Abstract
Determining how growth and differentiation are coordinated is key to understanding normal development, as well as disease states such as cancer, where that control is lost. We have previously shown that growth and neuronal differentiation are coordinated by the insulin receptor/target of rapamycin (TOR) kinase (InR/TOR) pathway. Here we show that the control of growth and differentiation diverge downstream of TOR. TOR regulates growth by controlling the activity of S6 kinase (S6K) and eIF4E. Loss of s6k delays differentiation, and is epistatic to the loss of tsc2, indicating that S6K acts downstream or in parallel to TOR in differentiation as in growth. However, loss of eIF4E inhibits growth but does not affect the timing of differentiation. We also show, for the first time in Drosophila, that there is crosstalk between the InR/TOR pathway and epidermal growth factor receptor (EGFR) signaling. InR/TOR signaling regulates the expression of several EGFR pathway components including pointedP2 (pntP2). In addition, reduction of EGFR signaling levels phenocopies inhibition of the InR/TOR pathway in the regulation of differentiation. Together these data suggest that InR/TOR signaling regulates the timing of differentiation through modulation of EGFR target genes in developing photoreceptors.
A fundamental challenge during the development of any complex organism is the coordination of proliferation and differentiation. The rate of proliferation is not constant during development (Neufeld et al. 1998) and depends on the developmental stage as well as hormonal and nutritional cues (Britton et al. 2002). Coordinating growth and differentiation is a particular challenge in complex tissues, such as the nervous system. Neurogenesis is preceded by a period of proliferation, which generates a pool of precursor cells. Selected cells from this pool exit the cell cycle and initiate a complex program of gene expression that will result in the formation of the mature neuron.
The Drosophila retina is a highly tractable model for studying the relationship between growth and neuronal differentiation (Wolff and Ready 1993). Photoreceptor (PR) differentiation in Drosophila is initiated at the beginning of the third larval instar when a physical indentation, known as the morphogenetic furrow (MF), develops at the posterior of the eye imaginal disc. Over a period of ∼48 hr the MF sweeps anteriorly leading to the formation of PR preclusters. The MF is initiated by the morphogen Hedgehog (Hh) and is propagated anteriorly through a combination of Hh and Decapentapalegic (Dpp) signaling (Voas and Rebay 2004). Posterior to the MF, PRs are specified sequentially through reiterative use of the Notch and EGFR pathways (Brennan and Moses 2000; Voas and Rebay 2004).
As in other neurogenic contexts, neuronal differentiation in the Drosophila eye is a temporally restricted process. Patterning of the mature cluster of eight PRs is highly stereotyped with each row forming about every 2 hr (Figure 1A) (Wolff and Ready 1993). The mechanism underlying the temporal control of PR differentiation has proven elusive. Several models have been proposed including control by receptor-mediated cell–cell interactions and intrinsic or extrinsic cellular clocks (Freeman 1997; Brennan and Moses 2000; Voas and Rebay 2004). We found that the conserved InR/TOR pathway plays a key role in controlling the timing of neuronal differentiation in Drosophila (Bateman and McNeill 2004). Using mutants in various components of the InR/TOR pathway, we showed that activation of this pathway causes precocious differentiation of neurons. Conversely, inhibition of InR/TOR signaling significantly delays neurogenesis. How the InR/TOR pathway regulates neuronal differentiation is unclear.
Figure 1.—
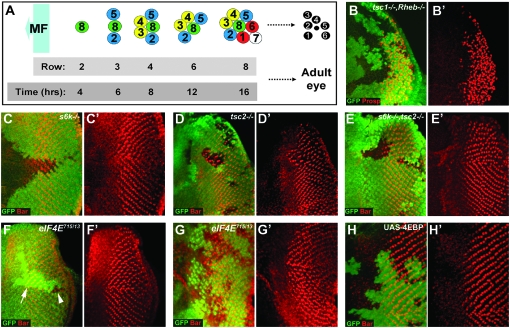
InR and TOR signaling act through S6K, but not eIF4E to control the timing of neuronal differentiation. (A) Schematic showing the spatiotemporal nature of PR differentiation in the Drosophila eye imaginal disc. MF, morphogenetic furrow. (B and B′) tsc12G3, Rheb2D1 double-mutant clones have an identical delay in differentiation (stained for Prospero expression, shown in red) to Rheb2D1 clones (Figure 2C). (C and C′) Loss of S6k causes a slight delay in the differentiation of PR 7 and cone cells (stained for Bar expression, shown in red). (D and D′) tsc2 (gig56) clones cause precocious differentiation of PRs 1 and 6 (stained for Bar expression, shown in red). (E and E′) The precocious differentiation phenotype of tsc2 cells is suppressed in tsc2 (gig192), s6k11 clones (Bar staining in red). (F and G) eIF4E715/13 LOF clones inhibit growth resulting in small clones, compare clone (arrowhead) to twin spot (arrow) size in F, but do not affect differentiation in posterior clones generated using hs-flp (F and F′) or clones close to the MF, generated using ey-flp (G and G′), (Bar staining in red). (H and H′) overexpression of 4EBP (shown by the presence of GFP staining) does not have any affect on differentiation of PRs 1 and 6 (stained for Bar expression, shown in red). LOF clones in B–G are marked by the loss of GFP (shown in green). Anterior is to the left in all panels.
Temporal control of neuronal differentiation is a property of the entire InR/TOR pathway. Ligand binding to the InR causes recruitment and phosphorylation of the insulin receptor substrate (IRS) and subsequent activation of PI3K, which catalyzes the production of phosphatidylinositol 3,4,5-trisphosphate (PIP3) at the membrane (Leevers and Hafen 2004). PDK1 and PKB/AKT, both PH domain-containing kinases, become membrane localized by their interaction with PIP3 where PKB/AKT can be fully activated. InR signaling controls growth and proliferation through the inhibition of the GTPase activating protein (GAP) TSC2 (Gao and Pan 2001; Potter et al. 2001; Tapon et al. 2001; Cai et al. 2006). TSC2 inhibits the activity of the small GTPase Rheb, which activates TOR (Long et al. 2005). TOR is a phosphatidylinositol kinase-related kinase that is part of a complex (TORC1) that controls growth through the regulation of ribosome biogenesis and translation via S6K and eIF4E, respectively (Inoki and Guan 2006; Wullschleger et al. 2006). TOR is also a component of the TORC2 complex. TORC2 is insensitive to rapamycin and has recently been shown to phosphorylate AKT at Ser473 (Sarbassov et al. 2005; Guertin et al. 2006b). TOR has other functions including the regulation of microautophagy and fat metabolism (Rusten et al. 2004; Scott et al. 2004). In addition, inhibition of TOR by treatment with rapamycin elicits a transcriptional response involving several hundred genes (Peng et al. 2002; Guertin et al. 2006a). Recently a negative feedback loop in which S6K regulates IRS, both transcriptionally and by phosphorylation, has been shown to exist in both Drosophila (Radimerski et al. 2002) and mammalian systems (Harrington et al. 2004; Shah et al. 2004; Um et al. 2004).
What is the mechanism by which InR signaling controls the timing of neuronal differentiation? In mammalian systems activation of insulin/IGF receptor tyrosine kinases causes activation of both PI3K and Ras/mitogen-activated protein kinase (MAPK) pathways (Baltensperger et al. 1993; Skolnik et al. 1993; Downward 2003). Ligand binding to the InR results in tyrosine phosphorylation of IRS proteins and/or Shc which, through the adaptor protein Grb2, results in recruitment to the membrane of SOS for the activation of Ras (Baltensperger et al. 1993; Skolnik et al. 1993). However, flies expressing a version of the Drosophila IRS chico, in which the putative Drk (the Drosophila ortholog of Grb2) binding site had been mutated, are able to fully rescue the growth defects of chico flies (Oldham et al. 2002). Therefore it is currently unclear whether the InR activates MAPK signaling in Drosophila (Bateman and McNeill 2006).
In the current study we find that differentiation is temporally regulated by TOR and S6K, but not by 4EBP or eIF4E, thus providing the first branch in the differentiation pathway downstream of InR signaling in the eye. We also show that activation of the InR/TOR pathway regulates the expression, at the transcriptional level, of the EGFR pathway components Argos, rhomboid (rho), and pointedP2 (pntP2). Moreover, reducing the level of EGFR signaling, by using a pntP2 hypomorphic allele, causes a cell-type-specific delay in differentiation, which is identical to that in mutants that inhibit the InR/TOR pathway. Finally we show that the EGFR and InR/TOR pathways genetically interact in controlling the timing of PR differentiation.
MATERIALS AND METHODS
To generate loss-of-function clones, 48- to 72-hr-old larvae were heat-shocked for 1–2 hr at 37°. Overexpression clones were generated using the “flp-out” technique (Neufeld et al. 1998), where 48- to 60-hr-old larvae were heat-shocked for 2.5 hr at 37°. Third instar eye discs were fixed in PBSA/4% formaldehyde (EMS Scientific) for 45 min, washed in PBSA/0.1% TritonX100 (Sigma, St. Louis) and incubated overnight with primary antibody. Primary antibodies were used as follows: mouse and rabbit anti-GFP (Molecular Probes, Eugene, OR; 1:1000), rabbit anti-Bar (a gift from K. Saigo; 1:200), mouse anti-Prospero (DHSB; 1:10), guinea pig anti-Senseless (a gift from H. Bellen; 1:1000), mouse anti-β-galactosidase (Roche, Indianapolis; 1:1000), rabbit anti-Spalt (a gift from R. Barrio; 1:500), mouse anti-Rough (DSHB; 1:100), mouse anti-Cut (DSHB; 1:20), and mouse anti-Argos (DSHB; 1:100). Secondary antibodies were from Jackson Laboratories (West Grove, PA). After staining, discs were mounted in Vectastain (Vector Laboratories, Burlingame, CA) and analyzed with a Zeiss confocal microscope or a Zeiss Apotome.
To quantify eIF4E mutant growth rates the mutant clone area relative to the twin-spot area was quantified using ImageJ and in three independent clones for each genotype.
The following stocks were kindly provided to us: The pten flies were from Sally Leevers and tsc1 flies from Nic Tapon. The s6k, tsc2 stock was from D. J. Pan. The Rheb stocks were from Ernst Hafen. The UAS-4EBP stock was from Nahum Sonenberg. pnt stocks were from Christian Klämbt. The rhoX81 stock was from Matthew Freeman. eIF4E (11720), aosW11 (2513), and TOR (7014) mutants were from The Bloomington Stock Center. Genotypes for generating clones were as follows:
tsc1, Rheb mutant clones: y, w, hs-flp; FRT82, dRheb2D1, tsc12G3/FRT82B, Ubi-GFP.
tsc2 mutant clones: y, w, hs-flp; gig56, FRT80/FRT80, Ubi-GFP.
tsc2 mutant clones with pntP2-LacZ: y, w, hs-flp; gig56, FRT80, pnt1277/FRT80, Ubi-GFP.
s6k mutant clones: y, w, hs-flp; s6kl1, FRT80B/FRT80, P{LacW}RpL14, eGFP.
s6k, tsc2 mutant clones: y, w, hs-flp; gig192, s6kl1, FRT80/FRT80, Ubi-GFP.
eIF4E mutant clones: y, w, hs-flp; eIF4E 07238, FRT80/FRT80, arm-LacZ or y, w, hs-flp; eIF4E715/13, FRT80/FRT80, arm-LacZ.
eIF4E, tsc2 mutant clones: y, w, hs-flp; eIF4E 07238, gig56, FRT80/FRT80, P{LacW}RpL14, eGFP.
4EBP overexpression clones: y, w, hs-flp; UAS-4EBP/act>y>Gal4, UAS-GFP.
tsc1 mutant clones: y, w, hs-flp; tsc1Q87X, FRT82B/FRT82B, Ubi-GFP.
Rheb mutant clones: hs-flp; Rheb2D1, FRT82/82FRT, Ubi-GFP, M[95A], Rps63.
pten mutant clones: y, w, hs-flp; pten1, FRT40/FRT40, Ubi-GFP.
pten mutant clones with aos-LacZ: y, w, hs-flp; pten1, FRT40/FRT40, Ubi-GFP; aosW11/+.
Rheb mutant clones with aos-LacZ: hs-flp; aosW11, Rheb2D1, FRT82/82FRT, Ubi-GFP, M[95A], Rps63.
pten mutant clones with rho-LacZ: y, w, hs-flp; pten1, FRT40/FRT40, Ubi-GFP; rhoX81/+.
pten mutant clones with pntP2-LacZ: y, w, hs-flp; pten1, FRT40/FRT40, Ubi-GFP; pnt1277/+.
TOR mutant clones: y, w, hs-flp; TORΔD, FRT40A/FRT40, Ubi-GFP; pnt1277/+.
pntP2 hypomorph clones: y, w, hs-flp; FRT82, pnt1230/FRT8, Ubi-GFP.
Rheb, pntP2 mutant clones: hs-flp; pnt1230, Rheb2D1, FRT82/82FRT, Ubi-GFP, M[95A], Rps63.
UAS-Dp110, pntP2 clones: hs-flp, UAS-GFP; UAS-Dp110; tub-Gal80, FRT82, pnt1230/FRT82, tub-Gal80.
UAS-pntP2 clones: hs-flp; act>y>Gal4, UASGFP; UAS-pntP2.
EGFRACT clones: hs-flp; act>y>Gal4, UASGFP; UAS-EGFRACT.
RESULTS
The InR controls differentiation through a pathway including TOR and S6K, but not 4EBP/eIF4E:
We have shown previously that tsc1 loss-of-function (LOF) clones cause precocious differentiation of PRs in the developing eye (Bateman and McNeill 2004). TSC1, together with TSC2, functions as a GAP for the small GTPase Rheb. We found that loss of Rheb causes a strong delay in differentiation suggesting that TSC1/2 acts upstream of Rheb in controlling differentiation as it does in growth (Saucedo et al. 2003; Zhang et al. 2003). However, TSC1 has targets other than Rheb and can activate RhoGTPase and inhibit Rac1 through interaction with the ERM family of actin binding proteins (Lamb et al. 2000; Astrinidis et al. 2002; Goncharova et al. 2004). Therefore we asked whether TSC1 is able to affect differentiation independently of Rheb. To do this we generated Rheb, tsc1 double-mutant clones and observed the differentiation phenotype by staining with anti-Prospero (Xu et al. 2000). If Rheb is absolutely required for regulation of differentiation by TSC1 then Rheb, tsc1 double-mutant clones should have a similar phenotype to Rheb clones. Alternatively, if the TSC1/2 complex is able to regulate differentiation independent of Rheb, then the delayed differentiation phenotype caused by loss of Rheb should be abrogated in Rheb, tsc1 clones. Rheb, tsc1 double-mutant clones show a strong delay in differentiation (Figure 1B), similar to that seen in Rheb clones (Figure 2, C and D). This result suggests that the primary target of TSC1/2 in controlling the timing of neuronal differentiation is Rheb.
Figure 2.—
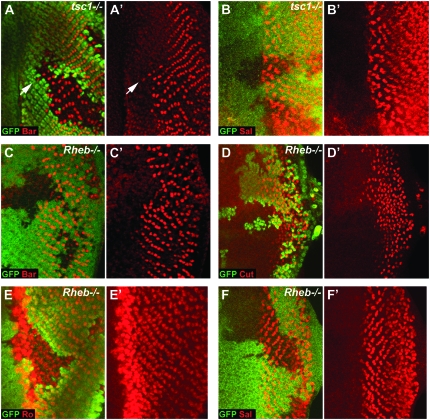
InR/TOR signaling controls the differentiation of specific cell types in the developing eye. (A and A′) Cells mutant for tsc1 (tsc1Q87X) show precocious differentiation of PRs 1 and 6 (stained for Bar expression, shown in red) ahead of the wild-type differentiation front; arrow indicates an example of a precociously differentiated PR. (B and B′) Differentiation of PRs 3/4 (stained for Spalt expression, shown in red) is unaffected in tsc1Q87X clones. (C and D) Differentiation of PRs 1 and 6 (stained for Bar expression, shown in red in C and C′) and cone cells (stained with Cut, shown in red in D and D′) is strongly delayed in Rheb2D1 clones. (E and F) PR 2/5 (stained for Rough expression, shown in red in E and E′) and PR 3/4 (stained for Spalt expression, shown in red in F and F′) differentiation is unaffected in Rheb2D1 clones. LOF clones in all panels are marked by the loss of GFP staining (shown in green). Anterior is to the left in all panels.
The TSC1/2 complex and Rheb regulate TOR (Leevers and Hafen 2004). TOR is part of the TORC1 complex, controls growth by phosphorylation of S6K and 4EBP, which in turn affect translation and ribosome biogenesis by regulating RpS6 and eIF4E, respectively (Inoki and Guan 2006; Wullschleger et al. 2006). We asked whether S6K and 4EBP are also able to control neuronal differentiation. s6k LOF clones do cause a slight delay in differentiation (Figure 1C), which is much weaker than the delay seen in Rheb or TOR LOF clones (Figure 2, C and D; (Bateman and McNeill 2004). To determine whether S6K mediates the precocious differentiation phenotype seen in tsc2 clones (Figure 1D) we generated s6k, tsc2 double-mutant clones. These clones have a wild-type differentiation phenotype (Figure 1E), indicating that S6K acts either downstream or in parallel to TSC2 in controlling differentiation.
TOR also controls growth via the translation initiation factor eIF4E and its inhibitory binding partner 4EBP. Homozygous eIF4E Drosophila arrest growth during larval development (Lachance et al. 2002). Lachance et al. (2002) however did not determine whether eIF4E mutant cells have a growth defect. To assess this we made LOF clones of cells using either weak (eIF4E07238) or strong (eIF4E715/13) eIF4E alleles. Clones made using eIF4E07238 had a mild but significant growth defect (mean clone size = 67% ± 1% size of twin spot, n = 3; supplemental Figure 1), while clones made using eIF4E715/13 had a severe growth defect (Figure 1F, compare clone to twin-spot size; mean clone size = 8.7% ± 2% size of twin spot, n = 3). Control clones made using a wild-type FRT chromosome were a similar size to the twin spot (mean clone size = 98% ± 1% size of twin spot, n = 3) as expected. Surprisingly, neither eIF4E07238 (supplemental Figure 1) nor eIF4E715/13 LOF clones have any effect on differentiation in posterior (Figure 1F) or anterior clones close to the MF (Figure 1G). Also, eIF4E07238, tsc2 mutant clones have a similarly strong precocious differentiation phenotype to tsc2 clones (supplemental Figure 1), further suggesting that eIF4E is not required for InR/TOR-dependent control of PR differentiation. We also analyzed the differentiation phenotype of the eIF4E inhibitory binding partner 4EBP. In accordance with our results with eIF4E, overexpression of 4EBP also has no effect on differentiation (Figure 1H). In addition, we do not observe any differentiation phenotype in clones of wild-type cells generated in a background heterozygous for a ribosomal subunit dominant mutation (a Minute mutant; data not shown), confirming that alteration of the overall translation rate does not affect differentiation. Taken together these data suggest that the control of the timing of neuronal differentiation is regulated by S6K and is independent of 4EBP/eIF4E, while growth is controlled by both these factors.
InR/TOR signaling controls the timing of the differentiation of a subset of photoreceptors:
Each ommatidium in the Drosophila eye consists of eight photoreceptor (PR) neurons and 12 accessory cells. We have shown that the InR/TOR pathway controls the timing of differentiation of PRs 1, 6, and 7 and cone cells, but does not affect PR 8 (Bateman and McNeill 2004). The differentiation of PR 8 is followed by the sequential differentiation of PRs 2/5, then PRs 3/4, and finally PRs 1, 6, and 7 (Figure 1A). To determine whether the differentiation of PRs 2–5 is also regulated by the InR/TOR pathway we used antibodies against the transcription factors Rough (Kimmel et al. 1990) and Spalt (Barrio et al. 1999) to analyze the differentiation of PRs 2/5 and 3/4, respectively. If InR/TOR signaling does regulate the differentiation of PRs 2–5 we would expect activation of the pathway by loss of tsc1 to cause precocious differentiation of these PRs. Both Rough and Spalt staining appeared normal within tsc1 clones (Figure 2B and data not shown), suggesting that the InR/TOR pathway does not affect the timing of differentiation of PRs 2/5 or PRs 3/4.
We were concerned that since PRs 2/5 and 3/4 differentiate close to the morphogenetic furrow (rows 3 and 4, respectively, Figure 1A), that it might be difficult to resolve cells which are precociously differentiating. To overcome this issue we made Rheb LOF clones to determine whether there is any delay in the differentiation of PRs 2–5 when InR/TOR signaling is inhibited. Differentiation of PRs 1, 6, and 7 and cone cells is strongly delayed in Rheb clones (Figure 2, C and D and (Bateman and McNeill 2004), however, both Rough (PRs 2 and 5) and Spalt (PRs 3 and 4) staining is unaffected in these clones (Figure 2, E and F). Therefore temporal control of differentiation by the InR/TOR pathway in the developing eye is stage/cell type specific: the late differentiating PRs 1, 6, and 7 and cone cells are dependent on the InR/TOR pathway, while the early differentiating PRs 2–5 and 8 are independent of InR/TOR signaling.
Transcription of Argos, a reporter of EGFR signaling activity, is regulated by the InR/TOR pathway:
The stage/cell-type-specific nature of the temporal control of differentiation suggests that the InR/TOR pathway achieves this regulation through a novel mechanism. To investigate this we asked whether any of the pathways known to be important for PR differentiation are affected by changes in InR/TOR signaling. Since the passage of the MF is unaffected by the InR/TOR pathway it seemed unlikely that Dpp, Hh, or Wingless signaling were being affected. Next we asked whether EGFR signaling is regulated by the InR/TOR pathway in the developing eye. We had previously analyzed EGFR signaling activity in two ways. First we stained with an antibody against dual phosphorylated MAPK (dpERK), which gives a direct readout of EGFR signaling levels (Gabay et al. 1997). Second we analyzed the level of the E26 transformation-specific sequence (ETS) protein Yan, whose accumulation in the nucleus is dependent on its phosporylation state and hence the level of EGFR activity (Tootle et al. 2003; Song et al. 2005). Neither dpERK nor Yan staining are affected by activation of InR/TOR signaling (Bateman and McNeill 2004). However, we had not tested whether EGFR signaling is being affected downstream or in parallel to MAPK and Yan.
To test whether there is any overall activation of EGFR signaling by the InR/TOR pathway we looked at the expression of Argos. Argos is a secreted protein that functions as an inhibitory ligand of the EGFR (Freeman et al. 1992b). argos expression is induced by EGFR activation in differentiating cells and is thought to result in a feedback loop that inhibits the differentiation of surrounding cells (Golembo et al. 1996). As a consequence of its dependence on EGFR activation Argos is strongly expressed in developing PRs as they differentiate (Freeman et al. 1992b). To analyze the expression of Argos in cells in which InR/TOR signaling is activated we stained pten LOF clones with an Argos monoclonal antibody. Although Argos stains poorly in imaginal discs we see a consistent increase in Argos accumulation in pten clones (Figure 3A).
Figure 3.—
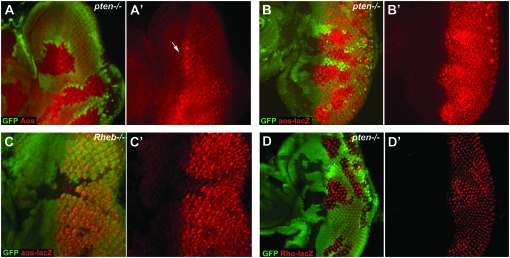
argos and rho expression is regulated by InR/TOR signaling in developing neurons. (A and A′) The level of Argos protein (detected using an anti-Argos monoclonal antibody, shown in red) is increased in pten1 mutant cells (marked by loss of GFP staining). Note how Argos staining is seen ahead of the normal expression front (marked with an arrow). (B and C) argos expression is regulated by InR/TOR signaling at the level of transcription. Expression of β-galactosidase (stained with an anti-β-galactosidase antibody, shown in red) from the P{lwB}-element insertion in argos (aosw11) is upregulated in pten1 clones (B and B′) and downregulated in Rheb2D1 clones (C and C′). (D and D′) rho expression (using the rhoX81 reporter, detected by staining with a anti-β-galactosidase antibody, shown in red) is upregulated in pten1 clones. Clones are marked by loss of GFP staining and anterior is to the left in all panels.
Next we asked whether the ability of the InR/TOR pathway to modulate Argos levels is caused by changes in argos gene expression. This result would indicate that EGFR signaling is being affected, rather than a stabilization of Argos post-transcriptionally. To address this we used the argosW11 lacZ reporter line (Freeman et al. 1992a,b). Using argosW11 we observed a strong increase in argos expression in pten LOF clones (Figure 3B). Interestingly, in pten clones that cross the MF, strong precocious expression of argos is seen in the mutant cells (Figure 3B). To determine whether inhibition of the InR/TOR pathway can regulate argos expression we generated Rheb clones in larvae carrying the argosW11 allele. Loss of Rheb causes a strong decrease in argos expression in differentiating cells (Figure 3C). Thus both positive and negative regulators of the InR/TOR signaling pathway lead to alterations in argos expression.
Since Argos is also an inhibitory ligand of the EGFR (Freeman et al. 1992b), we analyzed the expression of rhomboid-1 (rho) as an independent readout of EGFR activity. rho expression was monitored using the X81 enhancer trap line which is expressed strongly in PRs 2/5 and 8 (Freeman et al. 1992a). In accordance with the argos data, rho expression is upregulated in pten LOF clones (Figure 3D). These changes appear to be specific since the expression of several other cell fate genes is unaffected by changes in InR/TOR signaling (Bateman and McNeill 2004), including the Notch ligand Delta (supplemental Figure 2). In conclusion, these data suggest that there is crosstalk between InR/TOR signaling and the EGFR pathway and that this occurs downstream of MAPK.
Expression of pntP2 is regulated by InR/TOR signaling:
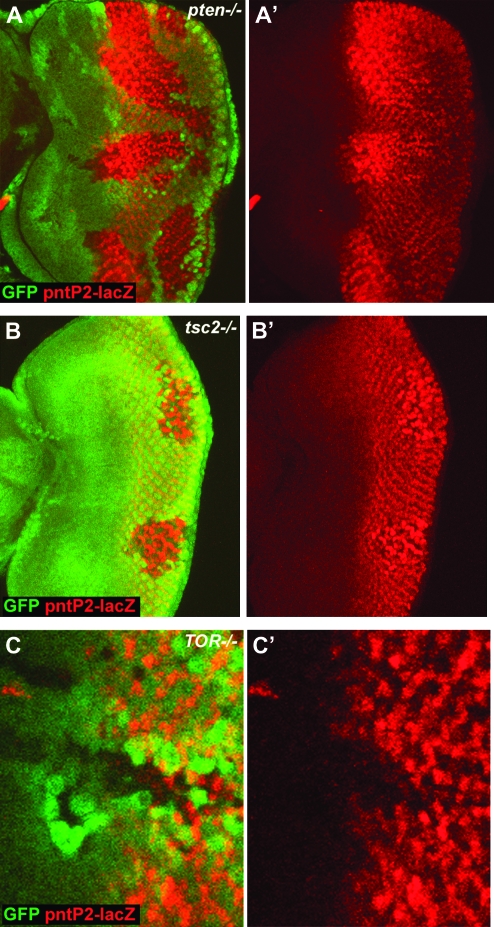
argos expression is activated by the ETS transcription factor pointed (pnt). pnt is expressed as two alternatively spliced isoforms, P1 and P2, which share a C-terminal region that contains the ETS motif (Scholz et al. 1993). pntP2 is expressed specifically in the embryonic midline glial cells (Klambt 1993), and argos expression is lost in these cells in pointed (pnt) mutant embryos (Scholz et al. 1997). Activation of the EGFR results in phosphorylation of MAPK, which enters the nucleus and phosphorylates pntP2 (Brunner et al. 1994; O'Neill et al. 1994). In the eye imaginal disc pntP2 is expressed in precursor cells posterior to the MF and in PRs 1, 6, and 7 and cone cells (Brunner et al. 1994). Since argos is a transcriptional target of pntP2 we wondered whether pntP2 expression might also be regulated by InR/TOR signaling. To test whether pntP2 expression is regulated by InR/TOR signaling we used the pnt1277 allele which contains a P{LacW} element within the first, noncoding exon of pntP2 (Scholz et al. 1993). Using pnt1277 we observe a strong increase in pntP2 expression in pten LOF clones (Figure 4A). Interestingly, the increase in pntP2 expression differs spatiotemporally across the field of differentiating cells. pntP2 expression is increased most strongly in cells as they differentiate, but this increase is lost once the cells become more mature. Moreover, dramatic precocious expression of pntP2 is observed in pten clones that span the MF (Figure 4A). Importantly, pntP2 expression is also upregulated in undifferentiated cells around the MF, suggesting that the increase in expression is not simply an indirect consequence of the precocious differentiation of PRs. We also observe a similar upregulation of pntP2 expression in clones that have activated InR/TOR signaling due to loss of tsc2 (Figure 4B). The increase in pntP2 expression is not a result of a general increase in transcription due to increased growth, since we do not see increased expression of several other markers of PR cell fate (Bateman and McNeill 2004). To examine the effect of blocking InR/TOR signaling we examined pntP2 expression in cells mutant for TOR. LOF clones of TOR show decreased expression of pntP2 (Figure 4C). Therefore pntP2 expression is sensitive to both activation and inhibition of InR/TOR signaling. To determine whether this property is specific to the eye we looked at pntP2 expression in pten clones in the leg and eye discs. We did not observe any change in pntP2 expression in these clones (supplemental Figure 3), suggesting either that InR/TOR regulation of pntP2 is specific to the developing eye (perhaps requiring specific factors expressed close to the MF) or that the spatiotemporal nature of eye development in Drosophila makes it possible to observe changes that cannot be resolved in other imaginal discs.
Figure 4.—
pntP2 expression is regulated by InR/TOR signaling. (A and B) pntP2 transcription, detected by staining for β-galactosidase in flies carrying a P{LacW} element in pntP2 (pnt1277), is upregulated and precocious in pten1 (A and A′) and tsc2 (gig56) clones (B and B′). Note that the disc shown in B is a younger disc and so pntP2 is upregulated more posteriorly. Conversely pntP2 transcription is downregulated in TORΔD clones (C and C′). LOF clones in A–C are marked by the loss of GFP staining (shown in green). Anterior is to the left in all panels.
Reducing EGFR signaling phenocopies loss of Rheb or TOR in developing PRs:
Argos, rho, and pntP2 expression levels are all regulated by InR/TOR signaling, suggesting crosstalk between InR/TOR and EGFR pathways. However, complete loss of EGFR or pntP2 activity (using null alleles) completely blocks the differentiation of all PRs except PR 8 (data not shown; (Baonza et al. 2001, 2002; Yang and Baker 2003), whereas inhibition of the InR/TOR pathway causes a delay only in the differentiation of PRs 1, 6, and 7 and cone cells (Figure 2). To reconcile these observations we wondered whether a reduction, rather than a complete loss in EGFR activity would cause the same cell-type-specific delay in differentiation as inhibition of the InR/TOR pathway.
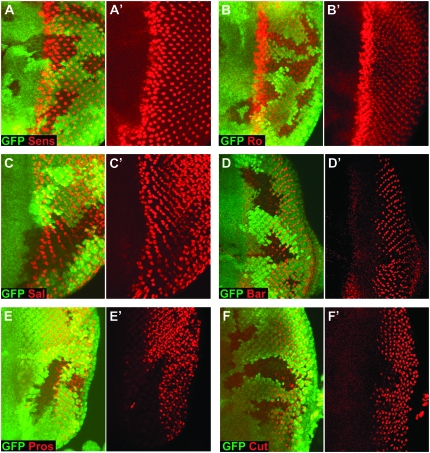
To determine the affect of reducing EGFR signaling levels we used a hypomorphic allele of pntP2 (pntP21230), which was generated by the imprecise excision of a P element in the first, noncoding exon of pntP2 (Klambt 1993). We stained pntP21230 clones with the same panel of markers that we had used to analyze the differentiation phenotype of InR/TOR pathway mutants (Figure 2). Interestingly the PR differentiation phenotype in pntP21230 clones is identical to that in Rheb or TOR LOF clones (compare Figure 5 to Figure 2). Specifically, PR 8 (stained for Senseless expression; Figure 5A), PRs 2/5 (stained for Rough expression; Figure 5B), and PRs 3/4 (stained for Spalt expression; Figure 5C) differentiate normally in pntP21230 clones. In contrast the differentiation of PRs 1 and 6 (stained for Bar expression; Figure 5D), PR 7 (stained for Prospero expression; Figure 5E), and cone cells (stained for Prospero expression; Figure 5E and Cut expression; Figure 5F) are strongly delayed but not completely blocked. The phenotypic similarity between PRs with reduced EGFR signaling and PRs in which InR/TOR signaling is inhibited is consistent with InR/TOR signaling modulating EGFR transcriptional outputs to control neuronal differentiation.
Figure 5.—
Reducing EGFR signaling phenocopies the differentiation phenotype of loss of Rheb or TOR. pntP2 hypomorphic clones made using the allele pntP21230 show cell-type-specific delays in PR differentiation identical to those seen in LOF clones of positive effectors of InR/TOR signaling such as Rheb and TOR. (A–C) pntP21230 clones have no effect on the differentiation of PR 8 (stained for Senseless expression, shown in red in A and A′), PRs 2/5 (stained for Rough expression, shown in red in B and B′) or PRs 3/4 (stained for Spalt expression, shown in red in C and C′). Note that Spalt staining shows a delay toward the posterior of the disc where the antibody also stains PRs 1 and 6. In contrast pntP21230 clones show a strong delay in the differentiation of PRs 1 and 6 (stained for Bar expression, shown in red in D and D′), PR 7 (stained for Prospero expression, which is also expressed in cone cells, shown in red in E and E′) and cone cells (stained for Cut expression, shown in red in F and F′ and Prospero expression, shown in red in E and E′). LOF clones in all panels are marked by the loss of GFP staining (shown in green). Anterior is to the left in all panels.
InR/TOR and EGFR signaling interact genetically:
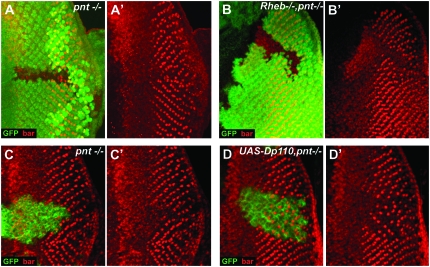
Since reducing EGFR pathway activity through pntP2 phenocopied inhibition of the InR/TOR pathway we wondered whether these two pathways could interact genetically. To test this we generated clones that were double mutant for pntP21230 and Rheb2D1. Inhibition of differentiation in these clones (Figure 6B) was much more severe than in pntP2 (or Rheb) single mutant clones (Figure 6A). pntP21230, Rheb2D1 double-mutant clones block rather than delay the differentiation of PRs 1 and 6 (Figure 6B). Conversely, when we overexpressed Dp110 in pnt1230 clones using the mosaic analysis with a repressible cell marker (MARCM) technique (Lee and Luo 1999), the delay in the differentiation of PRs 1 and 6 was much less severe (Figure 6D) than in pnt1230 clones alone (Figure 6C) and the precocious differentiation normally seen with Dp110 overexpression was completely suppressed, strongly suggesting that pntP2 acts downstream of Dp110. These data demonstrate that the InR/TOR and EGFR pathways can interact genetically and are consistent with the regulation of neuronal differentiation by the InR/TOR through modulation of EGFR transcriptional output.
Figure 6.—
InR/TOR and EGFR signaling interact genetically. (A and B) Differentiation of PRs 1 and 6 is delayed in pnt1230 clones (A and A′) and blocked in pnt1230, Rheb2D1 clones (B and B′). (C and D) Using the MARCM system differentiation of PRs 1 and 6 is delayed in pnt1230 clones (C and C′), whereas the delay is significantly weaker in pnt1230 clones overexpressing Dp110 (D and D′). Mutant cells are marked by the absence of GFP in A and B, but by the presence of GFP in C and D. PRs 1 and 6 are shown by Bar staining (red) in all panels.
DISCUSSION
Tight coordination of growth and differentiation is essential for normal development. We have previously shown that InR/TOR signaling controls the timing of neuronal differentiation (Bateman and McNeill 2004) in the eye and leg in Drosophila. Here we demonstrate that the InR/TOR pathway regulates neuronal differentiation in an S6K-dependent, but 4EBP/eIF4E-independent manner. Previously we were unable to determine whether InR/TOR signaling was acting downstream or in parallel to the EGFR/MAPK pathway. Using argos and rho as reporters we have shown that the InR/TOR pathway is able to regulate EGFR/MAPK signaling downstream of MAPK. Moreover, pntP2 expression is up- and downregulated by activation or inhibition of InR/TOR signaling, respectively, and InR/TOR and EGFR pathways interact through pntP2. Taken together our data suggest that temporal control of differentiation by the InR/TOR pathway is achieved by modulation of EGFR pathway transcriptional targets in differentiating PRs.
TOR is part of two multimeric complexes (TORC1 and TORC2) and is a core component of the InR pathway (Inoki and Guan 2006; Wullschleger et al. 2006). TORC1 activity is regulated by nutrient and energy levels (Hara et al. 1998; Inoki et al. 2003) providing a conduit for hormonal and catabolic cellular inputs. Growth is regulated by two downstream targets of TORC1: S6K and 4EBP. Our data demonstrate that upstream of TORC1, differentiation and growth are regulated by the same factors. Downstream of TORC1, differentiation and growth differ significantly in that loss of s6k, but not eIF4E (or overexpression of 4EBP) affects differentiation. eIF4E regulates 7-methyl-guanosine cap-dependent translation and is the rate-limiting factor in translation initiation (Richter and Sonenberg 2005). Our finding that eIF4E does not affect differentiation suggests that the temporal control of differentiation is not based on a translation initiation-dependent mechanism. Strikingly, we show that loss of s6k blocks the precocious differentiation induced by loss of tsc2. Given the relatively weak effects of loss of s6k this may seem surprising. However, the degree of suppression is similar to the effect of loss of s6k on the overgrowth phenotype caused by loss of tsc2, namely, tsc2, s6k double-mutant cells are the same size as wild-type cells (Gao et al. 2002). Although loss of eIF4E has no affect on differentiation it may act redundantly with another factor, such as s6k. Testing this hypothesis though is technically challenging since the Drosophila genome contains eight different eIF4E isoforms (Hernandez et al. 2005). It will be interesting in future to test whether any of these isoforms regulate differentiation or alternatively whether eIF4E and s6k act redundantly. Although further work is required to determine the precise relationship between S6K and the InR/TOR pathway, our data point to a critical role of S6K in coordinating neuronal differentiation and growth.
As in other neuronal systems, differentiation of PRs in the Drosophila eye occurs in a stereotyped manner. The advantage of the Drosophila retina as an experimental system is that the PRs differentiate spatiotemporally. Using this feature, as well as a series of cell-type-specific antibodies, we have demonstrated that InR/TOR signaling is selective in the cell-types that it affects. The differentiation of PRs 2/5, 3/4, and 8 are unaffected by perturbations in InR/TOR signaling, whereas PRs 1, 6, and 7 and cone cells are dependent on this pathway for temporal control of differentiation. Interestingly the affected cells all differentiate after the second mitotic wave. However, we have shown that regulators of the cell cycle do not affect the temporal control of differentiation (Bateman and McNeill 2004). Why then are PRs 1, 6, and 7 and cone cells specifically affected? In cells with increased InR/TOR signaling, the expression of argos, rho, and pntP2 is precocious and increased throughout the clone, suggesting that the upregulation of EGFR signaling occurs in all cells. However, decreasing EGFR activity using a hypomorphic pntP2 allele specifically affects the differentiation of PRs 1, 6, and 7 and cone cells. Interestingly, pntP2 expression in differentiated cells is also restricted to PRs 1, 6, and 7 and cone cells. These observations suggest that differentiation of PRs 1, 6, and 7 and cone cells is critically dependent on EGFR levels signaling through pntP2. Therefore, although activation of InR/TOR signaling causes upregulation of EGFR transcriptional targets in all cells as they differentiate, the phenotypic effect is only seen in PRs 1, 6, and 7 and cone cells since these cells are highly sensitive to EGFR activity signaling through pntP2. This possibility is supported by the fact that precocious differentiation caused by overexpression of Dp110 can be suppressed by the simultaneous reduction of pntP2 levels (Figure 6). The complete suppression of the Dp110 differentiation phenotype by simultaneous reduction of pntP2 strongly suggests that pntP2 acts downstream of Dp110 and InR/TOR signaling in a pathway that regulates the temporal control of differentiation. It has been suggested that later differentiating PRs require higher levels of EGFR activity than their earlier differentiating neighbors. In particular, the activation of PR 7 requires both EGFR and Sevenless RTKs (Freeman 1996). In the case of InR/TOR pathway activation it may be that, through its regulation of EGFR downstream targets, the “second burst” of RTK activity is enhanced causing PRs 1, 6, and 7 and cone cells to differentiate precociously. There may also be other as yet unidentified factors through which the InR/TOR pathway controls the expression of Aos and rho in PRs 2–5 and 8.
Activation of insulin and insulin-like growth factor receptors in mammalian systems is well known to elicit a response via the Ras/MAPK pathway (Baltensperger et al. 1993; Skolnik et al. 1993; Downward 2003;). However, loss of the InR in the Drosophila eye does not result in a loss of PRs, a hallmark of the Ras pathway (Brogiolo et al. 2001), nor does mutation of the putative Drk binding site in chico affect the function of the Drosophila IRS (Oldham et al. 2002). In accordance with these data we do not observe any change in dpERK staining when the InR/TOR pathway is activated in the eye disc. Rather than a direct activation of Ras signaling by the InR, our data suggest that in the developing eye crosstalk between these pathways occurs at the level of regulation of the expression of EGFR transcriptional outputs. The most proximal component of the EGFR pathway that is regulated by InR/TOR signaling is pntP2. However, our data suggest that temporal control of PR differentiation requires concerted regulation of EGFR transcriptional outputs, since overexpression of pntP2 alone is not sufficient to cause precocious differentiation, whereas overexpression of activated EGFR is sufficient (supplemental Figure 4). Interestingly, microarray analyses of Drosophila and human cells have shown that the InR/TOR pathway regulates the expression of hundreds of genes (Peng et al. 2002; Guertin et al. 2006a). The mechanism by which this transcriptional control is exerted has yet to be elucidated. It will be interesting in future to determine the extent of transcriptional crosstalk between InR/TOR and EGFR pathways in developing neurons.
Acknowledgments
We gratefully acknowledge the generous gifts of antibodies or fly stocks from Sally Leevers, Nic Tapon, Hugo Bellen, Ernst Hafen, D. J. Pan, Nahum Sonenberg, Christian Klämbt, Frank Pichaud, and Matthew Freeman. This work was supported by Cancer Research United Kingdom, the United States Department of Defense, and the Tuberous Sclerosis Alliance.
References
- Astrinidis, A., T. P. Cash, D. S. Hunter, C. L. Walker, J. Chernoff et al., 2002. Tuberin, the tuberous sclerosis complex 2 tumor suppressor gene product, regulates Rho activation, cell adhesion and migration. Oncogene 21 8470–8476. [DOI] [PubMed] [Google Scholar]
- Baltensperger, K., L. M. Kozma, A. D. Cherniack, J. K. Klarlund, A. Chawla et al., 1993. Binding of the Ras activator son of sevenless to insulin receptor substrate-1 signaling complexes. Science 260 1950–1952. [DOI] [PubMed] [Google Scholar]
- Baonza, A., T. Casci and M. Freeman, 2001. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr. Biol. 11 396–404. [DOI] [PubMed] [Google Scholar]
- Baonza, A., C. M. Murawsky, A. A. Travers and M. Freeman, 2002. Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell Biol. 4 976–980. [DOI] [PubMed] [Google Scholar]
- Barrio, R., J. F. de Celis, S. Bolshakov and F. C. Kafatos, 1999. Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev. Biol. 215 33–47. [DOI] [PubMed] [Google Scholar]
- Bateman, J. M., and H. McNeill, 2004. Temporal control of differentiation by the insulin receptor/tor pathway in Drosophila. Cell 119 87–96. [DOI] [PubMed] [Google Scholar]
- Bateman, J. M., and H. McNeill, 2006. Insulin/IGF signalling in neurogenesis. Cell. Mol. Life Sci. 63 1701–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, C. A., and K. Moses, 2000. Determination of Drosophila photoreceptors: timing is everything. Cell. Mol. Life Sci. 57 195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton, J. S., W. K. Lockwood, L. Li, S. M. Cohen and B. A. Edgar, 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2 239–249. [DOI] [PubMed] [Google Scholar]
- Brogiolo, W., H. Stocker, T. Ikeya, F. Rintelen, R. Fernandez et al., 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11 213–221. [DOI] [PubMed] [Google Scholar]
- Brunner, D., K. Ducker, N. Oellers, E. Hafen, H. Scholz et al., 1994. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature 370 386–389. [DOI] [PubMed] [Google Scholar]
- Cai, S.-L., A. R. Tee, J. D. Short, J. M. Bergeron, K. Kim et al., 2006. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 173 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward, J., 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3 11–22. [DOI] [PubMed] [Google Scholar]
- Freeman, M., 1996. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87 651–660. [DOI] [PubMed] [Google Scholar]
- Freeman, M., 1997. Cell determination strategies in the Drosophila eye. Development 124 261–270. [DOI] [PubMed] [Google Scholar]
- Freeman, M., B. E. Kimmel and G. M. Rubin, 1992. a Identifying targets of the rough homeobox gene of Drosophila: evidence that rhomboid functions in eye development. Development 116 335–346. [DOI] [PubMed] [Google Scholar]
- Freeman, M., C. Klambt, C. S. Goodman and G. M. Rubin, 1992. b The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell 69 963–975. [DOI] [PubMed] [Google Scholar]
- Gabay, L., R. Seger and B. Z. Shilo, 1997. In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277 1103–1106. [DOI] [PubMed] [Google Scholar]
- Gao, X., and D. Pan, 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15 1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X., Y. Zhang, P. Arrazola, O. Hino, T. Kobayashi et al., 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4 699–704. [DOI] [PubMed] [Google Scholar]
- Golembo, M., R. Schweitzer, M. Freeman and B. Z. Shilo, 1996. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development 122 223–230. [DOI] [PubMed] [Google Scholar]
- Goncharova, E., D. Goncharov, D. Noonan and V. P. Krymskaya, 2004. TSC2 modulates actin cytoskeleton and focal adhesion through TSC1-binding domain and the Rac1 GTPase. J. Cell Biol. 167 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin, D. A., K. V. Guntur, G. W. Bell, C. C. Thoreen and D. M. Sabatini, 2006. a Functional genomics identifies TOR-regulated genes that control growth and division. Curr. Biol. 16 958–970. [DOI] [PubMed] [Google Scholar]
- Guertin, D. A., D. M. Stevens, C. C. Thoreen, A. A. Burds, N. Y. Kalaany et al., 2006. b Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 11 859–871. [DOI] [PubMed] [Google Scholar]
- Hara, K., K. Yonezawa, Q. P. Weng, M. T. Kozlowski, C. Belham et al., 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273 14484–14494. [DOI] [PubMed] [Google Scholar]
- Harrington, L. S., G. M. Findlay, A. Gray, T. Tolkacheva, S. Wigfield et al., 2004. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, G., M. Altmann, J. M. Sierra, H. Urlaub, R. Diez del Corral et al., 2005. Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech. Dev. 122 529–543. [DOI] [PubMed] [Google Scholar]
- Inoki, K., and K. L. Guan, 2006. Complexity of the TOR signaling network. Trends Cell Biol. 16 206–212. [DOI] [PubMed] [Google Scholar]
- Inoki, K., T. Zhu and K. L. Guan, 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115 577–590. [DOI] [PubMed] [Google Scholar]
- Kimmel, B. E., U. Heberlein and G. M. Rubin, 1990. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev. 4 712–727. [DOI] [PubMed] [Google Scholar]
- Klambt, C., 1993. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development 117 163–176. [DOI] [PubMed] [Google Scholar]
- Lachance, P. E., M. Miron, B. Raught, N. Sonenberg and P. Lasko, 2002. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell. Biol. 22 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, R. F., C. Roy, T. J. Diefenbach, H. V. Vinters, M. W. Johnson et al., 2000. The TSC1 tumour suppressor hamartin regulates cell adhesion through ERM proteins and the GTPase Rho. Nat. Cell Biol. 2 281–287. [DOI] [PubMed] [Google Scholar]
- Lee, T., and L. Luo, 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22 451–461. [DOI] [PubMed] [Google Scholar]
- Leevers, S. J., and E. Hafen, 2004. Growth regulation by insulin and TOR signalling in Drosophila, pp. 167–192 in Cell Growth, edited by M. N. Hall, R. Raff and G. Thomas. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Long, X., Y. Lin, S. Ortiz-Vega, K. Yonezawa and J. Avruch, 2005. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15 702–713. [DOI] [PubMed] [Google Scholar]
- Neufeld, T. P., A. F. de la Cruz, L. A. Johnston and B. A. Edgar, 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93 1183–1193. [DOI] [PubMed] [Google Scholar]
- O'Neill, E. M., I. Rebay, R. Tjian and G. M. Rubin, 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78 137–147. [DOI] [PubMed] [Google Scholar]
- Oldham, S., H. Stocker, M. Laffargue, F. Wittwer, M. Wymann et al., 2002. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development 129 4103–4109. [DOI] [PubMed] [Google Scholar]
- Peng, T., T. R. Golub and D. M. Sabatini, 2002. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol. Cell. Biol. 22 5575–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, C. J., H. Huang and T. Xu, 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105 357–368. [DOI] [PubMed] [Google Scholar]
- Radimerski, T., J. Montagne, M. Hemmings-Mieszczak and G. Thomas, 2002. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 16 2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, J. D., and N. Sonenberg, 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433 477–480. [DOI] [PubMed] [Google Scholar]
- Rusten, T. E., K. Lindmo, G. Juhasz, M. Sass, P. O. Seglen et al., 2004. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell 7 179–192. [DOI] [PubMed] [Google Scholar]
- Sarbassov, D. D., D. A. Guertin, S. M. Ali and D. M. Sabatini, 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307 1098–1101. [DOI] [PubMed] [Google Scholar]
- Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan et al., 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5 566–571. [DOI] [PubMed] [Google Scholar]
- Scholz, H., J. Deatrick, A. Klaes and C. Klambt, 1993. Genetic dissection of pointed, a Drosophila gene encoding two ETS-related proteins. Genetics 135 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz, H., E. Sadlowski, A. Klaes and C. Klambt, 1997. Control of midline glia development in the embryonic Drosophila CNS. Mech. Dev. 64 137–151. [DOI] [PubMed] [Google Scholar]
- Scott, R. C., O. Schuldiner and T. P. Neufeld, 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7 167–178. [DOI] [PubMed] [Google Scholar]
- Shah, O. J., Z. Wang and T. Hunter, 2004. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14 1650–1656. [DOI] [PubMed] [Google Scholar]
- Skolnik, E. Y., A. Batzer, N. Li, C. H. Lee, E. Lowenstein et al., 1993. The function of GRB2 in linking the insulin receptor to Ras signaling pathways. Science 260 1953–1955. [DOI] [PubMed] [Google Scholar]
- Song, H., M. Nie, F. Qiao, J. U. Bowie and A. J. Courey, 2005. Antagonistic regulation of Yan nuclear export by Mae and Crm1 may increase the stringency of the Ras response. Genes Dev. 19 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon, N., N. Ito, B. J. Dickson, J. E. Treisman and I. K. Hariharan, 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105 345–355. [DOI] [PubMed] [Google Scholar]
- Tootle, T. L., P. S. Lee and I. Rebay, 2003. CRM1-mediated nuclear export and regulated activity of the receptor tyrosine kinase antagonist YAN require specific interactions with MAE. Development 130 845–857. [DOI] [PubMed] [Google Scholar]
- Um, S. H., F. Frigerio, M. Watanabe, F. Picard, M. Joaquin et al., 2004. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431 200–205. [DOI] [PubMed] [Google Scholar]
- Voas, M. G., and I. Rebay, 2004. Signal integration during development: insights from the Drosophila eye. Dev. Dyn. 229 162–175. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. Ready, 1993. Pattern formation in the Drosophila retina, pp. 1277–1326 in The Development of Drosophila melanogaster, edited by M. Bates and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Wullschleger, S., R. Loewith and M. N. Hall, 2006. TOR signaling in growth and metabolism. Cell 124 471–484. [DOI] [PubMed] [Google Scholar]
- Xu, C., R. C. Kauffmann, J. Zhang, S. Kladny and R. W. Carthew, 2000. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103 87–97. [DOI] [PubMed] [Google Scholar]
- Yang, L., and N. E. Baker, 2003. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev. Cell 4 359–369. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., X. Gao, L. J. Saucedo, B. Ru, B. A. Edgar et al., 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5 578–581. [DOI] [PubMed] [Google Scholar]