WESTERN medicine is in crisis. Continually increasing resources are being expended to combat the age-related diseases that include diabetes and metabolic syndrome, Alzheimer's disease, Parkinson's disease, cardiovascular disease, and cancer. Yet the causes of these diseases remain a mystery, while their incidence and morbidity either remain constant or are increasing (Wallace 2005b).
Huge investments in biomedical research in the recent past have resulted in some striking accomplishments, including the sequencing of the human chromosomal DNA (Lander et al. 2001; Venter et al. 2001), the identification of hundreds of thousands of human chromosomal single nucleotide polymorphisms (SNPs), and the identification of regional clusters of chromosomal SNPs (the HapMap) (International HapMap Consortium et al. 2007). However, these accomplishments have failed to reveal the anticipated genetic causes for the common age-related diseases. For example, a series of “whole-genome scans” encompassing hundreds of thousands of chromosomal SNPs and >32,000 subjects has revealed nine polymorphic loci associated with type II diabetes, yet the aggregate risk for all nine loci accounts for only a small proportion of the overall diabetes risk (Saxena et al. 2007; Scott et al. 2007; Sladek et al. 2007; Zeggini et al. 2007).
Thomas Kuhn, in his book The Structure of Scientific Revolutions (Kuhn 1996), argued that when the scientific effort expended on a problem increases—yet productivity declines—then the difficulty may lie with the assumptions (paradigms) on which the research is based. For the past 100 years, Western biomedical science has stood on two philosophical pillars: the anatomical paradigm of medicine and the Mendelian paradigm of genetics. The anatomical paradigm of medicine has at its foundation the work of Vesalius, who first described the organs of the human body 450 years ago. Since then, physicians and medical scientists have specialized in individual organs and their associated disease manifestations, leading to the fields of neurology, ophthalmology, nephrology, cardiology, endocrinology, etc. The organ-specific compartmentalization of medicine has also led to several generally accepted corollaries: organ-associated symptoms are the result of organ-specific problems, organ-specific problems are the result of tissue-specific protein and gene defects, and tissue-specific protein defects should be treated with chemicals that specifically interact with the defective tissue-specific protein.
The Mendelian paradigm of genetics argues that genetic traits are transmitted across generations according to the laws of Gregor Mendel. The associated medical corollary is that if a clinical trait is transmitted in a Mendelian fashion, it is genetic, but if it is not, then the trait must be the consequence of environmental factors. This corollary is formalized through the estimation of heritability by dividing the frequency that a phenotypic trait is shared by identical twins with the frequency that it is shared by fraternal twins. However, since Mendelian genetics is the result of chromosomal dynamics, the Mendelian paradigm is specific for nuclear DNA (nDNA) genes.
While the anatomical paradigm of medicine and the Mendelian paradigm of genetics have been powerful predictors of medical relationships for the past century, they are failing to direct us toward solutions for the common age-related diseases. According to Kuhn, when a prevailing paradigm fails to make productive predictions, then hypothesis-based research begins to fail. To resolve the crisis and return to productive “normal science,” a new paradigm must be generated that encompasses the strengths of the previous paradigm but adds new elements that address the current problems being confronted. Assuming that this Kuhnian analysis is applicable to the biomedical sciences today, what could be the missing components of the anatomical and Mendelian paradigms necessary for understanding the age-related diseases?
The first suggestion of an answer to this question came with the publication of three articles in 1988—20 years ago. The first article reported that deletions in the extra-nuclear mitochondrial DNA (mtDNA) could be associated with a characteristic muscle pathology involving ragged red muscle fibers and abnormal mitochondria, designated mitochondrial myopathy (Holt et al. 1988). Mitochondrial DNA deletions and mitochondrial myopathy have subsequently been associated with the spontaneously occurring chronic external progressive ophthalmopelgia. The second article reported that a missense mutation at nucleotide (nt) 11,778 (G > A) in the mtDNA ND4 polypeptide (R340H) was the cause of maternally inherited Leber hereditary optic neuropathy (LHON) (Wallace et al. 1988a). The third article used maternal inheritance to link a familial brain and muscle disease called myoclonic epilespy and ragged red fiber to the mtDNA (Wallace et al. 1988b), a conclusion that was subsequently confirmed by the identification of the causal mutation in the mtDNA tRNALys gene at nt 8344 (A > G) (Shoffner et al. 1990). Since the mtDNA encodes genes for proteins of mitochondrial energy metabolism, these articles had two major implications. First, human diseases affecting a wide range of organs could result from systemic defects in energy metabolism and, second, hereditary human diseases could result from mutations in the non-Mendelian mtDNA. Consequently, mitochondrial biology and genetics become excellent candidates for expanding the anatomical and Mendelian paradigms to address the complexities of the age-related diseases, aging, and cancer (Wallace 1992b).
Life involves the interplay between structure and energy. For the eukaryotic cell, this duality was cemented ∼2 billion years ago by the symbiosis of what appears to have been a glycolytic motile cell, which gave rise to the nucleus–cytosol, and an oxidative α-proteobacterium, which evolved into the mitochondrion (Margulis 1981; Lang et al. 1997; Gray et al. 1999). Initially, each organism was free living and contained all of the genes for an independent life form. However, over the subsequent 1.2 billion years, the single-cell descendants of the initial symbiosis experimented with many alternative arrangements of biochemical interdependence and genomic reorganization. Ultimately, however, an arrangement was achieved in which the mitochondrion became specialized in energy production and the nucleus–cytosol became specialized in structure. This final design provided the impetus for the development of multicellularity and the evolution of higher plants and animals, including humans (Wallace 2007).
The restructuring of the proto-mitochondrial genome included the transfer of virtually all of the genes of the mitochondrial genome, ∼1500, into the chromosomal nDNA. Yet the mtDNA persisted and today still retains 13 polypeptide-encoding genes plus a small and large rRNA gene and 22 tRNA genes. All of the mtDNA-encoded polypeptides are core subunits of the enzyme complexes of the mitochondrial energy-generating apparatus, oxidative phosphorylation (OXPHOS). In OXPHOS, reducing equivalents (electrons) derived from the calories of our diet are transferred down a series of redox enzyme complexes located within the mitochondrial inner membrane, collectively known as the electron transport chain. The electrons enter at either complex I or II and are transferred through coenzyme Q to complex III, then to cytochrome c, on to complex IV, and finally to oxygen to generate H2O. The energy that is released as the electrons traverse complexes I, III, and IV is used to pump protons out of the mitochondrial matrix across the inner membrane, resulting in an electrochemical gradient, the biological equivalent of a capacitor. This capacitance is used as a source of potential energy to drive a variety of activities. For example, the protons can flow back across the inner membrane into the matrix through a proton channel in complex V, the ATP synthase. In the process, potential energy is converted into the high-energy γ-phosphate bond of ATP, which can be used to drive chemical work. If mitochondrial OXPHOS is efficient in converting caloric energy to ATP, it is said to be tightly coupled, and these mitochondria will generate the maximum ATP and thus work for the minimum calories burned. However, if the mitochondria are less efficient at generating ATP, partially uncoupled, then more calories must be burned to generate the same amount of ATP. The energetic difference is dissipated as heat. Thus, in endotherms such as humans, changes in the mitochondrial coupling efficiency determine the relative allocation of calories between ATP for work and heat to maintain the body temperature (Wallace 2007).
Another product of OXPHOS is reactive oxygen species (ROS). Mitochondrial ROS provides a signaling system from the mitochondrion to the nucleus (Burdon 1995; Hansen et al. 2006; Jones 2006). However, when mitochondrial ROS production becomes excessive, the mitochondria and mtDNAs can be damaged. While each cell contains hundreds of mitochondria and thousands of mtDNAs, as the cellular mtDNAs become mutated by oxidative damage, the mtDNA information necessary for repairing damaged mitochondria is depleted and the mitochondrial energy output declines. Ultimately, there is insufficient mitochondrial energy for the cell to carry out its normal function and it malfunctions. The malfunctioning cell can then disrupt normal tissue function and integrity. To resolve this deleterious state, the mitochondria-deficient cell must be removed by apoptosis. This is achieved by the activation of the mitochondrial permeability transition pore (mtPTP), which senses increased oxidative stress, reduced electrochemical potential, reduced high-energy phosphates, and the mitochondrial uptake of excessive calcium.
The 13 polypeptides of the mtDNA include 7 of the ∼45 polypeptides of complex I (ND1, -2, -3, -4L, -4, -5, -6), 1 of the 11 polypeptides of complex III (cytochrome b), 3 of the 13 polypeptides of complex IV (COI, -II, -III), and 2 of the ∼15 polypeptides of complex V (ATP6 and -8). All of the other genes of the mitochondrial genome are dispersed across the chromosomes and include the mitochondrial DNA polymerase γ (POLG), RNA polymerase, ribosomal proteins, metabolic enzymes, etc.
If it was beneficial for the first 1500 mitochondrial genes to be transferred to the nucleus, why not the last 13? After all, transfer of the final 13 proteins would have permitted the elimination of an entire redundant mitochondrial genetic information system. Yet every oxidative organism retains an mtDNA and virtually all organisms of the fungal–animal lineage retain the same mtDNA genes. Hence, the retention of these genes in the mtDNA must be important (Wallace 2007).
For those mtDNA-encoded proteins for which the function is known (cytochrome b, COI, COII, COIII, and ATP6), the protein is either an electron or a proton carrier of OXPHOS. Moreover, all of these charge carriers interact in the generation, maintenance, or utilization of the same entity, the mitochondrial inner membrane electrochemical gradient. Thus the polypeptide genes of the mtDNA encode the wiring diagram for the mitochondrial capacitor in a single integrated mitochondrial circuit. As a consequence, a mutation in any one of the mtDNA polypeptides within a mtDNA will have physiological consequences for all of the other polypeptides in that mtDNA, effectively shifting the energetic balance of the entire circuit. The new aggregate metabolic state will then be tested for local genetic fitness by natural selection. The accrual of mtDNA mutations over many generations will then result in the divergence of mtDNA sequences and the development of new metabolic strategies for coping with changing environments. Because of the functional coevolution of the genes of an individual mtDNA, all of the genetic polymorphisms for the proteins of that mtDNA must be intercompatible; i.e., they must match. Therefore, the random mixing of the protein polymorphisms between two different mtDNA lineages could result in combining incompatible genetic elements, thus shorting the capacitor and resulting in energetic failure. To prevent such random mixing of divergent circuit elements, the genes of different mtDNA lineages must be prohibited from undergoing recombination. This is accomplished by having the mtDNA inherited from only one parent—the mother in the case of humans (Giles et al. 1980) and most other species.
Because mitochondrial OXPHOS impinges on many cellular functions, including energy allocation, ROS generation, redox control, calcium homeostasis, and programmed cell death, different mitochondrial energy circuits can be beneficial in a wide spectrum of environmental contexts. For example, mtDNA polymorphisms that produced tightly coupled mitochondria could be advantageous in the tropics where calories would produce maximum ATP and minimum heat. By contrast, more loosely coupled mitochondria could be advantageous in the arctic where the oxidation of additional calories to generate heat would increase the resistance to cold (Wallace 1994, 2005b, 2007).
Because the energetic demands of the environment can change rapidly, it is advantageous for an endothermal species to maintain a diverse array of mtDNA genotypes and thus energetic solutions. This would ensure that some individuals can survive a sudden environmental energetic change. However, the lack of recombination limits the ability of the mitochondrial system to generate an array of genetic combinations and thus energetic solutions. This dilemma is resolved by the mtDNA having a high mutation rate, such that new mitochondrial energy solutions are generated de novo each generation. Presumably, the mtDNA mutation rate is regulated by modulating mitochondrial ROS production and detoxification rates as well as by mtDNA repair (Wallace 2007). Rapid segregation of variant mtDNAs within the female germline results in maternal lineages that approach homoplasmic (purely mutant) for variant mtDNAs (Jenuth et al. 1996). Individuals harboring these variant mtDNA genotypes can differ in mitochondrial physiologies. This provides the needed variation among the individuals within the population to increase the probability that some individuals may survive if the environment changes suddenly (Wallace 2007).
That human mtDNA variation is extensive and adaptive has been demonstrated by the analysis of the regional mtDNA variation in indigenous populations from different parts of the world. This has revealed that the human mtDNA tree has discrete branches with each branch encompassing a group of related mtDNA sequences (haplotypes) called a haplogroup. Moreover, the haplogroups correlate with the geographic distribution of indigenous populations and consequently with their environmental niche. Macro-haplogroup L, which encompasses haplogroups L0, L1, L2, and L3, is found almost exclusively in sub-Saharan Africa. Two derivatives of African L3 founded macro-haplogroups M and N, the only two mtDNAs to successfully leave Africa to colonize all of Eurasia. Macro-haplogroup N radiated into Europe, giving rise to haplogroups H, I, J, Uk, T, U, V, W, and X. Both macro-haplogroups M and N radiated into Asia, M giving rise to haplogroups C, D, G, and many others and N to haplogroups A, B, F, and others. Of the Asian haplogroups, only A, C, and D became enriched in northeastern Siberia and were in a position to cross the Bering land bridge to be the first human inhabitants of the Americas. A, C, and D were subsequently joined in the Americas by haplogroups B and X (Wallace et al. 1999). Generally, each haplogroup is founded by one of more functional variants. Moreover, a number of the functional mtDNA variants have arisen multiple times in different populations, demonstrating convergent evolution and confirming adaptive selection (Wallace et al. 1999, 2003; Mishmar et al. 2003; Ruiz-Pesini et al. 2004; Ruiz-Pesini and Wallace 2006).
That this ancient mtDNA variation affects human health has been demonstrated through the identification of multiple associations between mtDNA haplogroups and various clinical conditions. The first such association revealed that European haplogroup J increases the penetrance of the milder LHON pathogenic mutations (Brown et al. 1995, 1997, 2002; Torroni et al. 1997). Subsequently, haplogroup T was associated with increased risk for bipolar affective disorder (McMahon et al. 2000). Haplogroup J was then correlated with longevity in Europeans (Ivanova et al. 1998; De Benedictis et al. 1999; Rose et al. 2001; Niemi et al. 2003) and D with longevity in Asians (Tanaka et al. 1998, 2000). Haplogroup H has been associated with increased risk, and haplogroups J and Uk with decreased risk, for developing Parkinson's disease (Van Der Walt et al. 2003; Ghezzi et al. 2005; Khusnutdinova et al. 2008). The nt 4336 sublineage of haplogroup H has been associated with increased Alzheimer's disease risk, while haplogroups U and T are associated with decreased risk in certain contexts (Shoffner et al. 1993; Chagnon et al. 1999; Carrieri et al. 2001; Van Der Walt et al. 2004). Haplogroup H has also been correlated with reduced risk of age-related macular degeneration, while haplogroups J and U are associated with increased drusen levels and retinal pigment abnormalities (Jones et al. 2007). Haplogroup J has been associated with increased risk of diabetes in certain European descent populations (Mohlke et al. 2005; Crispim et al. 2006; Saxena et al. 2006), while haplogroup N9a is protective of diabetes, metabolic syndrome, and myocardial infarction in Asians (Fuku et al. 2007; Nishigaki et al. 2007; Wallace et al. 2007a). Haplogroup H has been associated with protection against sepsis (Baudouin et al. 2005) and U with increased serum IgE levels (Raby et al. 2007). Finally, various haplogroups have been correlated with altered risk for particular cancers (Booker et al. 2006; Bai et al. 2007; Darvishi et al. 2007), which augments recent observations of associations between mutations in mitochondrial genes, both nDNA and mtDNA, and cancer (Gottlieb and Tomlinson 2005; Wallace 2005a; Brandon et al. 2006).
These haplogroup clinical associations are now being complemented by evidence of physiological differences between mtDNA haplogroups. Studies of elite Finnish athletes have revealed differences in haplogroup distributions between long distance runners and sprinters (Niemi and Majamaa 2005). Different haplogroups have been correlated with differences in sperm motility, which is determined by the energy output of the mitochondria in the sperm mid-piece (Ruiz-Pesini et al. 1998; Montiel-Sosa et al. 2006). Finally, direct physiological alterations have been associated with the missense mutations that define macro-haplogroup N (Kazuno et al. 2006).
While the high mtDNA mutation rate has been successful in generating extensive adaptive mtDNA variation, random mutations are much more likely to have an adverse effect on a protein than a beneficial one. Therefore, diseases resulting from mtDNA mutations should be very common. The frequency of mtDNA diseases is indeed high, currently estimated as having an incidence of 1.65/10,000 (Schaefer et al. 2004, 2008). Moreover, in the past 20 years, mtDNA mutations have been linked to a broad spectrum of clinical problems affecting the central nervous system including forms of blindness, deafness, dementia, and movement disorders; the heart and cardiovascular system; the musculoskeletal system; and the renal and endocrine systems—many of the same systems that are affected by aging and the age-related diseases (Wallace et al. 2007b).
The appearance of a new mtDNA mutation in a cell results in an intracellular mosaic of mutant and normal mtDNAs, a state known as heteroplasmy. As the heteroplasmic cell undergoes cytokinesis, the mutant and normal mtDNAs are thought to be randomly partitioned into the daughter cells such that the percentage of mutant mtDNAs can drift during both mitotic and meiotic cell division, a process known as replicative segregation. As a result, identical twins derived from the same heteroplasmic egg can have different mtDNA genotypes and different clinical phenotypes. Also, the members of a family harboring a heteroplasmic mtDNA mutation can each inherit a different percentage of mtDNA mutants and thus have completely different clinical manifestations. The heteroplasmic nt 14,459 (A > G) missense mutation in the ND6 protein (L60S) can manifest as LHON when heteroplasmic but as generalized dystonia when homoplasmic (purely mutant) (Jun et al. 1994). The heteroplasmic nt 8993 (T > G) ATP6 missense mutation (L156R) can present as retinitis pigmentosa at 75% mutant, but can cause macular degeneration, olivopontocerebellar atrophy, and lethal childhood Leigh syndrome as the percentage of mutant mtDNA increases to 95% (Ortiz et al. 1993). Thus, the same mtDNA mutation can result in totally different symptoms, each seen by a different clinical subspecialist, simply owing to chance fluctuations in mtDNA heteroplasmy.
The high mtDNA mutation rate necessary to maintain a spectrum of functional energetic alternatives should generate a diverse array mutations. Most of these mutations should be deleterious due to the high evolutionarily conservation of the mtDNA polypeptides (Neckelmann et al. 1987; Wallace et al. 1987). While pathogenic mtDNA tRNA mutations such as the mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes mutation in tRNALeu(UUR) at nt 3243 (A > G) are quite common (Goto et al. 1990; Schaefer et al. 2004, 2008), there is a striking dearth of the most severe polypeptide mutations in patients with mitochondrial disease (Wallace et al. 2007b). This puzzle is explained in mammals by the presence within the female germline of a mitochondrial mutant selection system that preferentially eliminates the most deleterious mtDNA polypeptide mutations prior to ovulation (Fan et al. 2008; Stewart et al. 2008). Thus a wide diversity of mild functional polypeptide mutations can be generated without excessively burdening the fitness of the species with a high frequency of debilitating polypeptide mutants.
The female germline's mtDNA selective system encompasses two processes acting in concert: replicative segregation of mtDNA genotypes into different cells and selection against the cells with the most severe mtDNA mutant genotypes. The segregation of heteroplasmic genotypes is accomplished because the female primordial germ cells have a reduced number of “mtDNA segregating units,” generated from the estimated minimum of 953–1561 mtDNAs per primordial germ cell via either nucleoid clustering or differential replication (Cao et al. 2007). The primordial germ cells have been proposed to undergo ∼20 mitotic cell divisions generating several million proto-oocytes. During this process, the mutant and normal mtDNA segregating units are dispersed through repeated cytokineses, resulting in the proto-oocytes segregating their mtDNA genotypes toward predominantly normal or mutant mtDNAs by genetic drift (Jenuth et al. 1996). Once the mtDNA genotypes have sorted out, then intra-ovarian selection eliminates those proto-oocytes that have the most severe mitochondrial defects. Hence, only those proto-oocytes with more normal functions survive and are ovulated. The mechanism by which oocytes harboring the more defective mitochondria are recognized is unknown. However, one possibility is that the defective mitochondria generate more ROS and this leads to preferential cell death, perhaps through activation of the mtPTP (Fan et al. 2008; Stewart et al. 2008).
The mtDNA mutation rate is high not only in the female germline, but also in the body's somatic tissues. Consequently, mtDNA rearrangement and base substitution mutations have been found to accumulate with age in multiple tissues (Wallace 2005b, 2007; Wallace et al. 2007b). As a result, the age-related accumulation of mtDNA mutations with an associated decline in mitochondrial function is thought to be an important factor in the aging clock. Two lines of evidence suggest that the accumulation of somatic mutations is a cause of aging in mammals. Introduction of an error-prone POLG into the mouse resulted in an increased mtDNA mutation rate and a premature aging phenotype (Trifunovic et al. 2004; Kujoth et al. 2005). Also the generation of a transgenic mouse in which the peroxisomal antioxidant enzyme catalase was redirected into the mitochondrial matrix resulted in an extended life span in conjunction with reduced mtDNA oxidative damage and somatic mutation levels (Schriner et al. 2005).
A synthesis of these specific mitochondrial genomic concepts provides a plausible model for the predisposition toward and development of age-related diseases. In this scenario, an individual is born with an initial mitochondrial energetic capacity based on inherited variation in nDNA- and mtDNA-encoded mitochondrial genes. Relevant nDNA variation could affect mitochondrial energetic output, antioxidant defenses, apoptotic thresholds, mitochondrial biogenesis and turnover, etc. As the individual ages, somatic mtDNA mutations arise in post-mitotic cells and stem cells (Wallace 2005b; Wallace et al. 2007b), and the individual somatic mtDNA mutations become clonally amplified within each affected cell, both post-mitotic (Muller-Hocker et al. 1993; Khrapko et al. 1999; Wang et al. 2001; Herbst et al. 2007) and stem (Michikawa et al. 1999; McDonald et al. 2006). The clonal amplification within the cell either is the result of intracellular genetic drift during the turnover of mitochondria during successive cell divisions or, alternatively, is the result of the selective amplification of the mutant mtDNA (Wallace 2005b). As the mutant mtDNAs accumulate, they progressively erode the individual cell's energetic capacity. Ultimately, cellular energetics drops below the minimal output necessary for normal cellular and tissue function and survival. This leads to a decline in organ function, loss of cell numbers, tissue failure, and an aging phenotype (Wallace 1992a,b).
While mitochondrial defects are systemic, the clinical manifestations are often organ specific. This is because different organs and tissues in the body have different needs and roles in energy homeostasis, the body's energy anatomy. Certain tissues require high levels of mitochondrial ATP, such as the retina of the eye, the cochlea of the ear, the other components of the central nervous system, the heart, the muscles, and the renal system. These organs are preferentially affected as mitochondrial energy production declines. Other tissues store energy in fat. The white adipose tissues store fat for later ATP production while the brown adipose tissues store fat for later thermal regulation. The liver is an energy homeostasis tissue, maintaining the serum glucose level within acceptable limits. The pancreatic α- and β-cells are energy-sensing tissues. They monitor calorie type and availability and send the appropriate signals, glucagon or insulin, to the energy-utilizing, storage, and homeostasis tissues (Wallace 2007).
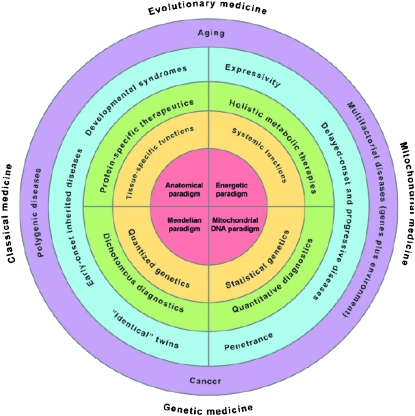
The eukaryotic cell nucleus–cytosol and mitochondrial duality now completes the human biomedical paradigm (Figure 1). The nucleus–cytosol organism specializes in elaborating new structures by changing the expression of tissue and organ-specific genes and proteins through developmental regulators. The nuclear genes are biparental, permitting recombination between the mother's and the father's alleles to maximize their capacity to generate structural diversity. The two copies for each gene also result in quantized biochemical and phenotypic manifestations of biallelic loci (+/+, +/−, and −/−), facilitating the expression of new genetic combinations. In contradistinction, the mitochondrial organism specializes in energetics. Its mtDNA genes are uniparental, present in thousands of copies per cell, and have a high sequence evolution rate. Thus, a continuous distribution of percentages of mutant mtDNAs are possible for a biallelic mtDNA locus, resulting in graded biochemical changes and a quantitative genetics advantageous for coping with graded changes in the environment. Therefore, the structural adaptation of the nDNA genes and the energetic adaptation of the mtDNA genes provide the animal with a multidimensional capacity for adapting to new environments, providing a coherent evolutionary medicine. Similarly, the quantized nDNA and statistical mtDNA genetics generate both stepwise and graded genotypic and consequent phenotypic changes, providing a coherent medical genetics. Acting together, these two systems provide all of the parameters necessary for understanding the biology and genetics of the common age-related diseases.
Figure 1.—
A new biomedical paradigm that combines the anatomical paradigm of disease and the Mendelian quantized paradigm of genetics from the eukaryotic nucleus–cytosol organism (left quadrants) with the energetic paradigm of health and the mtDNA quantitative paradigm of genetics of the eukaryotic mitochondrial organism (right quadrants). When combined, the top two quadrants provide an integrated perspective of environmental interactions (evolutionary medicine) and the lower two quadrants provide a coherent explanation of human inheritance (genetic medicine). Figure reprinted with permission (Wallace 2007).
While the bipartite eukaryotic cell provides an explanation for many of the unexplained phenomena of age-related diseases, will it be useful for making predictions that result in effective new treatments for these diseases? Unfortunately, very limited resources have been invested in understanding mitochondrial biology and genetics. Still, a few drugs have been shown to increase mitochondrial function. These include those that upregulate the transcription of nDNA-encoded mitochondrial genes through acting as agonists of the transcription factor peroxisome-proliferator-activated receptor γ (PPARγ) (e.g., rosiglitazone) (Strum et al. 2007) or by the activation through deacetylation of the mitochondrial PPARγ transcriptional coactivator-1α (PGC-1α) via agonist-activated SIRT1 (e.g., resveratrol and derivatives) (Lagouge et al. 2006; Milne et al. 2007). Mitochondrial fatty acid oxidation has been enhanced using bezafibrate (Gobin-Limballe et al. 2007) and attempts have been made to ameliorate the adverse effects of mitochondrial ROS production using combinations of natural antioxidants (lipoic acid, vitamins A and C, CoQ10, etc.) (Milgram et al. 2007) or synthetic catalytic antioxidants (MnTDEIP, EUK134, etc.) (Melov et al. 1998, 2001; Tong et al. 2007). Apopotosis is being modulated by regulating the mtPTP using cyclosporin A and its analogs (e.g., N-methyl-4-isoleucine cyclosporine) (Waldmeier et al. 2002; Irwin et al. 2003) and efforts are being directed toward regulating mitochondrial turnover by autophagy (Kim et al. 2007).
Still, the number of therapeutic approaches of relevance to the mitochondrion are severely limited by our lack of basic knowledge about mitochondrial biology. How might we jump-start the search for mitochondrially active backbone compounds to treat the age-related diseases? One promising approach might be traditional Asian herbal medications. Unlike drug development in Western pharmacology, which requires a known drug target around which the drug is developed, traditional Asian pharmaceuticals were discovered by trial and error, on the basis of what made the patient better. This trial-and-error approach to drug development is inefficient but it is paradigm blind. If mitochondrial dysfunction is as important a factor in age-related diseases as proposed in this essay, then Asian herbal medications should be as likely to have targeted a mitochondrial energetic function as a tissue-specific structural function. If so, we might be able to identify active mitochondrial backbone drugs by screening traditional Asian therapeutics for those that modulate mitochondrial function. To detect mitochondrially active compounds, we have assembled a mitochondrial cDNA expression array, the MITOCHIP, which interrogates ∼1000 genes involved in mitochondrial energy production, ROS biology, and apoptosis. We have tested the veracity of our hypothesis that Asian medications might target mitochondrial function by testing the effects of Ginkgo biloba leaf extract on mitochondrial function in cultured cells. G. biloba did indeed alter mitochondrial gene expression, appearing to modulate mitochondrially associated apoptosis (Smith et al. 2002). The association between Asian herbal medications and mitochondria has been further enhanced by the discovery that the potent antimalarial Artemisinin (quinghaosu) acts on the mitochondrion (Li et al. 2005) and the observation that, after screening 2490 compounds for the effects on mitochondrial gene expression and physiology, the Chinese herbal derivative deoxysppanone B was found to act through microtubules to increase OXPHOS and decrease mitochondrial ROS (Wagner et al. 2008).
Perhaps, then, a systematic survey of Asian herbal medications using a variety of mitochondrial functional readouts may reveal previously unrecognized mitochondrial pathways and new therapeutic strategies to manipulate them. These could then be applied to treating the common age-related diseases. If this strategy proves successful, then it may have been prescient that a major concept in the parlance of traditional Asian medicine is “chi,” which loosely translates as “vital force or energy.”
Acknowledgments
This work has been supported by National Institutes of Health grants AG24373, NS21328, AG13154, DK73691, and AG16573 and by a California Institute for Regenerative Medicine comprehensive grant (RC1-00353-1) and a Doris Duke Clinical Interfaces Award.
References
- Bai, R. K., S. M. Leal, D. Covarrubias, A. Liu and L. J. Wong, 2007. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 67 4687–4694. [DOI] [PubMed] [Google Scholar]
- Baudouin, S. V., D. Saunders, W. Tiangyou, J. L. Elson, J. Poynter et al., 2005. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet 366 2118–2121. [DOI] [PubMed] [Google Scholar]
- Booker, L. M., G. M. Habermacher, B. C. Jessie, Q. C. Sun, A. K. Baumann et al., 2006. North American white mitochondrial haplogroups in prostate and renal cancer. J. Urol. 175 468–472. [DOI] [PubMed] [Google Scholar]
- Brandon, M., P. Baldi and D. C. Wallace, 2006. Mitochondrial mutations in cancer. Oncogene 25 4647–4662. [DOI] [PubMed] [Google Scholar]
- Brown, M. D., A. Torroni, C. L. Reckord and D. C. Wallace, 1995. Phylogenetic analysis of Leber's hereditary optic neuropathy mitochondrial DNAs indicates multiple independent occurrences of the common mutations. Hum. Mutat. 6 311–325. [DOI] [PubMed] [Google Scholar]
- Brown, M. D., F. Sun and D. C. Wallace, 1997. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am. J. Hum. Genet. 60 381–387. [PMC free article] [PubMed] [Google Scholar]
- Brown, M. D., E. Starikovskaya, O. Derbeneva, S. Hosseini, J. C. Allen et al., 2002. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup. J. Hum. Genet. 110 130–138. [DOI] [PubMed] [Google Scholar]
- Burdon, R. H., 1995. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Rad. Biol. Med. 18 775–794. [DOI] [PubMed] [Google Scholar]
- Cao, L., H. Shitara, T. Horii, Y. Nagao, H. Imai et al., 2007. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat. Genet. 39 386–390. [DOI] [PubMed] [Google Scholar]
- Carrieri, G., M. Bonafe, M. De Luca, G. Rose, O. Varcasia et al., 2001. Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer's disease. Hum. Genet. 108 194–198. [DOI] [PubMed] [Google Scholar]
- Chagnon, P., M. Gee, M. Filion, Y. Robitaille, M. Belouchi et al., 1999. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am. J. Med. Genet. 85 20–30. [DOI] [PubMed] [Google Scholar]
- Crispim, D., L. H. Canani, J. L. Gross, B. Tschiedel, K. E. Souto et al., 2006. The European-specific mitochondrial cluster J/T could confer an increased risk of insulin-resistance and type 2 diabetes: an analysis of the m.4216T > C and m.4917A > G variants. Ann. Hum. Genet. 70 488–495. [DOI] [PubMed] [Google Scholar]
- Darvishi, K., S. Sharma, A. K. Bhat, E. Rai and R. N. Bamezai, 2007. Mitochondrial DNA G10398A polymorphism imparts maternal haplogroup N, a risk for breast and esophageal cancer. Cancer Lett. 249 249–255. [DOI] [PubMed] [Google Scholar]
- De Benedictis, G., G. Rose, G. Carrieri, M. De Luca, E. Falcone et al., 1999. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 13 1532–1536. [DOI] [PubMed] [Google Scholar]
- Fan, W., K. Waymire, N. Narula, P. Li, C. Rocher et al., 2008. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 319 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuku, N., K. S. Park, Y. Yamada, Y. Nishigaki, Y. M. Cho et al., 2007. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am. J. Hum. Genet. 80 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi, D., C. Marelli, A. Achilli, S. Goldwurm, G. Pezzoli et al., 2005. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson's disease in Italians. Eur. J. Hum. Genet. 13 748–752. [DOI] [PubMed] [Google Scholar]
- Giles, R. E., H. Blanc, H. M. Cann and D. C. Wallace, 1980. Maternal inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA 77 6715–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobin-Limballe, S., F. Djouadi, F. Aubey, S. Olpin, B. S. Andresen et al., 2007. Genetic basis for correction of very-long-chain acyl-coenzyme A dehydrogenase deficiency by bezafibrate in patient fibroblasts: toward a genotype-based therapy. Am. J. Hum. Genet. 81 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, Y., I. Nonaka and S. Horai, 1990. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348 651–653. [DOI] [PubMed] [Google Scholar]
- Gottlieb, E., and I. P. Tomlinson, 2005. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat. Rev. Cancer 5 857–866. [DOI] [PubMed] [Google Scholar]
- Gray, M. W., G. Burger and B. F. Lang, 1999. Mitochondrial evolution. Science 283 1476–1481. [DOI] [PubMed] [Google Scholar]
- Hansen, J. M., Y. M. Go and D. P. Jones, 2006. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu. Rev. Pharmacol. Toxicol. 46 215–234. [DOI] [PubMed] [Google Scholar]
- Herbst, A., J. W. Pak, D. McKenzie, E. Bua, M. Bassiouni et al., 2007. Accumulation of mitochondrial DNA deletion mutations in aged muscle fibers: evidence for a causal role in muscle fiber loss. J. Gerontol. A Biol. Sci. Med. Sci. 62 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, I. J., A. E. Harding and J. A. Morgan-Hughes, 1988. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331 717–719. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium, K. A. Frazer, D. G. Ballinger, D. R. Cox, D. A. Hinds et al., 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin, W. A., N. Bergamin, P. Sabatelli, C. Reggiani, A. Megighian et al., 2003. Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat. Genet. 35 367–371. [DOI] [PubMed] [Google Scholar]
- Ivanova, R., V. Lepage, D. Charron and F. Schachter, 1998. Mitochondrial genotype associated with French Caucasian centenarians. Gerontology 44 349. [DOI] [PubMed] [Google Scholar]
- Jenuth, J. P., A. C. Peterson, K. Fu and E. A. Shoubridge, 1996. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 14 146–151. [DOI] [PubMed] [Google Scholar]
- Jones, D. P., 2006. Disruption of mitochondrial redox circuitry in oxidative stress. Chem. Biol. Interact. 163 38–53. [DOI] [PubMed] [Google Scholar]
- Jones, M. M., N. Manwaring, J. J. Wang, E. Rochtchina, P. Mitchell et al., 2007. Mitochondrial DNA haplogroups and age-related maculopathy. Arch. Ophthalmol. 125 1235–1240. [DOI] [PubMed] [Google Scholar]
- Jun, A. S., M. D. Brown and D. C. Wallace, 1994. A mitochondrial DNA mutation at np 14459 of the ND6 gene associated with maternally inherited Leber's hereditary optic neuropathy and dystonia. Proc. Natl. Acad. Sci. USA 91 6206–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuno, A. A., K. Munakata, T. Nagai, S. Shimozono, M. Tanaka et al., 2006. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2 e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko, K., N. Bodyak, W. G. Thilly, N. J. van Orsouw, X. Zhang et al., 1999. Cell-by-cell scanning of whole mitochondrial genomes in aged human heart reveals a significant fraction of myocytes with clonally expanded deletions. Nucleic Acids Res. 27 2434–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusnutdinova, E., I. Gilyazova, E. Ruiz-Pesini, O. Derbeneva, R. Khusainova et al., 2008. A mitochondrial etiology of neurodegenerative diseases: evidence from Parkinson disease. Ann. NY Acad. Sci. (in press). [DOI] [PubMed]
- Kim, I., S. Rodriguez-Enriquez and J. J. Lemasters, 2007. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, T. S., 1996. The Structure of Scientific Revolutions. University of Chicago Press, Chicago.
- Kujoth, G. C., A. Hiona, T. D. Pugh, S. Someya, K. Panzer et al., 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309 481–484. [DOI] [PubMed] [Google Scholar]
- Lagouge, M., C. Argmann, Z. Gerhart-Hines, H. Meziane, C. Lerin et al., 2006. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127 1109–1122. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Lang, B. F., G. Burger, C. J. O'Kelly, R. Cedergren, G. B. Golding et al., 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387 493–497. [DOI] [PubMed] [Google Scholar]
- Li, W., W. Mo, D. Shen, L. Sun, J. Wang et al., 2005. Yeast model uncovers dual roles of mitochondria in action of artemisinin. PLoS Genet. 1 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis, L., 1981. Symbiosis in Cell Evolution: Life and Its Environment on the Early Earth. W. H. Freeman, San Francisco.
- McDonald, S. A., S. L. Preston, L. C. Greaves, S. J. Leedham, M. A. Lovell et al., 2006. Clonal expansion in the human gut: mitochondrial DNA mutations show us the way. Cell Cycle 5 808–811. [DOI] [PubMed] [Google Scholar]
- McMahon, F. J., Y. S. Chen, S. Patel, J. Kokoszka, M. D. Brown et al., 2000. Mitochondrial DNA sequence diversity in bipolar affective disorder. Am. J. Psychiatry 157 1058–1064. [DOI] [PubMed] [Google Scholar]
- Melov, S., J. A. Schneider, B. J. Day, D. Hinerfeld, P. Coskun et al., 1998. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat. Genet. 18 159–163. [DOI] [PubMed] [Google Scholar]
- Melov, S., S. R. Doctrow, J. A. Schneider, J. Haberson, M. Patel et al., 2001. Lifespan extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J. Neurosci. 21 8348–8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michikawa, Y., F. Mazzucchelli, N. Bresolin, G. Scarlato and G. Attardi, 1999. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science 286 774–779. [DOI] [PubMed] [Google Scholar]
- Milgram, N. W., J. A. Araujo, T. M. Hagen, B. V. Treadwell and B. N. Ames, 2007. Acetyl-L-carnitine and alpha-lipoic acid supplementation of aged beagle dogs improves learning in two landmark discrimination tests. FASEB J. 21 3756–3762. [DOI] [PubMed] [Google Scholar]
- Milne, J. C., P. D. Lambert, S. Schenk, D. P. Carney, J. J. Smith et al., 2007. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar, D., E. E. Ruiz-Pesini, P. Golik, V. Macaulay, A. G. Clark et al., 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA 100 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlke, K. L., A. U. Jackson, L. J. Scott, E. C. Peck, Y. D. Suh et al., 2005. Mitochondrial polymorphisms and susceptibility to type 2 diabetes-related traits in Finns. Hum. Genet. 118 245–254. [DOI] [PubMed] [Google Scholar]
- Montiel-Sosa, F., E. Ruiz-Pesini, J. A. Enriquez, A. Marcuello, C. Diez-Sanchez et al., 2006. Differences of sperm motility in mitochondrial DNA haplogroup U sublineages. Gene 368C 21–27. [DOI] [PubMed] [Google Scholar]
- Muller-Hocker, J., P. Seibel, K. Schneiderbanger and B. Kadenbach, 1993. Different in situ hybridization patterns of mitochondrial DNA in cytochrome c oxidase-deficient extraocular muscle fibres in the elderly. Virchows Arch A Pathol. Anat. Histopathol. 422 7–15. [DOI] [PubMed] [Google Scholar]
- Neckelmann, N., K. Li, R. P. Wade, R. Shuster and D. C. Wallace, 1987. cDNA sequence of a human skeletal muscle ADP/ATP translocator: lack of a leader peptide, divergence from a fibroblast translocator cDNA, and coevolution with mitochondrial DNA genes. Proc. Natl. Acad. Sci. USA 84 7580–7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi, A. K., and K. Majamaa, 2005. Mitochondrial DNA and ACTN3 genotypes in Finnish elite endurance and sprint athletes. Eur. J. Hum. Genet. 13 965–969. [DOI] [PubMed] [Google Scholar]
- Niemi, A. K., A. Hervonen, M. Hurme, P. J. Karhunen, M. Jylha et al., 2003. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum. Genet. 112 29–33. [DOI] [PubMed] [Google Scholar]
- Nishigaki, Y., Y. Yamada, N. Fuku, H. Matsuo, T. Segawa et al., 2007. Mitochondrial haplogroup N9b is protective against myocardial infarction in Japanese males. Hum. Genet. 120 827–836. [DOI] [PubMed] [Google Scholar]
- Ortiz, R. G., N. J. Newman, J. M. Shoffner, A. E. Kaufman, D. A. Koontz et al., 1993. Variable retinal and neurologic manifestations in patients harboring the mitochondrial DNA 8993 mutation. Arch. Ophthalmol. 111 1525–1530. [DOI] [PubMed] [Google Scholar]
- Raby, B. A., B. Klanderman, A. Murphy, S. Mazza, C. A. Camargo, Jr. et al., 2007. A common mitochondrial haplogroup is associated with elevated total serum IgE levels. J. Allergy Clin. Immunol. 120 351–358. [DOI] [PubMed] [Google Scholar]
- Rose, G., G. Passarino, G. Carrieri, K. Altomare, V. Greco et al., 2001. Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. Eur. J. Hum. Genet. 9 701–707. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini, E., and D. C. Wallace, 2006. Evidence for adaptive selection acting on the tRNA and rRNA genes of the human mitochondrial DNA. Hum. Mutat. 27 1072–1081. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini, E., C. Diez, A. C. Lapena, A. Perez-Martos, J. Montoya et al., 1998. Correlation of sperm motility with mitochondrial enzymatic activities. Clin. Chem. 44 1616–1620. [PubMed] [Google Scholar]
- Ruiz-Pesini, E., D. Mishmar, M. Brandon, V. Procaccio and D. C. Wallace, 2004. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303 223–226. [DOI] [PubMed] [Google Scholar]
- Saxena, R., P. I. de Bakker, K. Singer, V. Mootha, N. Burtt et al., 2006. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am. J. Hum. Genet. 79 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, R., B. F. Voight, V. Lyssenko, N. P. Burtt, P. I. de Bakker et al., 2007. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316 1331–1336. [DOI] [PubMed] [Google Scholar]
- Schaefer, A. M., R. W. Taylor, D. M. Turnbull and P. F. Chinnery, 2004. The epidemiology of mitochondrial disorders: past, present and future. Biochim. Biophys. Acta 1659 115–120. [DOI] [PubMed] [Google Scholar]
- Schaefer, A. M., R. McFarland, E. L. Blakely, L. He, R. G. Whittaker et al., 2008. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 63 35–39. [DOI] [PubMed] [Google Scholar]
- Schriner, S. E., N. J. Linford, G. M. Martin, P. Treuting, C. E. Ogburn et al., 2005. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 308 1909–1911. [DOI] [PubMed] [Google Scholar]
- Scott, L. J., K. L. Mohlke, L. L. Bonnycastle, C. J. Willer, Y. Li et al., 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoffner, J. M., M. T. Lott, A. M. Lezza, P. Seibel, S. W. Ballinger et al., 1990. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell 61 931–937. [DOI] [PubMed] [Google Scholar]
- Shoffner, J. M., M. D. Brown, A. Torroni, M. T. Lott, M. R. Cabell et al., 1993. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics 17 171–184. [DOI] [PubMed] [Google Scholar]
- Sladek, R., G. Rocheleau, J. Rung, C. Dina, L. Shen et al., 2007. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445 881–885. [DOI] [PubMed] [Google Scholar]
- Smith, J. V., A. J. Burdick, P. Golik, I. Khan, D. Wallace et al., 2002. Anti-apoptotic properties of Ginkgo biloba extract EGb 761 in differentiated PC12 cells. Cell. Mol. Biol. 48 699–707. [PubMed] [Google Scholar]
- Stewart, J. B., C. Freyer, J. L. Elson, A. Wredenberg, Z. Cansu et al., 2008. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 6 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strum, J. C., R. Shehee, D. Virley, J. Richardson, M. Mattie et al., 2007. Rosiglitazone induces mitochondrial biogenesis in mouse brain. J. Alzheimers Dis. 11 45–51. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., J. S. Gong, J. Zhang, M. Yoneda and K. Yagi, 1998. Mitochondrial genotype associated with longevity. Lancet 351 185–186. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., J. Gong, J. Zhang, Y. Yamada, H. J. Borgeld et al., 2000. Mitochondrial genotype associated with longevity and its inhibitory effect on mutagenesis. Mech. Ageing Dev. 116 65–76. [DOI] [PubMed] [Google Scholar]
- Tong, J., S. E. Schriner, D. McCleary, B. J. Day and D. C. Wallace, 2007. Life extension through neurofibromin mitochondrial regulation and antioxidant therapy for neurofibromatosis-1 in Drosophila melanogaster. Nat. Genet. 39 476–485. [DOI] [PubMed] [Google Scholar]
- Torroni, A., M. Petrozzi, L. D'Urbano, D. Sellitto, M. Zeviani et al., 1997. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am. J. Hum. Genet. 60 1107–1121. [PMC free article] [PubMed] [Google Scholar]
- Trifunovic, A., A. Wredenberg, M. Falkenberg, J. N. Spelbrink, A. T. Rovio et al., 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429 417–423. [DOI] [PubMed] [Google Scholar]
- van der Walt, J. M., K. K. Nicodemus, E. R. Martin, W. K. Scott, M. A. Nance et al., 2003. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am. J. Hum. Genet. 72 804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt, J. M., Y. A. Dementieva, E. R. Martin, W. K. Scott, K. K. Nicodemus et al., 2004. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci. Lett. 365 28–32. [DOI] [PubMed] [Google Scholar]
- Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural et al., 2001. The sequence of the human genome. Science 291 1304–1351. [DOI] [PubMed] [Google Scholar]
- Wagner, B. K., T. Kitami, T. J. Gilbert, D. Peck, A. Ramanathan et al., 2008. Large-scale chemical dissection of mitochondrial function. Nat. Biotechnol. 26 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmeier, P. C., J. J. Feldtrauer, T. Qian and J. J. Lemasters, 2002. Inhibition of the mitochondrial permeability transition by the nonimmunosuppressive cyclosporin derivative NIM811. Mol. Pharmacol. 62 22–29. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., 1992. a Diseases of the mitochondrial DNA. Annu. Rev. Biochem. 61 1175–1212. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., 1992. b Mitochondrial genetics: A paradigm for aging and degenerative diseases? Science 256 628–632. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., 1994. Mitochondrial DNA sequence variation in human evolution and disease. Proc. Natl. Acad. Sci. USA 91 8739–8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, D. C., 2005. a Mitochondria and cancer: Warburg address. Cold Spring Harbor Symp. Quant. Biol. 70 363–374. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., 2005. b A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, D. C., 2007. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76 781–821. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., J. H. Ye, S. N. Neckelmann, G. Singh, K. A. Webster et al., 1987. Sequence analysis of cDNAs for the human and bovine ATP synthase b-subunit: mitochondrial DNA genes sustain seventeen times more mutations. Curr. Genet. 12 81–90. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., G. Singh, M. T. Lott, J. A. Hodge, T. G. Schurr et al., 1988. a Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242 1427–1430. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., X. Zheng, M. T. Lott, J. M. Shoffner, J. A. Hodge et al., 1988. b Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell 55 601–610. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., M. D. Brown and M. T. Lott, 1999. Mitochondrial DNA variation in human evolution and disease. Gene 238 211–230. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., E. Ruiz-Pesini and D. Mishmar, 2003. mtDNA variation, climatic adaptation, degenerative diseases, and longevity. Cold Spring Harbor Symp. Quant. Biol. 68 479–486. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., L.-M. Chuang, P. H. Wang, Y.-C. Chang, O. Derbeneva et al., 2007. a Asian mitochondrial DNA (mtDNA) lineages are associated with altered risk of developing type 2 diabetes and metabolic syndrome. American Society of Human Genetics 57th Annual Meeting, Abstract 1549/F.
- Wallace, D. C., M. T. Lott and V. Procaccio, 2007. b Mitochondrial genes in degenerative diseases, cancer and aging, pp. 194–298 in Emery and Rimoin's Principles and Practice of Medical Genetics, Ed. 5, edited by D. L. Rimoin, J. M. Connor, R. E. Pyeritz and B. R. Korf. Churchill Livingstone Elsevier, Philadelphia.
- Wang, Y., Y. Michikawa, C. Mallidis, Y. Bai, L. Woodhouse et al., 2001. Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc. Natl. Acad. Sci. USA 98 4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini, E., M. N. Weedon, C. M. Lindgren, T. M. Frayling, K. S. Elliott et al., 2007. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]