Abstract
A change in chromosome number that is not the exact multiple of the haploid karyotype is known as aneuploidy. This condition interferes with growth and development of an organism and is a common characteristic of solid tumors. Here, we review the history of studies on aneuploidy and summarize some of its major characteristics. We will then discuss the molecular basis for the defects caused by aneuploidy and end with speculations as to whether and how aneuploidy, despite its deleterious effects on organismal and cellular fitness, contributes to tumorigenesis.
ALTERATIONS in a species' karyotype that involve changes in chromosome number are classified as either aneuploidies or polyploidies. Aneuplopidy is defined as a chromosome number that is not an exact multiple of the usually haploid number. This condition is distinct from the condition of polyploidy, which is defined as having a chromosome number that is a multiple greater than two of the monoploid number. The biological consequences of aneuploidy are also markedly different from those of polyploidy. Polyploidy is frequently found in nature; can be part of the normal physiology of plants and animals, including a few types of human cells; and generally does not lead to gross defects in the development of an organism or its physiology (Otto and Whitton 2000). In addition, duplications of an entire genome have taken place in the evolution of several groups such as plants and yeasts and may be a natural event necessary for this process (Wolfe and Shields 1997; Kellis et al. 2004; Adams and Wendel 2005). In contrast, aneuploidy frequently causes lethality and has been associated with disease, sterility, and tumor formation.
Why are entire genome duplications generally well tolerated whereas aneuploidy has severe effects on organismal growth and development? In this article, we will first review the long history of the aneuploidy field and then provide a summary of the evidence to suggest that it is imbalances in gene dosage that cause the severe defects associated with aneuploidy. Finally, we will discuss why a condition that generally interferes with cell proliferation and decreases fitness is frequently associated with the disease of uncontrolled proliferation, cancer.
THE STUDIES OF ANEUPLOIDY: A LONG TRADITION
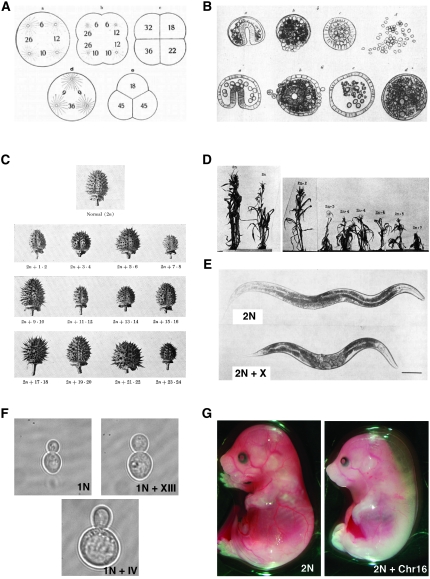
The first systematic analysis of the effects of aneuploidy on cell and organismal physiology was performed more than a century ago in the sea urchins species Paracentrotus lividus, Echinus microtuberculatus, and Strongylocentrotus purpuratus by Theodor Boveri (Boveri 1902, 1904). He examined the development of sea urchin eggs that were fertilized by two sperm and hence were triploid and, more importantly, therefore formed four (and sometimes three), rather than two, centrosomes during the first embryonic mitosis. This leads to the formation of a tetrapolar (or sometimes tripolar) spindle during the first mitosis; chromosomes are segregated to the four poles, generating a four-celled embryo skipping the two-celled stage (Figure 1A). The result of this division is massive aneuploidy. Boveri observed that the embryos that resulted from dispermic fertilizations exhibited developmental defects and died (examples of these abnormal blastulae are shown in Figure 1B). Only those doubled-fertilized embryos that by chance received the species-typical chromosomal complement (∼8%) developed into normal larvae. Boveri concluded that chromosome gain or loss leads to abnormal development and lethality. Thus, among Boveri's many seminal contributions to biology is the discovery that an abnormal number of chromosomes disrupts development.
Figure 1.—
Aneuploidy interferes with growth and development. (A) Schematics by Theodor Boveri of the first division of doubly fertilized sea urchin embryos that form four (top) or three centrosomes (bottom) during the first embryonic mitosis. The figure was reproduced from Figure 2.12-5 in Cremer (1985). (B) Drawings by Boveri showing abnormally developing dispermic sea urchin embryos. These embryos were obtained from dispermic fertilizations that were subsequently dissociated at the four-celled stage using calcium-free seawater. Each of the dissociated cells then develops into a larva. Each row represents larvae from one dissociated embryo. The first embryos at the left (top and bottom) represent relatively normal larvae, and the other larvae show varying degrees of developmental abnormalities. This drawing was reproduced from Figure 2.12-6 in Cremer (1985). (C) Drawing of the capsules of diploid D. stramonium (top) and the 12 autosomal 2n + 1 trisomies. Datura has 12 pairs of chromosomes that are referred to with numbers associated with their ends. The largest chromosome is 1·2, the next largest is 3·4, and so on. The drawings were obtained from Avery (1959). (D) Photographs of diploid maize (left) and seven 2n + 1 trisomic plants. The maize genome consists of 10 chromosomes where chromosome 1 is the smallest and 10 is the largest. The images were reproduced from McClintock (1929). (E) Photographs of an adult diploid worm (2n; top) and a worm trisomic for the X chromosome (2n + X; bottom). The images were reproduced from Hodgkin et al. (1979). (F) Haploid S. cerevisiae that is euploid (top left) and carries an extra copy of chromosome XIII (top right) or an extra copy of chromosome IV (bottom). (G) Mouse embryos that are euploid (left) or trisomic for chromosome 16 (right) at day 14.5 of development. Trisomic mouse is reduced in size and displays nucal edemas.
Twenty years later, Calvin Bridges reported the first characterization of an aneuploid fruit fly (Bridges 1921a,b). Drosophila melanogaster contains three pairs of autosomes (II, III, and IV) and a pair of sex chromosomes (females are XX, males XY). Bridges showed that a Drosophila mutant known as “Diminished” was monosomic for the smallest fourth chromosome (Bridges 1921a,b). As the name of the mutant indicates, flies lacking one copy of chromosome IV are smaller in size, are sterile, and exhibit a number of developmental abnormalities. In the 1970s, Lindsley, Sandler and co-workers created thousands of Drosophila mutant lines that either carried additional fragments of different chromosomes (segmental trisomies) or lacked different chromosomal regions (segmental monosomies) by crossing flies containing translocations between the Y chromosome and autosomes (Lindsley et al. 1972). A detailed description of their work is beyond the scope of this article but several general conclusions can be drawn. First, segmental monosomies are less well tolerated than segmental trisomies. On average, only ≤0.5% of the haploid genome is tolerated as heterozygous deficient. Second, extensive hyperploidy (that is, large parts of the genome being trisomic) is lethal whereas triploid flies are viable. Third, intermediate hyperploidy results in a set of traits that is independent of the identity of the triploid segment. These characteristics include lower viability, reduced size, and developmental deficiencies such as abnormal eye, wing, and abdominal structures. Finally, the viability of flies decreases as the size of the trisomic region increases, with the largest tolerated segment including 66% of chromosome II. Together, these results showed that aneuploidy causes a series of defects that become more pronounced as the size of the trisomic segment increases.
At about the same time as Bridges studies on chromosome IV monosomy in Drosophila were reported, Albert Blakeslee and colleagues at the Smith College Genetics Experiment Station (Northampton, MA) characterized the consequences of aneuploidy in the jimson weed, Datura stramonium (Blakeslee et al. 1920). The first spontaneous aneuploid Datura, although not recognized as such, was described in 1915 in the Botanical Garden of Storrs, Connecticut, and noted for its atypical globose fruit and unusual pattern of inheritance (Avery 1959). Subsequently, several other plants were identified that exhibited different traits but were also inherited in an unusual manner. In 1920, Blakeslee et al. showed that the gametes of these variants harbored 13 rather than 12 chromosomes and suggested that different chromosome duplications were responsible for the different phenotypes exhibited by these Datura variants. Subsequently, all possible 2n + 1 plants were isolated as spontaneously occurring variants or generated from crosses between triploid and diploid plants or selfed triploids. The 2n + 1 aneuploid plants were shown to display an array of phenotypes (an example is shown in Figure 1C). However, all 2n + 1 plants grew more slowly than euploid plants and were outcompeted quickly when co-cultivated with wild-type plants. As in Drosophila, the larger the extra chromosome, the more severe the consequences on fitness were. It appears that some plant species tolerate aneuploidy better than animals. In some instances, chromosome gains or losses are even part of their evolution. There are, however, several well-studied examples in maize, rice, and Arabidopsis where aneuploid plants grow significantly more poorly than their wild-type counterparts (McClintock 1929; Singh et al. 1996; Henry et al. 2005). Particularly, in maize and rice, the degree of poor growth correlates with the size of the additional chromosome (Figure 1D).
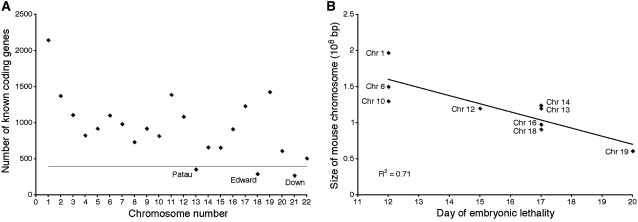
In 1959, Lejeune et al. showed that the condition known as Down's syndrome was due to the presence of an additional copy of chromosome 21 (Lejeune et al. 1959). Trisomy 21 is the only autosomal trisomy that is viable in humans. Two other trisomies, trisomy 13 (Patau's syndrome) and trisomy 18 (Edward's syndrome), can survive to birth but die within the first few months of life (Pai et al. 2003). Chromosome 13, 18, and 21 are the smallest chromosomes in humans with respect to the number of transcripts that they encode (Figure 2A). All other autosomal trisomies are embryonic lethal (as are all autosomal monosomies), again supporting the idea that the amount of additional genetic material determines the severity of the defects associated with the chromosome imbalance. Interestingly, the viable trisomies share a number of defects. Cardiovascular and craniofacial defects, developmental abnormalities of the nervous system, as well as growth retardation are observed in patients with Down's, Edward's and Patau's syndromes (Pai et al. 2003). The consequences of trisomy were also examined in the mouse. Using Robertsonian translocations, all possible trisomies were generated (Dyban and Baranov 1987). Only trisomy 19 animals develop to birth but die soon thereafter; all other trisomies die during embryogenesis. A comparison between chromosome size and the time when embryos die reveals a striking correlation (Figure 2B), indicating that in this organism, an inverse correlation also exists between the size of genome present in three copies and organismal fitness. Furthermore, in the mouse, cardiovascular, neurological, and craniofacial defects as well as growth retardation are also common among the different trisomies (see Figure 1G).
Figure 2.—
A correlation between degree of aneuploidy and organismal fitness in humans and mice. (A) The number of known human transcripts/chromosome in humans. Trisomies below the line develop to birth. Trisomies above the line are embryonic lethal. Only those chromosomes containing the least amount of transcripts survive birth. (B) In mice, survival of the embryo is inversely correlated with the size of the chromosome that is present in three copies. Linear regression analysis fits the data with an R2 of 0.71.
Caenorhabditis elegans contains five pairs of similarly sized autosomes and a pair of X chromosomes in hermaphrodites and a single X chromosome in males. In 1979, Jonathan Hodgkin, H. Robert Horvitz, and Sydney Brenner generated the first aneuploid round worm that contained three copies of the X chromosome (Figure 1E) (Hodgkin et al. 1979). These worms were viable but were morphologically abnormal and exhibited slower growth as well as subfertility (Hodgkin et al. 1979). Subsequent efforts led to the generation of worms trisomic for chromosome IV (Sigurdson et al. 1986). These animals also are subfertile and exhibit morphological defects. Other trisomies have not been recovered from screens for duplications, suggesting that they are lethal. Monosomies have never been recovered in any mutant or deficiency screen either. This finding, together with the observation that worms carrying large autosomal deficiencies are unhealthy and exhibit numerous morphological defects, implies that monosomies are lethal in C. elegans. However, large free duplications and deficiencies ranging from 2.4 to 11.6 Mbp (2.4–11.9% of the genome) and from 1.1 to 4.6 Mbp (1.1–4.7%), respectively, have been obtained, indicating that C. elegans is able to tolerate some levels of genomic imbalance (Hodgkin 2005).
The first aneuploid Saccharomyces cerevisiae strains were generated by Mortimer and Hawthorne in 1966 for mapping purposes (Mortimer and Hawthorne 1966). In 1970, Parry and Cox generated a series of disomic strains by sporulating triploid yeast strains (Parry and Cox 1970). They recovered a large number of offspring and identified strains disomic for at least five chromosomes, leading them to suggest that aneuploidy is well tolerated in yeast. Subsequent studies employed disomic and monosomic yeast strains for the purpose of measuring chromosome loss rates (Hartwell et al. 1982), but a systematic characterization of all disomic strains was not conducted until recently. We analyzed 13 of 16 possible 1n + 1 yeast strains and found them to be impaired in proliferation and sensitive to a number of conditions interfering with protein synthesis and turnover (Torres et al. 2007; Figure 1F). The genome size of Schizosaccharomyces pombe is similar to that of S. cerevisiae but is contained on only three chromosomes. Yanagida and coworkers showed in 1985 that only strains disomic for the smallest chromosome (chromosome III) are viable but severely growth retarded (Niwa and Yanagida 1985). More recent studies on whole chromosomal aneuploidy as well as segmental aneuploidy indicate that aneuploidy in S. pombe hampers cell proliferation (Niwa et al. 2006).
In summary, aneuploidy causes developmental abnormalities and reduces organismal fitness in all species where this condition was examined. It is also clear that loss of genetic information due to monosomies is less well tolerated than the gain of genetic information due to trisomies or, in the case of haploid organisms, disomies. The molecular bases for the reduced fitness are discussed in the following section.
WHY DOES ANEUPLOIDY REDUCE ORGANISMAL FITNESS?
To understand the basis for the reduced fitness caused by aneuploidy, we must first ask whether the effects of chromosome gains and losses are the same. Many studies indicate that the answer to this question is “no.” Imbalances in protein stoichiometry are likely to be the cause of defects in organisms with extra chromosomes. In the case of chromosome losses, two reasons are likely to be responsible for the defects associated with this condition: (1) a reduction in protein activity due to the reduction in gene dosage, which is known as haplo-insufficiency, and (2) as in the case of chromosome gains, protein stoichiometry imbalances. We will summarize the evidence supporting this notion with a focus on the effects of protein imbalances on organismal fitness. Furthermore, we propose that it is the additive effects of many protein stoichiometry imbalances that are responsible for many of the cellular defects and some of the organismal and developmental abnormalities associated with aneuploidy.
Defects caused by chromosome loss—reduction in gene dosage:
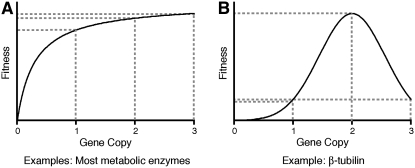
Organisms have much less tolerance for chromosome losses than for gains. The likely reason for this is that, in addition to protein imbalances caused by an incorrect karyotype (discussed below), a reduction in net protein levels occurs. Burns and Kacser proposed that a reduction in the dosage of genes encoding most metabolic enzymes will not affect fitness because the enzyme activity is dictated by the flux of the pathway and a change of 0.5-fold in enzyme concentration will have minimal effects (Figure 4A, left; Kacser and Burns 1981; see also Veitia 2002). Nevertheless, budding yeast encodes 184 genes (3% of the yeast genome), which, when in the heterozygous state, lead to a reduction in fitness due to decreases in protein levels and not protein imbalances (Deutschbauer et al. 2005). Deutschbauer et al. probed the yeast knock-out collection for genes that, when in the heterozygous state, lead to a reduction in proliferative capacity. They then reasoned that if protein imbalances were responsible for the decreased fitness in these heterozygous strains, overexpression of the gene should also lead to a reduction in proliferation capacity. Overexpression of 13 of the 16 genes identified as haplo-insufficient, however, did not lead to a detectable reduction in fitness, indicating that it is not imbalances in protein stoichiometry but reduced protein levels that are responsible for this decrease in organismal fitness. In Drosophila, loss of one copy of 64 out of a total of 79 cytoplasmic ribosomal proteins leads to the “Minute” phenotype, with flies being small and exhibiting poor fertility and viability (Marygold et al. 2007). In humans, several dozens of genes have been identified which, when present in only one copy, result in disease (Fisher and Scambler 1994).
Figure 4.—
Two of the simplest possible effects of changes in gene copy number on organismal fitness. (A) Changes in copy number of genes that encode for enzymes should follow the Kacser and Burns hypothesis. That is, the effects on fitness are dictated by the activity of the enzymatic pathway rather than by the enzyme concentration. On the other hand (B), changes in copy number of structural genes such β-tubulin have much more deleterious effect on fitness.
Defects caused by chromosome gains or losses—protein stoichiometry imbalances:
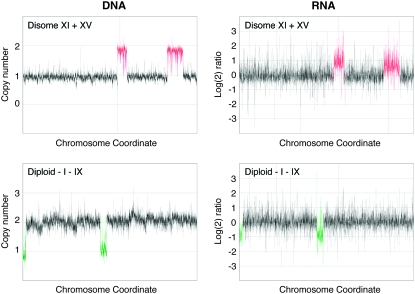
Dosage compensation at the transcriptional level occurs for many sex chromosomes (Straub and Becker 2007), but autosomes do not appear to be regulated in this fashion. Although dosage compensation at the transcriptional level occurs for individual genes (Birchler et al. 1990; Guo and Birchler 1994), microarray-based expression analyses indicate that, overall, transcript levels correlate with gene copy number in aneuploids. Haploid and diploid yeast strains containing an extra chromosome show a corresponding increase in transcript levels (Torres et al. 2007; Figure 3); diploid yeast strains lacking a chromosome show a matching decrease (Figure 3). Transcript levels of genes encoded on chromosome 21 parallel the increase in gene copy number in patients with Down's syndrome (Mao et al. 2003). Similar results were obtained in mice with a partial trisomy for chromosome 16 or 17 (Kahlem et al. 2004; Lyle et al. 2004; Vacik et al. 2005). In addition, changes in gene expression appear to correlate with changes in gene copy number in aneuploid cancer cells (Pollack et al. 2002; Tsafrir et al. 2006). If we assume that translation also occurs according to gene copy number—there is some evidence that this is the case at least for some proteins in yeast and trisomic mouse embryos (Klose and Putz 1983; Torres et al. 2007)—aneuploidy would result in deviations from the normal stoichiometry of protein complex subunits. These changes in intracellular protein composition would then cause defects in many cellular processes, ultimately leading to developmental defects and a decrease in organismal fitness. This theory is known as the balance theory and can explain the defects associated with aneuploidy due to chromosome gain and, in part, those due to chromosome loss (Veitia 2002; Papp et al. 2003).
Figure 3.—
Changes in RNA expression correlate with changes in DNA copy number in aneuploid S. cerevisiae. Each box represents the genome of an aneuploid yeast strain, with data points from the microarray analysis ordered according to their chromosomal coordinates. The left end of the left arm of chromosome I is shown at the left end of the graph, and the right end of the right arm of chromosome XVI is on the right end. DNA copy number and the log2 ratio of changes in gene expression of haploid yeast cells disomic for chromosomes XI and XV (top) and diploid cells mosomic for chromosome I and IX (bottom) are shown as normalized to the wild type. Data are courtesy of E. Torres and M. Dunham.
Defects caused by chromosome gains—additive effects of imbalance:
While haplo-insufficiency is not uncommon in eukaryotic genomes, the number of genes, for which a 50% increase in gene dosage leads to a severe defect, is likely to be small. Proteins that function exclusively in protein complexes are expected to fall into this class (Figure 4B). The gene encoding β-tubulin is one such example. In budding yeast, the presence of an extra copy of the β-tubulin-encoding genes in cells is lethal, and cells survive only when they harbor an additional copy of chromosome 13, on which the α-tubulin genes are located (Katz et al. 1990). Conversely, in diploid yeast cells, heterozygous deletion of the gene encoding α-tubulin results in haplo-insufficiency due to increased levels of β-tubulin (Schatz et al. 1988). In humans, duplication of a limited number of genes has been found to be associated with disease. For example, duplication of the SNCA gene, which encodes α-synuclein, leads to early onset Alzheimer's disease (reviewed in Farrer 2006); duplication of PMP22 leads to Charcot-Marie-Tooth 1A (CMT1A) neuropathy (reviewed in Hanemann and Muller 1998); and duplication of a fragment of the short arm of chromosome 7 (7p13-p22.1) causes severe developmental abnormalities (Papadopoulou et al. 2006).
Increasing the gene dosage of most genes, however, does not affect the fitness of an organism in a notable way. Does this mean that the imbalance of only a select number of genes is responsible for the severe phenotypes associated with chromosome gains? We propose that, while single genes certainly are responsible for some of the phenotypes (i.e., for developmental abnormalities unique to the gain of a certain chromosome), many of the traits, particularly those shared by different aneuploids (i.e., cellular defects), are due to additive effects of twofold increases of a large number of proteins leading to many imbalances. Together, they contribute to the significant decrease in fitness observed in aneuploid organisms. Predictions of this “additive effects of imbalance theory” are that:
Introduction of noncoding DNA, or DNA that produces proteins that do not interact with the host proteome, should not affect the fitness of an organism as severely as the introduction of additional DNA from the same species.
Organismal fitness should correlate with the fraction of the genome that is imbalanced.
In budding yeast, both these predictions appear to be met: (1) introduction of large chromosome-size amounts of mouse or human DNA does not significantly affect cell growth and proliferation in this organism (Torres et al. 2007) and (2) the phenotypes shared by aneuploid yeast strains increase in severity with the amount of additional DNA present in cells. Furthermore, the comparison between haploid yeast strains carrying an extra chromosome (1n + 1) and diploid yeast strains carrying an extra chromosome (2n + 1) showed that an increase in ploidy buffers the genetic imbalances of aneuploids with respect to sensitivities to compounds interfering with protein synthesis and folding because the ratio of imbalanced gene copy number is decreased (2/1 in disomic strains compared to 3/2 in trisomic strains; our unpublished observations). Additive effects of protein stoichiometry imbalances not only explain the discrepancy between the severity of the phenotype associated with chromosome gains and the small number of genes known to cause severe phenotypes when their copy number is increased by 50%, but also provide an explanation for the striking correlation between the amount of additional DNA present in aneuploids and the severity of the phenotype observed in these organisms.
In summary, the reduction in fitness of aneuploid organisms is likely due to a number of reasons, but protein stoichiometry imbalance is probably the major cause of defects associated with chromosome gains, and protein stoichiometry imbalances and reduction in gene dosage of a select number of genes are the major cause of defects associated with chromosome losses. We should, however, note that other factors could also contribute. Cells must replicate and maintain the extra chromosomal material, creating a higher demand on the DNA replication machinery and chromosome maintenance and segregation pathways. In fact, these pathways appear to be a cause of the reduced fitness of budding yeast strains that are polyploid (Storchova et al. 2006). Because the additional chromosomes in aneuploid cells are active, it is also possible that the transcription and translation machinery become rate limiting, which could also lead to a reduction in organismal fitness. Consistent with this idea is the observation that aneuploid yeast strains exhibit increased sensitivity to conditions that interfere with transcription and protein synthesis (Torres et al. 2007).
DO ORGANISMS RESPOND TO ANEUPLOIDY?
A key question that arises from the idea that many protein stoichiometry imbalances are at the heart of the aneuploidy-induced defects is whether cells respond to this state of imbalance and attempt to rectify it. This question has been addressed systematically only in budding yeast and only for strains that experienced chromosome gains. However, in this specific circumstance the answer appears to be “yes.” Dosage compensation seems to occur at the level of protein and not transcript abundance.
We compared the steady-state levels of 16 proteins whose transcripts were upregulated according to gene copy number. Thirteen of the 16 proteins did not appear to exhibit a corresponding increase in protein levels. Analysis of several proteins, whose encoding genes are located on chromosome 21 in cells of trisomy 21 patients, revealed similar results (Cheon et al. 2003a,b,c,d, 2007). Protein levels were not elevated in accordance with amounts of message. These results indicate that cells attempt to establish accurate protein stoichiometries in aneuploid cells either by downregulating translation or, more likely, by degrading the excess protein. There are several examples for the latter mode of regulation. α-Tubulin and histones are degraded if they are overexpressed or if their binding partners are missing (Gunjan and Verreault 2003; Lacefield et al. 2006). Perhaps protein complexes whose individual subunits serve different functions (i.e., one subunit contains the catalytic activity of the complex and others serve as substrate receptors) are controlled by this type of mechanism. Protein complexes that could fall into this class are the ribosome, the proteasome, and microtubules. The idea that dosage compensation occurs at the level of protein abundance and is at least in part mediated by protein degradation is also consistent with the finding in yeast that strains carrying extra chromosomes are sensitive to drugs that interfere with proteasome function (Torres et al. 2007) and with the observation that cancer cells, which are frequently aneuploid, depend on high proteasome activity since they are more sensitive to proteasome inhibitors than euploid cells (reviewed in Whitesell and Lindquist 2005).
Yeast cells carrying an extra chromosome exhibit other defects that could reflect a response to aneuploidy. These appear to be independent of the identity of the additional chromosome but dependent on the amount of additional yeast DNA present in cells (Torres et al. 2007). Aneuploid yeast cells show a transcriptional response similar to that described in yeast cells grown under many different stress conditions [called the environmental stress response (Gasch et al. 2000)], sensitivity to high temperature, and a delay in the G1 stage of the cell cycle. It is tempting to speculate that these traits also reflect the aneuploid cell's effort to reestablish protein stoichiometry. Slowing down cell proliferation in G1 and mounting a stress response that involves chaperones to shield the cell from adverse effects of unassembled protein complexes could all be part of a response that is not unlike a stress response and whose role is to promote survival when protein stoichiometries are imbalanced. The molecular mechanisms underlying this potential stress response and slowing of cell proliferation in G1, however, remain to be characterized.
Whether mechanisms exist that compensate for the loss of proteins due to monosomy is not known. In monosomic yeast cells, overall, transcription correlates with gene copy number, indicating that dosage compensation does not occur at the transcriptional level (Figure 3). It is possible that compensation occurs at the levels of protein translation. On a chromosome-wide level, this would, however, require substantial translational control. We speculate that eliminating proteins that are in excess is probably easier than upregulating protein production to compensate for a deficiency. An inability to compensate for gene losses could also explain why chromosome gains are better tolerated than chromosome losses.
WHY ARE MOST TUMORS ANEUPLOID?
Theodor Boveri was the first to raise the possibility of a link between aneuploidy and tumor formation. He noted that the disorganized cell clusters that accumulated in the center of aneuploid blastocysts resembled “Geschwuelste,” the undefined cell masses that are now known as tumors, and wondered whether multipolar mitoses and hence aneuploidy was the cause of tumor formation in humans (Boveri 1902). Boveri speculated that some aneuploid cells could proliferate better than wild-type cells or in ways that wild-type cells could not. Today, we know that aneuploidy interferes with growth and development in most, if not all, organisms. However, it is also clear that most solid tumors are aneuploid. If aneuploidy is so deleterious, why, then, are most solid tumors aneuploid?
Aneuploidy could be a late event in tumorigenesis, caused by the inactivation of the p53 pathway. Inactivation of p53 results in tetraploidization (Bunz et al. 2002), a state that might facilitate aneuploidy. Thus, aneuploidy would be a consequence rather than a cause of tumorigenesis and thus would not contribute to tumor development. Several lines of evidence indicate that aneuploidy suppresses rather than promotes tumorigenesis. First, individuals carrying an extra copy of chromosome 21 have a 50% lower probability of developing solid tumors than individuals with the correct chromosome number (Hasle et al. 2000; Satge et al. 2003). Second, mice carrying segmental trisomies exhibit a reduced incidence of neoplasia in the sensitized apcMin genetic background (Sussan et al. 2008). Third, a mouse model in which low-level aneuploidy was induced by interfering with the chromosome segregation machinery prevented tumor formation in most tissues and caused tumor formation only very late in the others (Weaver et al. 2007). Fourth, in humans, adenomas with mild-to-moderate dysplasia or atypical ductal hyperplastic lesions exhibit only low-grade aneuploidy (Bomme et al. 1998, 2001; Larson et al. 2006), indicating that tetraploidy and high-grade aneuploidy are not early occurrences in tumor formation. Finally, although cytogenetic analyses have identified many structural and numerical aberrations in solid tumors, relatively few are shared even among specific types of solid tumors or have been shown to contribute to tumor formation (reviewed in Albertson et al. 2003).
A few findings, however, argue for aneuploidy being an early and causative event during tumorigenesis. The observation that low-grade, small adenomas and atypical ductal hyperplastic cells do show a low degree of loss of heterozygosity (Bomme et al. 1998, 2001; Shih et al. 2001; Larson et al. 2006) of course can also be viewed as discrete chromosome gains playing a causative role early during tumorigenesis. Trisomy 21 patients, although less likely to develop solid tumors, are more prone to developing childhood leukemias (Hasle et al. 2000). Furthermore, even though tumors form late in mice carrying low-level aneuploidy-inducing mutations, they do arise with a statistically significant increased frequency in some tissues (Weaver et al. 2007). Thus, it is clear that under specific circumstances, i.e., in specific tissues or during certain developmental stages, an additional copy of specific chromosomes may accelerate some aspects of tumorigenesis. For example, gaining an additional copy of an oncogene or losing a copy of a tumor suppressor gene may promote inappropriate cell proliferation in differentiated nondividing tissues. In what follows, we propose a speculative model that could explain how aneuploidy, despite antagonizing proliferation, could promote tumorigenesis (Figure 5).
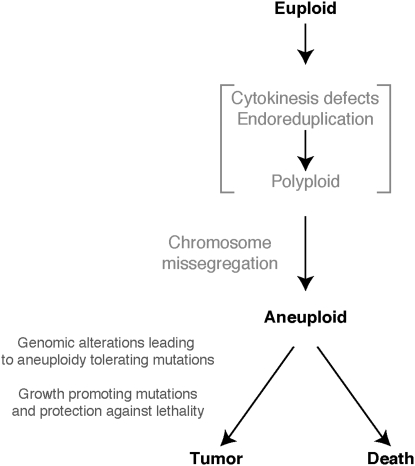
Figure 5.—
A model for how aneuploidy could promote tumorigenesis. See text for details.
During the lifetime of a multicellular organism, chromosome mis-segregation events will generate aneuploid cells at an approximate rate of 1 in 105 divisions (Rosenstraus and Chasin 1978; Hartwell et al. 1982). This process could occur in a euploid cell or in a polyploid intermediate that was precipitated by a cytokinesis defect or endoreduplication (Figure 5). Most of the aneuploid cells will die or proliferate so slowly that they are outcompeted by normal cells. However, if they have acquired proliferative potential under conditions in which the surrounding euploid cells in the tissue do not divide (i.e., through loss of growth or G1–S-phase transition control), even slowly proliferating aneuploid cells will have an advantage over euploid cells.
We further propose that the very events that cause aneuploid cells to proliferate slowly, and the stress caused by aneuploidy, are also responsible for aneuploid cells embarking on the path of tumorigenesis. The stress caused by aneuploidy precipitates an increase in mutation rate, in gene amplification and/or increased genomic instability. Precedence exists for all these scenarios. Several studies in Bacillus subtilis and Escherichia coli have shown that stress increases the mutation rate by promoting the utilization of error-prone DNA polymerases or by downregulation of the mismatch repair machinery (Sung et al. 2003; Ponder et al. 2005). In mouse fibroblasts, hypoxic stress causes decreased expression of the mismatch repair factor MLH1 and induces instability of a (CA)29 dinucleotide repeat (Mihaylova et al. 2003). Gene amplifications can also be induced by stress. In E. coli, amplification of the ampC genes promotes resistance to antibiotics (Edlund and Normark 1981). A partial aneuploid strain of Candida albicans that includes two copies of the left arm of chromosome 5 exhibits azole resistance (Selmecki et al. 2006). In yeast, growth under glucose limitation promotes amplification of the high-affinity glucose transporters (Dunham et al. 2002). Aneuploidy of course could also be a sign of genomic instability. This instability would further aid in the evolution of the tumor toward a state of high proliferative capacity.
We predict that among the first mutations that occur in aneuploid cells are those that allow cells to tolerate the adverse effects of aneuploidy. These combined with growth and proliferation-promoting genomic changes, such as amplification of oncogenes and loss of tumor suppressor genes, now promote growth and eventually lead to the selection of tumor cells with high proliferative capacity. Aneuploidy could also give tumors the ability to fine-tune gene dosages to promote growth in a particular environment within the body. Ciliates such as Tetrahymena utilize this strategy for generating cells with a range of gene dosages to create few cells that will survive under different kinds of stress (Yao and Chao 2005). We further propose that aneuploidy not only promotes genomic changes and provides mechanisms of adaptation in a particular environment, but also gives another important advantage to these evolving cells: it shields them from lethal mutations. By providing multiple copies of essential or haplo-insufficient genes, aneuploidy could protect cells from lethal events. Thus, we speculate that, in a rather counter-intuitive manner, the proliferation-inhibiting, stress-inducing properties of aneuploidy are the reasons why aneuploidy promotes tumor growth and development.
However, irrespective of whether aneuploidy causes tumorigenesis or whether it is an accidental by-product of the process, it is clear that aneuploidy interferes with cellular growth and proliferation. Single-celled organisms, such as fission and budding yeast, proliferate more slowly when they are aneuploid. Studies on trisomic mouse cell lines show that the presence of an extra chromosome hampers cell proliferation, and in some instances, immortalization (B. R. Williams, V. R. Prabhu, K. E. Hunter and A. Amon, personal communications). In cultures of the human cell lines RPE-1 and HCT116, nondiploid cells are also outcompeted by their euploid siblings (Thompson and Compton 2008). Thus, for an aneuploid tumor to reach high proliferative potential, it must acquire mutations that allow cells to tolerate the adverse effects of aneuploidy. Identifying such mutations will provide critical insights into tumor progression.
ANEUPLOIDY: CANCER'S ACHILLES HEAL?
Given that most solid tumors are aneuploid, the cellular consequences of aneuploidy could provide a novel target in cancer therapy. Aneuploid cells attempt to restore protein stoichiometries to the euploid state. This observation predicts that aneuploid cancer cells rely more heavily on the mechanisms that are employed to clear cells from excess protein or that shield cells from proteins that are not bound to their partners. Consistent with this idea are the observations that cancer cells exhibit increased sensitivity to proteasome inhibitors and the Hsp90 chaperone inhibitor geldanamycin (Whitesell and Lindquist 2005). Furthermore, inactivation of the transcription factor HSF1, which is required for the heat-shock response in mice, leads to a significant reduction in tumor incidence (Dai et al. 2007). Developing methodologies for detecting aneuploid cells in vivo could allow for early detection of cancerous lesions. Characterizing the phenotypes associated with aneuploidy in human cells as well as identifying small molecules that specifically kill aneuploid cells will provide new avenues in the treatment of cancer.
Acknowledgments
We are grateful to Steve Bell, Maitreya Dunham, David Housman, Doug Koshland, Frank Solomon, Alan Spradling, and members of the Amon lab for suggestions and their critical reading of this manuscript. Work in the Amon lab was supported by a grant from the National Institutes of Health grant GM56800. E.M.T. was supported by an Anna Fuller postdoctoral Fellowship. A.A. is also an Investigator of the Howard Hughes Medical Institute.
References
- Adams, K. L., and J. F. Wendel, 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8 135–141. [DOI] [PubMed] [Google Scholar]
- Albertson, D. G., C. Collins, F. McCormick and J. W. Gray, 2003. Chromosome aberrations in solid tumors. Nat. Genet. 34 369–376. [DOI] [PubMed] [Google Scholar]
- Avery, A. G., 1959. Blakeslee: The Genus Datura. Ronald Press, New York.
- Birchler, J. A., J. C. Hiebert and K. Paigen, 1990. Analysis of autosomal dosage compensation involving the alcohol dehydrogenase locus in Drosophila melanogaster. Genetics 124 679–686. [PMC free article] [PubMed] [Google Scholar]
- Blakeslee, A. F., J. Belling and M. E. Farnham, 1920. Chromosomal duplication and Mendelian phenomena in Datura mutants. Science 52 388–390. [DOI] [PubMed] [Google Scholar]
- Bomme, L., G. Bardi, N. Pandis, C. Fenger, O. Kronborg et al., 1998. Cytogenetic analysis of colorectal adenomas: karyotypic comparisons of synchronous tumors. Cancer Genet. Cytogenet. 106 66–71. [DOI] [PubMed] [Google Scholar]
- Bomme, L., R. A. Lothe, G. Bardi, C. Fenger, O. Kronborg et al., 2001. Assessments of clonal composition of colorectal adenomas by FISH analysis of chromosomes 1, 7, 13 and 20. Int. J. Cancer 92 816–823. [DOI] [PubMed] [Google Scholar]
- Boveri, T., 1902. Über mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. Verhandlungen der physikalisch-medizinischen Gesellschaft zu Würzburg. Neu Folge 35 67–90. [Google Scholar]
- Boveri, T., 1904. Ergebnisse über die Konstitution der Chromatischen Substanz des Zellkerns. Gustav Fischer, Jena, Germany.
- Bridges, C. B., 1921. a Genetical and cytological proof of non-disjunction of the fourth chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 7 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, C. B., 1921. b Proof of non-disjunction for the fourth chromosome of Drosophila melanogaster. Science 53 308. [DOI] [PubMed] [Google Scholar]
- Bunz, F., C. Fauth, M. R. Speicher, A. Dutriaux, J. M. Sedivy et al., 2002. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 62 1129–1133. [PubMed] [Google Scholar]
- Cheon, M. S., M. Bajo, S. H. Kim, J. O. Claudio, A. K. Stewart et al., 2003. a Protein levels of genes encoded on chromosome 21 in fetal Down syndrome brain: challenging the gene dosage effect hypothesis (Part II). Amino Acids 24 119–125. [DOI] [PubMed] [Google Scholar]
- Cheon, M. S., S. H. Kim, V. Ovod, N. Kopitar Jerala, J. I. Morgan et al., 2003. b Protein levels of genes encoded on chromosome 21 in fetal Down syndrome brain: challenging the gene dosage effect hypothesis (Part III). Amino Acids 24 127–134. [DOI] [PubMed] [Google Scholar]
- Cheon, M. S., S. H. Kim, M. L. Yaspo, F. Blasi, Y. Aoki et al., 2003. c Protein levels of genes encoded on chromosome 21 in fetal Down syndrome brain: challenging the gene dosage effect hypothesis (Part I). Amino Acids 24 111–117. [DOI] [PubMed] [Google Scholar]
- Cheon, M. S., K. S. Shim, S. H. Kim, A. Hara and G. Lubec, 2003. d Protein levels of genes encoded on chromosome 21 in fetal Down syndrome brain: challenging the gene dosage effect hypothesis (Part IV). Amino Acids 25 41–47. [DOI] [PubMed] [Google Scholar]
- Cheon, M. S., M. Dierssen, S. H. Kim and G. Lubec, 2007. Protein expression of BACE1, BACE2 and APP in Down syndrome brains. Amino Acids. http://www.springerlink.com/content/x5q6v728k3778n62/. [DOI] [PubMed]
- Cremer, T., 1985. Von der Zellenlehre zur Chromosomentheorie. Springer-Verlag, Berlin; Heidelberg, Germany; New York; Tokyo.
- Dai, C., L. Whitesell, A. B. Rogers and S. Lindquist, 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer, A. M., D. F. Jaramillo, M. Proctor, J. Kumm, M. E. Hillenmeyer et al., 2005. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown et al., 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyban, A. P., and V. S. Baranov, 1987. Cytogenetics of Mammalian Embryonic Development. Clarendon Press/Oxford University Press, Oxford/New York.
- Edlund, T., and S. Normark, 1981. Recombination between short DNA homologies causes tandem duplication. Nature 292 269–271. [DOI] [PubMed] [Google Scholar]
- Farrer, M. J., 2006. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat. Rev. Genet. 7 306–318. [DOI] [PubMed] [Google Scholar]
- Fisher, E., and P. Scambler, 1994. Human haploinsufficiency—one for sorrow, two for joy. Nat. Genet. 7 5–7. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen et al., 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan, A., and A. Verreault, 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115 537–549. [DOI] [PubMed] [Google Scholar]
- Guo, M., and J. A. Birchler, 1994. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266 1999–2002. [DOI] [PubMed] [Google Scholar]
- Hanemann, C. O., and H. W. Muller, 1998. Pathogenesis of Charcot-Marie-Tooth 1A (CMT1A) neuropathy. Trends Neurosci. 21 282–286. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. H., S. K. Dutcher, J. S. Wood and B. Garvik, 1982. The fidelity of mitotic chromosome reproduction in S. cerevisiae. Rec. Adv. Yeast Mol. Biol. 1 28–38. [Google Scholar]
- Hasle, H., I. H. Clemmensen and M. Mikkelsen, 2000. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet 355 165–169. [DOI] [PubMed] [Google Scholar]
- Henry, I. M., B. P. Dilkes, K. Young, B. Watson, H. Wu et al., 2005. Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170 1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 2005. Karyotype, ploidy, and gene dosage. WormBook, pp. 1–9. [DOI] [PMC free article] [PubMed]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser, H., and J. A. Burns, 1981. The molecular basis of dominance. Genetics 97 639–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlem, P., M. Sultan, R. Herwig, M. Steinfath, D. Balzereit et al., 2004. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res. 14 1258–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, W., B. Weinstein and F. Solomon, 1990. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol. Cell. Biol. 10 5286–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., B. W. Birren and E. S. Lander, 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428 617–624. [DOI] [PubMed] [Google Scholar]
- Klose, J., and B. Putz, 1983. Analysis of two-dimensional protein patterns from mouse embryos with different trisomies. Proc. Natl. Acad. Sci. USA 80 3753–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield, S., M. Magendantz and F. Solomon, 2006. Consequences of defective tubulin folding on heterodimer levels, mitosis and spindle morphology in Saccharomyces cerevisiae. Genetics 173 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, P. S., A. de las Morenas, S. R. Cerda, S. R. Bennett, L. A. Cupples et al., 2006. Quantitative analysis of allele imbalance supports atypical ductal hyperplasia lesions as direct breast cancer precursors. J. Pathol. 209 307–316. [DOI] [PubMed] [Google Scholar]
- Lejeune, J., M. Gautier and R. Turpin, 1959. Study of somatic chromosomes from 9 mongoloid children. C. R. Hebd. Seances Acad. Sci. 248 1721–1722. [PubMed] [Google Scholar]
- Lindsley, D. L., L. Sandler, B. S. Baker, A. T. Carpenter, R. E. Denell et al., 1972. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71 157–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle, R., C. Gehrig, C. Neergaard-Henrichsen, S. Deutsch and S. E. Antonarakis, 2004. Gene expression from the aneuploid chromosome in a trisomy mouse model of Down syndrome. Genome Res. 14 1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, R., C. L. Zielke, H. R. Zielke and J. Pevsner, 2003. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics 81 457–467. [DOI] [PubMed] [Google Scholar]
- Marygold, S. J., J. Roote, G. Reuter, A. Lambertsson, M. Ashburner et al., 2007. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8 R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1929. A cytological and genetical study of triploid maize. Genetics 14 180–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova, V. T., R. S. Bindra, J. Yuan, D. Campisi, L. Narayanan et al., 2003. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol. Cell. Biol. 23 3265–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer, R. K., and D. C. Hawthorne, 1966. Genetic mapping in Saccharomyces. Genetics 53 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, O., and M. Yanagida, 1985. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr. Genet. 9 463–470. [Google Scholar]
- Niwa, O., Y. Tange and A. Kurabayashi, 2006. Growth arrest and chromosome instability in aneuploid yeast. Yeast 23 937–950. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and J. Whitton, 2000. Polyploid incidence and evolution. Annu. Rev. Genet. 34 401–437. [DOI] [PubMed] [Google Scholar]
- Pai, G. S., R. C. Lewandowski and D. S. Borgaonkar, 2003. Handbook of Chromosomal Syndromes. John Wiley & Sons, New York.
- Papadopoulou, E., S. Sifakis, C. Sarri, J. Gyftodimou, T. Liehr et al., 2006. A report of pure 7p duplication syndrome and review of the literature. Am. J. Med. Genet. A 140 2802–2806. [DOI] [PubMed] [Google Scholar]
- Papp, B., C. Pal and L. D. Hurst, 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424 194–197. [DOI] [PubMed] [Google Scholar]
- Parry, E. M., and B. S. Cox, 1970. The tolerance of aneuploidy in yeast. Genet. Res. 16 333–340. [DOI] [PubMed] [Google Scholar]
- Pollack, J. R., T. Sorlie, C. M. Perou, C. A. Rees, S. S. Jeffrey et al., 2002. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. USA 99 12963–12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder, R. G., N. C. Fonville and S. M. Rosenberg, 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19 791–804. [DOI] [PubMed] [Google Scholar]
- Rosenstraus, M. J., and L. A. Chasin, 1978. Separation of linked markers in Chinese hamster cell hybrids: mitotic recombination is not involved. Genetics 90 735–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satge, D., A. J. Sasco and B. Lacour, 2003. Are solid tumours different in children with Down's syndrome? Int. J. Cancer 106 297–298. [DOI] [PubMed] [Google Scholar]
- Schatz, P. J., F. Solomon and D. Botstein, 1988. Isolation and characterization of conditional-lethal mutations in the TUB1 α-tubulin gene of the yeast Saccharomyces cerevisiae. Genetics 120 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki, A., A. Forche and J. Berman, 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, I. M., T. L. Wang, G. Traverso, K. Romans, S. R. Hamilton et al., 2001. Top-down morphogenesis of colorectal tumors. Proc. Natl. Acad. Sci. USA 98 2640–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson, D. C., R. K. Herman, C. A. Horton, C. K. Kari and S. E. Pratt, 1986. An X-autosome fusion chromosome of Caenorhabditis elegans. Mol. Gen. Genet. 202 212–218. [DOI] [PubMed] [Google Scholar]
- Singh, K., D. S. Multani and G. S. Khush, 1996. Secondary trisomics and telotrisomics of rice: origin, characterization, and use in determining the orientation of chromosome map. Genetics 143 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo, R., E. Hernando, E. Diaz-Rodriguez, J. Teruya-Feldstein, C. Cordon-Cardo et al., 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova, Z., A. Breneman, J. Cande, J. Dunn, K. Burbank et al., 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443 541–547. [DOI] [PubMed] [Google Scholar]
- Straub, T., and P. B. Becker, 2007. Dosage compensation: the beginning and end of generalization. Nat. Rev. Genet. 8 47–57. [DOI] [PubMed] [Google Scholar]
- Sung, H. M., G. Yeamans, C. A. Ross and R. E. Yasbin, 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 185 2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussan, T. E., A. Yang, F. Li, M. C. Ostrowski and R. H. Reeves, 2008. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down's syndrome. Nature 451 73–75. [DOI] [PubMed] [Google Scholar]
- Thompson, S. L., and D. A. Compton, 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, E. M., T. Sokolsky, C. M. Tucker, L. Y. Chan, M. Boselli et al., 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317 916–924. [DOI] [PubMed] [Google Scholar]
- Tsafrir, D., M. Bacolod, Z. Selvanayagam, I. Tsafrir, J. Shia et al., 2006. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 66 2129–2137. [DOI] [PubMed] [Google Scholar]
- Vacik, T., M. Ort, S. Gregorova, P. Strnad, R. Blatny et al., 2005. Segmental trisomy of chromosome 17: a mouse model of human aneuploidy syndromes. Proc. Natl. Acad. Sci. USA 102 4500–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia, R. A., 2002. Exploring the etiology of haploinsufficiency. BioEssays 24 175–184. [DOI] [PubMed] [Google Scholar]
- Weaver, B. A., A. D. Silk, C. Montagna, P. Verdier-Pinard and D. W. Cleveland, 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11 25–36. [DOI] [PubMed] [Google Scholar]
- Whitesell, L., and S. L. Lindquist, 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5 761–772. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387 708–713. [DOI] [PubMed] [Google Scholar]
- Yao, M. C., and J. L. Chao, 2005. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 39 537–559. [DOI] [PubMed] [Google Scholar]