.562 | 3.32 | 0.0659 |
| S × G | 1 | 2.981 | 4.17 | 0.0805 |
| F × G | 2 | 5.394 | 12.83 | 0.0007 |
| L × S × F | 14 | 0.77 | 1.22 | 0.3558 |
| L × S × G | 7 | 0.714 | 1.13 | 0.3962 |
| S × F × G | 2 | 1.436 | 2.28 | 0.1390 |
| L × F × G | 14 | 0.419 | 0.67 | 0.7715 |
| L × S × F × G | 14 | 0.63 | 2.39 | 0.0029 |
| Error | 864 | 0.264 | ||
| B. Transferrin 1 | ||||
| Line (L) | 7 | 4.273 | 1.81 | 0.1878 |
| Food (F) | 2 | 5.402 | 2.65 | 0.1055 |
| Genotype (G) | 1 | 30.805 | 26.53 | 0.0013 |
| L × F | 14 | 2.036 | 2.45 | 0.0523 |
| L × G | 7 | 1.16 | 1.4 | 0.2803 |
| F × G | 2 | 12.964 | 15.62 | 0.0003 |
| L × F × G | 14 | 0.829 | 2.42 | 0.0029 |
| Error | 432 | 0.343 | ||
Significant P-values are underlined. d.f., degrees of freedom; MS, mean squares.
TABLE 4.
ANOVA of olfactory behavior by line across all three larval food media for CG9664 (pooled both sexes) and Transferrin1 (females only)
| Line | Gene | Sex (S) | Food (F) | Genotype (G) | S × F | S × G | F × G | S × F × G |
|---|---|---|---|---|---|---|---|---|
| 158 | CG9664 | <0.0001 | 0.7912 | 0.0003 | 0.851 | 0.2726 | 0.0309 | 0.1523 |
| Tsf1 | — | 0.1107 | 0.0074 | — | — | 0.0119 | — | |
| 304 | CG9664 | 0.333 | 0.0492 | 0.0335 | 0.4837 | 0.3872 | 0.0491 | 0.0105 |
| Tsf1 | — | 0.0227 | 0.0254 | — | — | 0.0447 | — | |
| 362 | CG9664 | 0.3712 | <0.0001 | <0.0001 | <0.0001 | 0.3167 | 0.0285 | 0.1951 |
| Ttsf1 | — | 0.0301 | 0.4755 | — | — | 0.0028 | — | |
| 379 | CG9664 | 0.0474 | <0.0001 | 0.0938 | 0.1767 | <0.0001 | 0.0305 | 0.0105 |
| Tsf1 | — | 0.1017 | 0.016 | — | — | 0.0024 | — | |
| 380 | CG9664 | <0.0001 | 0.0001 | <0.0001 | 0.9179 | 0.0575 | 0.0405 | 0.0104 |
| Tsf1 | — | 0.0488 | 0.0243 | — | — | <0.0001 | — | |
| 774 | CG9664 | 0.0044 | 0.0125 | 0.3505 | 0.3827 | 1 | 0.4879 | 0.2615 |
| Tsf1 | — | <0.0001 | <0.0001 | — | — | <0.0001 | — | |
| 786 | CG9664 | 0.0764 | 0.4217 | 0.0487 | 0.356 | 0.9593 | 0.9032 | 0.0298 |
| Tsf1 | — | <0.0001 | <0.0001 | — | — | 0.2739 | — | |
| 859 | CG9664 | <0.0001 | 0.0118 | 0.0654 | 0.0081 | 0.9193 | <0.0001 | 0.4457 |
| Tsf1 | — | 0.0018 | <0.0001 | — | — | 0.7706 | — |
Numbers represent P-values for each of the ANOVA terms. Significant P-values are underlined.
A comparison of least square means for the L × G terms among the wild-derived lines in combination with CG9664 or Transferrin 1 shows substantial diversity in the nature of the GEI (Figure 5). First, the magnitude and direction of line means differ for both mutants crossed to the inbred lines. Second, the inbred lines differ widely in the manifestation of GEI with, in the case of CG9664, in some instances antagonistic effects between the sexes (Figure 5A). Our results demonstrate that genes that show transcriptional GEI for the trait can be implicated as candidate genes that contribute to its manifestation and that GEI at the level of gene expression can be correlated with GEI at the level of the trait phenotype.
Figure 5.—
Quantitative complementation tests for GEI for Transferrin 1 and CG9664 mutants. Least mean squares (mutant − control) calculated for the line × genotype interaction term is plotted along the y-axis. The numbers designate the eight lines used for expression microarray analysis (see also Figure 3). Blue, yellow, and red bars indicate behavior of flies reared on alcohol, standard, and tomato food sources. Solid bars indicate female olfactory behavior and thatched bars indicate male olfactory behavior.
DISCUSSION
GEI presents a confounding factor in human association studies. It is difficult to quantify the extent of GEI in human populations due to genetic background differences and uncontrolled environmental conditions. Consequently, only a handful of studies have demonstrated GEI in human genetics studies and these analyses were limited to allelic effects of single genes (Caspi et al. 2002, 2003; Hoffjan et al. 2005). Model organisms, in which genetic backgrounds and environmental conditions can be controlled, present a more favorable scenario for the analysis of GEI. QTL analyses have demonstrated GEI for life history traits, including age at maturity, fertility, egg size, and growth rate for Caenorhabditis elegans reared at different temperatures (Gutteling et al. 2007), inflorescence development in Arabidopsis thaliana in different photoperiod environments (Ungerer et al. 2003), GEI for fitness in D. melanogaster (Kondrashov and Houle 1994; Fernandez and Lopez-Fanjul 1996; Fry et al. 1996), sensory bristle number in D. melanogaster in different temperature environments (Gurganus et al. 1998), longevity in D. melanogaster under conditions of different temperatures (Vieira et al. 2000), and larval densities (Leips and Mackay 2000) and ovariole number in D. melanogaster (Wayne and Mackay 1998). Furthermore, a single nucleotide polymorphism at the Delta locus is associated with GEI in sensory bristle number in Drosophila (Geiger-Thornsberry and Mackay 2002).
GEI for olfactory behavior, an essential survival trait, has not been previously analyzed. In this study we sought not only to detect the presence of GEI, but also to quantify the extent of GEI in terms of both the fraction of total phenotypic variance contributed by GEI and the number of genes implicated in its manifestation. We found that GEI accounts for a substantial amount of the total phenotypic variation, as much as 50% in adult olfactory behavior, but that this variation is accompanied by transcriptional GEI of only a small number of genes. It should be noted, however, that genes that were not detected in our study because of low transcript levels might also contribute to GEI. Similarly, environment-dependent post-translational modifications could also affect GEI. Our results, however, are in concordance with a previous study on yeast, where six strains of Saccharomyces cerevisiae were grown in four different nutritional environments; transcriptional profiling showed that only 5% of the genes in the genome contributed to the observed GEI (Landry et al. 2006). Thus, a general underlying rule for the genetic architecture of complex traits may be that relatively few genes may give rise to extensive GEI.
The GEI effects on adult olfactory behavior detected in our study may depend not only on those genes with genotype dependent transcript plasticity which were detected using adult samples for our expression microarrays, but may also arise from genetic effects during the larval stage on adult development (or possibly during the pharate stage posteclosion before adult flies were collected). Thus, the subset of genes that gives rise to GEI may be an underestimate of the total fraction of the genome. Nonetheless, compared to the vast number of transcripts that vary in expression among the lines and the 329 transcripts that show environmental plasticity, our assessment that only a limited number of genes give rise to GEI is likely to remain valid.
The genes that give rise to GEI in olfactory behavior are not a priori associated with olfaction, but fulfill diverse functions, many of which appear to be related to defense responses aimed at metabolizing lipids, which would include most odorants. We have validated the accuracy of detection of our microarrays by quantitative RT–PCR studies on a sample of these genes. Furthermore, quantitative complementation tests show that at least two of these genes, CG9664 and Transferrin 1, influence the behavioral phenotype directly.
Previous studies have indicated that olfactory behavior is mediated by epistatic networks of pleiotropic genes (Fedorowicz et al. 1998) and that these epistatic interactions are sensitive to environmental conditions (Sambandan et al. 2006). The realization of pervasive pleiotropy raises two central questions for future studies. Do the same genes that contribute GEI for olfactory behavior also give rise to genotype-dependent phenotypic plasticity in other traits? And, would the same genes account for GEI in different populations? If indeed a core set of relatively few genes accounts for GEI for multiple traits across populations and possibly across species, identification of this subset of genes becomes feasible and may provide insights that will help manage the confounding effects of GEI both in studies on model organisms and in human populations.
Acknowledgments
We thank Richard Lyman for constructing the wild-derived inbred lines and for assistance with statistical analyses and Julien Ayroles for assistance with the cluster analysis. This is a publication of the W. M. Keck Center for Behavioral Biology at North Carolina State University. This work was supported by grant GM-59469 from the National Institutes of Health. D.S. was supported by a fellowship from a behavioral genomics training grant from the University of North Carolina Office of the President.
References
- Anholt, R. R. H., R. F. Lyman and T. F. C. Mackay, 1996. Effects of single P-element insertions on olfactory behavior in Drosophila melanogaster. Genetics 143 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. [Google Scholar]
- Caspi, A., J. McClay, T. E. Moffitt, J. Mill, J. Martin et al., 2002. Role of genotype in the cycle of violence in maltreated children. Science 297 851–854. [DOI] [PubMed] [Google Scholar]
- Caspi, A., K. Sugden, T. E. Moffitt, A. Taylor, I. W. Craig et al., 2003. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301 386–389. [DOI] [PubMed] [Google Scholar]
- Campesan, S., Y. Dubrova, J. C. Hall and C. P. Kyriacou, 2001. The nonA gene in Drosophila conveys species-specific behavioral characteristics. Genetics 158 1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Addison-Wesley Longman, Harlow, UK.
- Fanara, J. J., K. O. Robinson, S. M. Rollmann, R. R. H. Anholt and T. F. C. Mackay, 2002. Vanaso is a candidate quantitative trait gene for Drosophila olfactory behavior. Genetics 162 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorowicz, G. M., J. D. Fry, R. R. H. Anholt and T. F. C. Mackay, 1998. Epistatic interactions between smell-impaired loci in Drosophila melanogaster. Genetics 148 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, J., and C. Lopez-Fanjul, 1996. Spontaneous mutational variances and covariances for fitness-related traits in Drosophila melanogaster. Genetics 143 829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, J. D., S. L. Heinsohn and T. F. C. Mackay, 1996. The contribution of new mutations to genotype-environment interaction for fitness in Drosophila melanogaster. Evolution 50 2316–2327. [DOI] [PubMed] [Google Scholar]
- Ganguly, I., T. F. C. Mackay and R. R. H. Anholt, 2003. Scribble is essential for olfactory behavior in Drosophila melanogaster. Genetics 164 1447–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q., B. Yuan and A. Chess, 2000. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat. Neurosci. 3 780–785. [DOI] [PubMed] [Google Scholar]
- Greenspan, R. J., and J. F. Ferveur, 2000. Courtship in Drosophila. Annu. Rev. Genet. 34 205–232. [DOI] [PubMed] [Google Scholar]
- Geiger-Thornsberry, G. L., and T. F. C. Mackay, 2002. Association of single-nucleotide polymorphisms at the Delta locus with genotype by environment interaction for sensory bristle number in Drosophila melanogaster. Genet. Res. 79 211–218. [DOI] [PubMed] [Google Scholar]
- Gurganus, M. C., J. D. Fry, S. V. Nuzhdin, E. G. Pasyukova, R. F. Lyman et al., 1998. Genotype-environment interaction at quantitative trait loci affecting sensory bristle number in Drosophila melanogaster. Genetics 49 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteling, E. W., J. A. Riksen, J. Bakker and J. E. Kammenga, 2007. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity 98 28–37. [DOI] [PubMed] [Google Scholar]
- Hoffjan, S., D. Nicolae, I. Ostrovnaya, K. Roberg, M. Evans et al., 2005. Gene-environment interaction effects on the development of immune responses in the 1st year of life. Am. J. Hum. Genet. 76 696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., K. Suzuki, P. Estes, M. Ramaswami, D. Yamamoto et al., 1998. The organization of extrinsic neurons and their implications in the functional roles of the mushroom bodies in Drosophila melanogaster Meigen. Learn. Mem. 5 52–77. [PMC free article] [PubMed] [Google Scholar]
- Jefferis, G. S., E. C. Marin, R. F. Stocker, L. Luo et al., 2001. Target neuron prespecification in the olfactory map of Drosophila. Nature 414 204–208. [DOI] [PubMed] [Google Scholar]
- Kim, M. S., A. Repp and D. P. Smith, 1998. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics 150 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., and D. Houle, 1994. Genotype-environment interactions and the estimation of the genomic mutation rate in Drosophila melanogaster. Proc. Biol. Sci. 258 221–227. [DOI] [PubMed] [Google Scholar]
- Kulkarni, N. H., A. H. Yamamoto, K. O. Robinson, T. F. C. Mackay and R. R. H. Anholt, 2002. The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics 161 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume, K., S. Kume, S. K. Park, J. Hirsh and F. R. Jackson, 2005. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue, P. P., C. Reiter, P. R. Hiesinger, S. Halter, K. F. Fischbach et al., 1999. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J. Comp. Neurol. 405 543–552. [PubMed] [Google Scholar]
- Landry, C. R., J. Oh, D. L. Hartl and D. Cavalieri, 2006. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 366 343–351. [DOI] [PubMed] [Google Scholar]
- Leips, J., and T. F. C. Mackay, 2000. Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics 155 1773–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, E. C., G. S. Jefferis, T. Komiyama, H. Zhu and L. Luo, 2002. Representation of the glomerular olfactory map in the Drosophila brain. Cell 109 243–255. [DOI] [PubMed] [Google Scholar]
- Mertens, I., A. Vandingenen, E. C. Johnson, O. T. Shafer, W. Li et al., 2005. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 48 213–219. [DOI] [PubMed] [Google Scholar]
- Moore, M. S., J. DeZazzo, A. Y. Luk, T. Tully, C. M. Singh et al., 1998. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell 93 997–1007. [DOI] [PubMed] [Google Scholar]
- Mukai, M., Y. Kitadate, K. Arita, S. Shigenobu and S. Kobayashi, 2006. Expression of meiotic genes in the germline progenitors of Drosophila embryos. Gene Expr. Patterns 6 256–266. [DOI] [PubMed] [Google Scholar]
- Nichol, H., J. H. Law and J. J. Winzerling, 2002. Iron metabolism in insects. Annu. Rev. Entomol. 47 535–559. [DOI] [PubMed] [Google Scholar]
- Rollmann, S. M., T. F. C. Mackay and R. R. H. Anholt, 2005. Pinocchio, a novel protein expressed in the antenna, contributes to olfactory behavior in Drosophila melanogaster. J. Neurobiol. 63 146–158. [DOI] [PubMed] [Google Scholar]
- Rollmann, S. M., A. Yamamoto, T. Goossens, L. Zwarts, Z. Callaerts-Végh et al., 2007. The early developmental gene Semaphorin 5c contributes to olfactory behavior in adult Drosophila. Genetics 176 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J., H. Jiang, M. R. Kanost and Y. Wang, 2003. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene 304 117–131. [DOI] [PubMed] [Google Scholar]
- Sambandan, D., A. Yamamoto, J. J. Fanara, T. F. C. Mackay and R. R. H. Anholt, 2006. Dynamic genetic interactions determine odor-guided behavior in Drosophila melanogaster. Genetics 174 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandbhag, S., B. Muller and A. Steinbrecht, 1999. Atlas of olfactory organs of Drosophila melanogaster. 1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect Morphol. Embryol. 28 377–397. [Google Scholar]
- Shiraiwa, T., E. Nitasaka and T. Yamazaki, 2000. Geko, a novel gene involved in olfaction in Drosophila melanogaster. J. Neurogenet. 14 145–164. [DOI] [PubMed] [Google Scholar]
- Stocker, R. F., 1994. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 275 3–26. [DOI] [PubMed] [Google Scholar]
- Stoltzfus, J. R., W. J. Horton and M. S. Grotewiel, 2003. Odor-guided behavior in Drosophila requires Calreticulin. J. Comp. Physiol. A. 189 471–483. [DOI] [PubMed] [Google Scholar]
- Theopold, U., M. Rissler, M. Fabbri, O. Schmidt and S. Natori, 1999. Insect glycobiology: alectin multigene family in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 261 923–927. [DOI] [PubMed] [Google Scholar]
- Ungerer, M. C., S. S. Halldorsdottir, M. D. Purugganan and T. F. C. Mackay, 2003. Genotype-environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics 165 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, C., E. G. Pasyukova, Z. B. Zeng, J. B. Hackett, R. F. Lyman et al., 2000. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics 154 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall, L. B., A. M. Wong and R. Axel, 2000. An olfactory sensory map in the fly brain. Cell 102 147–159. [DOI] [PubMed] [Google Scholar]
- Wang, P., R. F. Lyman, S. A. Shabalina, T. F. C. Mackay and R. R. H. Anholt, 2007. Association of polymorphisms in odorant-binding protein genes with variation in olfactory response to benzaldehyde in Drosophila. Genetics 177 1655–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, M. L., and T. F. C. Mackay, 1998. Quantitative genetics of ovariole number in Drosophila melanogaster. II. Mutational variation and genotype-environment interaction. Genetics 148 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. A., H. S. Su, A. Bernards, J. Field and A. Sehgal, 2001. A circadian output in Drosophila mediated by Neurofibromatosis-1 and Ras/MAPK. Science 293 2251–2256. [DOI] [PubMed] [Google Scholar]
- Wong, A. M., J. W. Wang and R. Axel, 2002. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell 109 229–241. [DOI] [PubMed] [Google Scholar]