Abstract
We report a novel instance of negative interference during Saccharomyces cerevisiae meiosis, where Cre-mediated recombination between pairs of allelic loxP sites is more frequent than expected. We suggest that endogenous crossover recombination mediates cooperative pairing interactions between all four chromatids of a meiotic bivalent.
HOMOLOGOUS chromosomes pair in early meiotic prophase to ensure accurate segregation during anaphase I. Meiotic recombination plays an important role in homolog pairing for many organisms, including the budding yeast Saccharomyces cerevisiae (Page and Hawley 2003). The Cre/loxP site-specific recombination system provides a quantitative genetic probe of meiotic homolog pairing in living yeast cells (Peoples et al. 2002; Peoples-Holst and Burgess 2005; Lui et al. 2006). In an accompanying article, we show that crossover-associated meiotic recombination increases the probability of nearby Cre-mediated loxP recombination events (“collisions”) between homologous chromatids (Mell et al. 2008, this issue). These data suggest that formation of stable joint molecule recombination intermediates between homologs brings cis allelic sequences into close proximity.
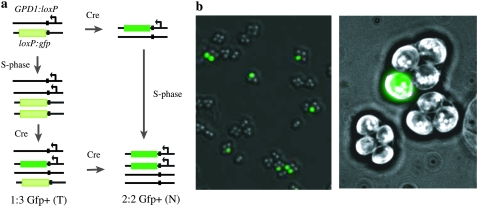
We asked whether Cre-mediated loxP recombination between allelic sites on homologous chromatids affects the frequency of loxP recombination between the two remaining chromatids. To analyze the segregation pattern of Cre/loxP crossovers in tetrads without resorting to tetrad dissection, we used the gene for green fluorescent protein (GFP) as a reporter of allelic loxP recombinants (Figure 1). One homologous chromosome bears a promoterless loxP:gfp contruct, while the other carries a reporterless GPD1:loxP contruct, bearing a promoter that is repressed during meiosis but constitutively expressed at other times. Cre-mediated recombination between allelic loxP sites places the GFP reporter behind the GPD1 promoter for one chromatid. Expression from the recombined GPD1:loxP:GFP reporter is sufficiently robust to allow visualization of GFP in single cells by fluorescence microscopy. When diploids carrying the GPD1:loxP:GFP reporter were sporulated, the Gfp+ phenotype was serendipitously spore autonomous: a GPD1:loxP:GFP heterozygote generated all 2:2 Gfp+ asci (n = 500 asci), while a Gfp+ homozygote produced all 4:0 Gfp+ asci (n = 500 asci).
Figure 1.—
Fluorescent tetrad analysis of allelic loxP collisions. (a) A schematic is shown for the Cre-mediated allelic loxP recombination system, where the frequency of reporter activation (“collision”) is proportional to the physical proximity of the two interacting loxP sites (Hildebrandt and Cozzarelli 1995; Burgess and Kleckner 1999). The loxP site has been incorporated into the ACT1 intron to allow for sufficient expression of the GFP protein to visualize the Gfp+ phenotype by fluorescence microscopy (plasmid and strain construction details available upon request). Presumably, expression from the GPD1 promoter in spores and the removal of the inverted repeat loxP site from the 5′-UTR of GFP mRNA by splicing allow for robust expression of GFP (K. Komachi and O. Hughes, unpublished data). Two paths are shown for generating 2:2 Gfp+ (N) tetrads, with Cre-mediated recombination occurring either before or after meiotic S-phase. (b) Two fields of asci sporulated with 0.03% galactose (which activates expression of Cre recombinase) added at t = 1 hr are shown. On the left, 1:3 Gfp+ (T) and 2:2 Gfp+ (N) four-spore asci are visible. On the right, a single tetrad showing 1:3 Gfp+ (T) segregation is shown at higher magnification.
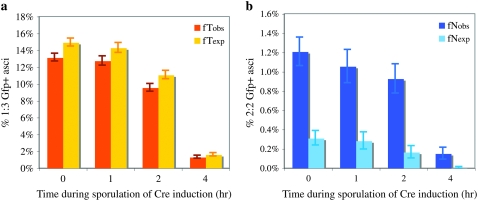
We analyzed four-spore asci from synchronized meiotic cultures of GPD1:loxP/loxP:gfp heterozygotes, in which Cre expression was induced at various time points after the start of sporulation (Figure 2; Table 1). The appearance of Gfp+ spores was used to measure the frequency of Cre-mediated loxP recombination between homologous chromatids. We interpreted 0:4, 1:3, and 2:2 Gfp+ asci as analogous to parental ditypes (P), tetratypes (T), and nonparental ditypes (N) as in a typical two-factor cross. We calculated the expected frequencies of 1:3 Gfp+ (T) and 2:2 Gfp+ (N) tetrads under the assumptions of Papazian (1952), using the “better” method to detect interference described in the Stahl Lab Online Tools at http://www.molbio.uoregon.edu/∼fstahl/, which assumes a Poisson-distributed number of events per meiotic cell, and a random distribution of two-, three-, and four-strand double events (i.e., “no chromatid interference”). Specifically, the appearance of 2:2 Gfp+ asci (N) indicates the occurrence of either a prereplicative Cre/loxP collision or a four-strand double-loxP collision involving all four chromatids of a meiotic bivalent (Figure 1).
Figure 2.—
Nonrandom segregation of Gfp+ in tetrads. The observed and expected frequencies of (a) 1:3 Gfp+ and (b) 2:2 Gfp+ are shown (indicating fT and fN, respectively). For expected values, the assumptions of Papazian (1952) and the method described at the Stahl Lab Online Tools were used: 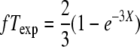 and
and 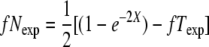 , where
, where 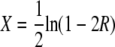 and R = fNobs + fTobs/2. (a) The observed and expected frequency of 1:3 Gfp+ tetrads (fTobs and fTexp) for different Cre induction times is illustrated. (b) The observed and expected frequency of 2:2 Gfp+ tetrads (fNobs and fNexp) is illustrated. Estimated standard error was used to size the error bars.
and R = fNobs + fTobs/2. (a) The observed and expected frequency of 1:3 Gfp+ tetrads (fTobs and fTexp) for different Cre induction times is illustrated. (b) The observed and expected frequency of 2:2 Gfp+ tetrads (fNobs and fNexp) is illustrated. Estimated standard error was used to size the error bars.
TABLE 1.
Segregation pattern of Gfp+ allelic loxP recombinants in tetrads
| Gal+ (hr) | Count of tetrad classes
|
||||
|---|---|---|---|---|---|
| 0:4 Gfp+ | 1:3 Gfp+ | 2:2 Gfp+ | Total | Nobs/Nexp | |
| 0 | 4683 | 718 | 66 | 5467 | 3.9 |
| 1 | 3025 | 447 | 37 | 3509 | 3.7 |
| 2 | 3574 | 381 | 37 | 3992 | 5.6 |
| 4 | 3915 | 52 | 6 | 3973 | 46.4 |
Cre recombinase was induced with 0.03% galactose in synchronized meiotic cultures at the time point indicated, as described in Peoples et al. (2002). Unambiguous four-spore tetrads were evaluated for the number of Gfp+ spores by fluorescence light microscopy (Figure 1). The Nobs/Nexp ratio was determined as the ratio of observed to expected 2:2 Gfp+ tetrads under the assumptions of Papazian (1952) as described in Figure 2. Using the χ2 method found at the Stahl Lab Online Tools, P-values for all time points were ≪0.001, indicating a significant deviation from random expectations.
When Cre recombinase was induced early in meiotic prophase I, we observed an approximately fourfold excess of 2:2 Gfp+ tetrads, indicative of “negative interference” (Figure 2; Table 1). When Cre was induced later in meiotic prophase I, there was an even stronger excess of doublet Gfp+ tetrads above that expected (Figure 2; Table 1). These results rule out the possibility that the excess four-strand double-loxP recombinants are due to Cre-mediated loxP recombination occurring at the two-strand stage (prior to DNA replication of the loxP sequences), since in this case we would expect negative interference to disappear when Cre is induced at later time points, as more of the culture transits meiotic S-phase.
These data show that the sister chromatids behave cooperatively, such that allelic loxP double crossovers involving all four chromatids occur more often than expected during late prophase I. This is in contrast to positive interference observed for Spo11p-initiated crossovers, where the frequency of nearby double crossovers is less than random expectations in wild-type yeast (Bishop and Zickler 2004).
Using a combination of endogenous and Cre/loxP recombination reporters, we have found that crossover-bound meiotic recombination intermediates increase interactions between loxP sites located on homologous chromatids (Mell et al. 2008). The results of the tetrad analysis presented here further suggest a constrained geometry of all four chromatids in a bivalent around these sites. We speculate that “synapsis initiation complexes” (Fung et al. 2004) stabilize local homologous chromatid interactions and also constrain interactions between the remaining two chromatids. Endogenous meiotic crossing over at the DNA level is coordinated with exchange between “homologous” axial elements, which involves both sister chromatids of a chromosome (Blat et al. 2002); thus the negative interference observed in Cre/loxP recombination may reflect this coupling between the sisters. Finally, the spore-autonomous segregation of Gfp+ using the reporter constructs described here may offer a useful tool for studies of meiotic recombination. Adding different fluorescent markers to linked positions in the genome will provide a rapid method of tetrad analysis that does not require tetrad dissection, similar to the visual tetrad analysis assay developed for Arabidopsis reported by Francis et al. (2007).
Acknowledgments
We thank Jeff Marblestone and Celia Shiau for technical assistance and Terry Orr-Weaver for suggesting experiments varying Cre induction. We also thank Dan Ohde and Ira Hall for comments on the manuscript. This work was supported by grants from the American Cancer Society (RSG-01-053-01-CCG) and the National Institutes of Health (NIH) (5RO1GM75119-2) to S.M.B. and a grant from the NIH/National Institute of Environmental Health Sciences (1R43ES010081-01) to O.H.
References
- Bishop, D. K., and D. Zickler, 2004. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117 9–15. [DOI] [PubMed] [Google Scholar]
- Blat, Y., R. U. Protacio, N. Hunter and N. Kleckner, 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111 791–802. [DOI] [PubMed] [Google Scholar]
- Burgess, S. M., and N. Kleckner, 1999. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev. 13 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, K. E., S. Y. Lam, B. D. Harrison, A. L. Bey, L. E. Berchowitz et al., 2007. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 3913–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, J. C., B. Rockmill, M. Odell and G. S. Roeder, 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116 795–802. [DOI] [PubMed] [Google Scholar]
- Hildebrandt, E. R., and N. R. Cozzarelli, 1995. Comparison of recombination in vitro and in E. coli cells: measure of the effective concentration of DNA in vivo. Cell 81 331–340. [DOI] [PubMed] [Google Scholar]
- Lui, D. Y., T. L. Peoples-Holst, J. C. Mell, H. Y. Wu, E. W. Dean et al., 2006. Analysis of close stable homolog juxtaposition during meiosis in mutants of Saccharomyces cerevisiae. Genetics 173 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mell, J. C., B. L. Wienholz, A. Salem and S. B. Burgess, 2008. Sites of recombination are local determinants of meiotic homolog pairing in Saccharomyces cerevisiae. Genetics 179 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2003. Chromosome choreography: the meiotic ballet. Science 301 785–789. [DOI] [PubMed] [Google Scholar]
- Papazian, H. P., 1952. The Analysis of Tetrad Data. Genetics 37 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples, T. L., E. Dean, O. Gonzalez, L. Lambourne and S. M. Burgess, 2002. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev. 16 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples-Holst, T. L., and S. M. Burgess, 2005. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 19 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]