Abstract
Wee1 kinases regulate the cell cycle through inhibitory phosphorylation of cyclin-dependent kinases (CDKs). Eukaryotic cells express multiple CDKs, each having a kinase subunit (Cdk) and a regulatory “cyclin” subunit that function at different stages of the cell cycle to regulate distinct processes. The cyclin imparts specificity to CDK–substrate interactions and also determines whether a particular CDK is subject to Wee1 regulation. Saccharomyces Wee1 (Swe1) inhibits Cdc28 (Cdk1) associated with the mitotic cyclin, Clb2, but not with the G1 (Cln1, -2, and -3) or the S-phase (Clb5 and -6) cyclins. Here, we show that this specificity depends on two amino acids associated with a conserved “hydrophobic patch” (HP) motif on the cyclin surface, which mediates specificity of CDK–substrate interactions. Mutation of Clb2 residues N260 and K270 largely abrogates Clb2-Cdc28 regulation by Swe1, and reciprocal mutation of the corresponding residues in Clb5 can subject Clb5-Cdc28 to regulation by Swe1. Swe1 phosphorylation by Clb2-Cdc28, which is thought to activate Swe1 kinase, depends on N260 and K270, suggesting that specific regulation of Clb2-Cdc28 by Swe1 derives from the specific ability of Clb2 to target Swe1 for activating phosphorylation. The stable association of Swe1 with Clb2-Cdc28 also depends on these residues, suggesting that Swe1 may competitively inhibit Clb2-Cdc28 interactions with substrates, in addition to its well-known function as a regulator of CDK activity through tyrosine phosphorylation.
CYCLIN-DEPENDENT kinases (CDKs) govern the execution of crucial events associated with the cell-division cycle of all eukaryotic cells, including chromosome replication and segregation, and cell growth and division (Morgan 1997). CDKs consist of a catalytic kinase subunit termed Cdk and a regulatory subunit termed cyclin, which is required for activity of the kinase. Eukaryotic cells typically express multiple, distinct, but related cyclins, and metazoans express multiple Cdk subunits as well. Cyclins are aptly named to describe their oscillating abundance during the cell-division cycle resulting from their cell cycle-regulated expression and proteolysis, which serves to control the level of specific CDK activities at different cell-cycle stages. Evolutionary divergence among the cyclins, manifested in their differential temporal expression, subcellular localization, and ability to interact with specific substrates, confers functional specificity to the different CDKs in governance of the cell cycle (Miller and Cross 2001; Murray 2004).
CDKs also are regulated by interaction with proteins of the p21/p27 family that function as tumor suppressors and/or developmental regulators in metazoans by broadly inhibiting CDK activities (Morgan 1996, 1997; Mendenhall and Hodge 1998). The crystal structure of cyclin A-Cdk2 bound to p27 has revealed multiple facets of this inhibitory mechanism, which include deformation of the Cdk2 catalytic domain and occlusion of the ATP binding pocket, as well as binding a conserved region of cyclin A that has also been implicated in mediating interaction with cyclin A-Cdk2 substrates (Russo et al. 1996; Schulman et al. 1998). This region of cyclin A contains a number of hydrophobic amino acids that form a hydrophobic patch motif (HP) on its surface and conserved glutamate and glutamine, all of which make contacts with p27. The HP region is conserved in B-type cyclins of budding yeast where it contributes to the specificity of different CDKs for their substrates (Archambault et al. 2004, 2005; Wilmes et al. 2004; Loog and Morgan 2005).
Additional regulation of CDK activity occurs through inhibitory phosphorylation of Cdk on a highly conserved tyrosine (Y15 in Cdc2) (and the adjacent threonine in vertebrates) near the catalytic site. Phosphorylation is carried out by a Wee1 tyrosine kinase, as well as Mik1 (in fission yeast) or Myt1 (in vertebrates) protein kinase (Gould and Nurse 1989; Lundgren et al. 1991; Gu et al. 1992; Mueller et al. 1995; Watanabe et al. 1995; Booher et al. 1997; Liu et al. 1997). In vertebrates, tyrosine (and threonine) phosphorylation negatively regulates the S phase promoting Cdk2 and the M phase promoting Cdc2 (Cdk1) and serves as an important target of cell-cycle-checkpoint mechanisms in response to genotoxic agents (Gu et al. 1992; Jin et al. 1996; Chow et al. 2003). Dephosphorylation of Cdk2 and Cdc2, which is required for progression through S phase and M phase, respectively, is accomplished by Cdc25 phosphatase(s) (Russell and Nurse 1986). Inhibition of Cdc25 activity is a critical mechanism of the checkpoints that regulate CDK function through Y15 phosphorylation.
In yeasts, Cdk tyrosine phosphorylation appears to impinge primarily on mitosis. In fission yeast, as in vertebrates, Cdk tyrosine phosphorylation by Wee1/Mik1 and dephosphorylation by Cdc25 are essential events that determine the timing of mitosis in relation to other cell-cycle events and mediate mitotic delay in response to DNA damage or replication blocks (Rhind and Russell 1998, 2001). In the budding yeast, Saccharomyces cerevisiae, the cyclin-dependent kinase Cdc28 (analogous to Cdc2) is also subject to a cycle of tyrosine (Y19) phosphorylation by Saccharomyces Wee1 (Swe1) and dephosphorylation by Cdc25 Mitotic inducer homolog 1 (Mih1), and genotoxic stress upregulates tyrosine phosphorylation (Russell et al. 1989; Booher et al. 1993; Liu and Wang 2006). However, phosphorylation/dephosphorylation of Cdc28 is not essential for cell-cycle progression and Cdc28 phosphorylation is dispensable for mitotic delay in response to genotoxic stresses (Amon et al. 1992; Sorger and Murray 1992). Nevertheless, Y19 phosphorylation of Cdc28 enforces a premitotic delay in response to defects in cellular morphogenesis, although a recent study has suggested that cell size rather than morphology is monitored by this checkpoint (Lew and Reed 1995; Harvey and Kellogg 2003; McNulty and Lew 2005). The finding that Cdk tyrosine phosphorylation exclusively regulates mitosis in yeasts suggests specificity in the interaction of Wee1/Swe1 with different CDKs in these organisms.
In S. cerevisiae, Cdc28 associates with one of nine cyclins to control the cell cycle (Mendenhall and Hodge 1998). Three G1 cyclins (Cln1–3) regulate Cdc28 activity in G1 phase, and six B-type cyclins (Clb1–6) regulate Cdc28 activity in S, G2, and M phases. Clb5 and Clb6 are expressed in late G1 and early S phases and are primarily responsible for initiating chromosomal DNA replication; Clb3 and Clb4 are expressed in late S phase and drive spindle assembly; and Clb1 and Clb2 are expressed in G2 phase and control chromosome segregation. Individual cyclins have evolved specific functions that allow them to carry out these distinct roles, and this has been particularly well studied with Clb5 and Clb2 as prototype S-phase and M-phase cyclins, respectively (Miller and Cross 2001; Murray 2004). Genetic studies have shown that these cyclins cannot fully substitute for each other's functions, even when expressed with identical timing (Cross et al. 1999; Cross and Jacobson 2000; Donaldson 2000). For example, early expression of Clb2 (or Clb4) from the CLB5 promoter does not activate S phase on schedule, arguing that Clb5 is inherently better at targeting one or more replication initiation factors. Subsequent biochemical studies comparing Clb2- and Clb5-Cdc28 identified preferred substrates of Clb5-Cdc28, many of which function in S phase, clearly indicating functional specificity (Archambault et al. 2004; Loog and Morgan 2005). This substrate preference depends on the HP of Clb5 and an RXL (or Cy) motif in the substrate.
Cyclins also confer specificity to regulation by Swe1 (Booher et al. 1993; Hu and Aparicio 2005; Keaton et al. 2007). Indeed, most of the delay in S-phase entry that occurs when Clb2, Clb3, or Clb4 is expressed early in the cell cycle results from Swe1 inhibition of these Clb-Cdc28 complexes, rather than from kinetic differences in their substrate-targeting (Hu and Aparicio 2005). This previous study shows that Swe1 inhibits Clb2-Cdc28, modestly inhibits Clb3- and Clb4-Cdc28, and does not inhibit Clb5- or Clb6-Cdc28 (or Cln) complexes. In addition, kinase activity of immunoprecipitated Clb2-Cdc28 (using histone H1 as substrate) is enhanced by SWE1 deletion, whereas Clb5-Cdc28-associated kinase activity is unaffected by SWE1 deletion (Hu and Aparicio 2005). This specificity is also reflected in more rapid Y19 phosphorylation of Cdc28 associated with Clb2 vs. Clb5 (Keaton et al. 2007). The structural basis for cyclin-specific inhibition of Cdc28 by Swe1 is unknown.
Because the Clb5 HP is required for its preferential interaction with certain substrates, we tested whether structural differences between the Clb5 and Clb2 HP regions account for the differential SWE1 sensitivity of the corresponding CDKs. Here we report that two amino acid positions in the Clb2 HP region are required for the inhibitory effect of SWE1 on DNA replication stimulated by early expressed CLB2. Stable association and mutual phosphorylations between Swe1 and Clb2-Cdc28 are disrupted by mutations of the HP region that mimic the Clb5 HP. We also show that mutation of the corresponding positions in Clb5 (to mimic Clb2) subjects Clb5-Cdc28 to inhibition by overexpressed Swe1, indicating that the HP region imparts specificity in this interaction. These findings emphasize and extend the importance of the HP region in determining the specificity of CDK regulation.
MATERIALS AND METHODS
Plasmid and strain constructions:
A QuikChange XL site-directed mutagenesis kit (Stratagene no.200516) was used according to the manufacturer's recommendations. Primer sequences used for mutagenesis are available upon request. Plasmids p314-C5p-CLB2 (K270A; K270E; N260M; K270A, N260M; and K270E, N260M) and p314-C5p-CLB5 (E207K; M197N; E207K, M197N) were generated by site-directed mutagenesis using p314-C5p-CLB2 (pC5C2-3NF; Cross et al. 1999) or p314-C5p-CLB5 as template, respectively. The 1.9-kb fragment including the CLB5 coding sequence (beginning immediately after the initiator ATG) plus 500 bp of downstream sequence (containing an EcoRI site) was amplified by PCR, digested with NotI, and partially digested with EcoRI to generate a 1.3-kb fragment; this 1.3-kb NotI–EcoRI fragment of CLB5 was ligated with the 6.5-kb fragment from p314-C5p-CLB2 digested with the same enzymes, yielding p314-C5p-CLB5. p316-C5p-CLB5 was constructed by inserting the 3.1-kb KpnI–SacI fragment of p314-C5p-CLB5 into KpnI–SacI-digested pRS316. p415-C5p-CLB5 was constructed by ligating the 2-kb SalI–NotI fragment of p314-C5p-CLB5 with the 7-kb SalI–NotI fragment of p415-C5p-CLB2. To create p415-C5p-CLB2, the 3.3-kb ApaI–SacII fragment of pC5C2-3NF was inserted into ApaI–SacII-digested pRS415. p415-C5p-CLB2 (N260M, K270A) and p415-C5p-CLB2 (N260M, K270E) were constructed by inserting the 3.3-kb ApaI–SacI fragment from p314-C5p-CLB2 (N260M, K270A) and p314-C5p-CLB2 (N260M, K270E), respectively, into ApaI–SacI-digested pRS415. The 2.5-kb NotI–PstI fragment containing the CLB2 ORF plus 600-bp upstream and 400-bp downstream sequences was amplified by PCR and cloned into NotI–PstI-digested pRS415, yielding p415-C2p-CLB2. The 637-bp XbaI–BglII C-terminal fragment of CLB2 from p314-C5p-CLB2 (N260M, K270A) and p314-C5p-CLB2 (N260M, K270E) was ligated with the 7.8-kb XbaI–BglII-digested p415-C2p-CLB2, yielding pRS415-C2p-CLB2 (N260M, K270A) and p415-C2p-CLB2 (N260M, K270E), respectively. p306-SWE1-MYC6 was constructed by inserting a 1.3-kb SacII–NotI-digested, PCR-produced C-terminal fragment of SWE1 into SacII–NotI-digested p306-ORC1-MYC6 (our unpublished data). Plasmid pΔclb2-URA3 was constructed by four-way ligation of a 560-bp PCR-amplified XbaI–XhoI 5′ CLB2 fragment, a 560-bp PCR-amplified BamHI–EcoRI 3′ CLB2 fragment, a 1.2-kb XhoI–BamHI URA3 fragment, and XbaI–EcoRI-digested pBluescript-KS+. The 1.6-kb XhoI–SacII fragment from p306-SWE1-MYC6 was inserted into XhoI–SacII-digested pRS405, yielding p405-SWE1-MYC6. All DNA sequences produced through PCR (including PCR mutagenesis) were sequenced to confirm that only desired mutations were introduced.
All strains are derived from W303-1a and are described in Table 1. Epitope-tagging, promoter replacement, and gene deletions were constructed as described (Longtine et al. 1998), with the following exceptions: SWE1 was deleted using pΔswe1-URA3 digested with NotI and EcoRI (Hu and Aparicio 2005); CLB2 was HA-tagged with plasmid pDK82B (TRP1) digested with XcmI (from D. R. Kellogg); SLD2 was MYC-tagged with plasmid p404-SLD2-MYC9 digested with MscI (Hu and Aparicio 2005); SWE1 was MYC-tagged with ClaI-digested p306-SWE1-MYC6 or SnaBI-digested p405-SWE1-MYC6; CLB2 was deleted using pΔclb2-URA3 digested with XbaI and HindIII. For introducing point mutations into the CLB5 or CLB2 locus, XhoI-digested p415-C5p-CLB2 (N260M; K270A; K270E; N260M, K270A; and N260M, K270E), or p415-C5p-CLB5 (E207K; M197N; and E207K, M197N), or p415-C2p-CLB2 (N260M, K270E) was cotransformed with pRS414 into a clb5Δ∷URA3 or clb2Δ∷URA3 host. Transformants were selected on −Trp plates and replica-plated onto FOA plates; FOA-resistant transformants were confirmed by PCR. We constructed other strains by standard mating and spore dissections. Strain genotypes are listed in Table 1.
TABLE 1.
Yeast strains
| Strain | Genotype (all are MATα unless otherwise indicated) | Source |
|---|---|---|
| DGy221 | bar1Δ∷hisG, clb6Δ∷LEU2 | Gibson et al. (2004) |
| FHy116 | bar1Δ∷URA3, clb5Δ∷CLB2, clb6Δ∷LEU2 | Hu et al. (2005) |
| FHy134 | bar1Δ∷URA3, clb5Δ∷CLB2, clb6Δ∷LEU2, swe1Δ∷HIS5 | Hu et al. (2005) |
| FHy136 | bar1Δ∷hisG, clb6Δ∷LEU2, swe1Δ∷HIS5 | Hu et al. (2005) |
| FHy259 | bar1Δ∷hisG, clb2Δ∷CLB2-3HA(TRP1) | Hu et al. (2005) |
| FHy284 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (A270) | This study |
| FHy285 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (E270) | This study |
| FHy302 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (M260, E270) | This study |
| FHy303 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (M260, E270), swe1Δ∷URA3 | This study |
| FHy336 | bar1Δ∷hisG, clb6Δ∷LEU2, sld2Δ∷SLD2-9MYC(TRP1) | Hu et al. (2005) |
| FHy340 | bar1Δ∷URA3, clb6Δ∷LEU2, clb5Δ∷CLB2, sld2Δ∷SLD2-9MYC(TRP1) | Hu et al. (2005) |
| FHy341 | bar1Δ∷URA3, clb6Δ∷LEU2, clb5Δ∷CLB2, swe1Δ∷HIS5, sld2Δ∷SLD2-9MYC(TRP1) | Hu et al. (2005) |
| FHy363 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (M260) | This study |
| FHy364 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (M260, A270) | This study |
| FHy366 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (M260, A270), swe1Δ∷URA3 | This study |
| FHy374 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB5 (N197, K207) | This study |
| FHy375 | bar1Δ∷hisG, swe1Δ∷SWE1-6MYC (URA3) | This study |
| YGy24 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB5(N197, K207), swe1Δ∷URA3 | This study |
| YGy29 | bar1Δ∷hisG, swe1Δ∷KanMX-GAL1-SWE1-6MYC (URA3) | This study |
| YGy54 | bar1Δ∷hisG, clb6Δ∷LEU2, swe1Δ∷URA3,clb5Δ∷KanMX-GAL1-CLB2 (M260, E270)-3HA (TRP1) | This study |
| YGy65 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷KanMX-GAL1-CLB2-3HA (TRP1), swe1Δ∷URA3 | This study |
| YGy69 | bar1Δ∷hisG, clb2Δ∷CLB2(M260, E270), swe1Δ∷KanMX-GAL1-SWE1-6MYC(LEU2) | This study |
| YGy72 | bar1Δ∷hisG, swe1Δ∷KanMX-GAL1-SWE1-6MYC(LEU2) | This study |
| YGy73 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (M260, E270), swe1Δ∷URA3, sld2Δ∷SLD2-9MYC(TRP1) | This study |
| YGy74 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB2 (M260, E270), sld2Δ∷SLD2-9MYC(TRP1) | This study |
| YGy78 | bar1Δ∷hisG, clb2Δ∷CLB2 (M260, E270)-3HA(TRP1) | This study |
| YGy80 | bar1Δ∷hisG, clb2Δ∷CLB2(M260, E270), swe1Δ∷SWE1-6MYC (LEU2) | This study |
| YGy81 | bar1Δ∷hisG, clb6Δ∷LEU2, swe1Δ∷KanMX-GAL1-SWE1 | This study |
| YGy82 | bar1Δ∷hisG, clb6Δ∷LEU2, clb5Δ∷CLB5 (N197,K207), swe1Δ∷KanMX-GAL1-SWE1 | This study |
Yeast methods:
α-Factor block-and-release experiments were performed as described previously (Aparicio et al. 2004). YEP medium was used for all experiments, with 2% glucose unless otherwise indicated. DNA content analysis (FACS analysis) has been described previously (Aparicio et al. 2004). Budding analysis was determined by microscopic analysis of 100 or 200 formaldehyde-fixed cells at each time point.
Protein analysis:
Gel electrophoresis was carried out at constant current, 8 mA in stacking gel and 18 mA in resolving mini-gel. Proteins were separated on 10% (75:1) SDS polyacrylamide gels. Antibodies for Western blot analysis were used as follows: anti-Myc (9E10) antibody (Covance-BAbCo, 1:2000); anti-Clb2 (Santa Cruz SC-9071, 1:500); anti-Cdc2 phosphotyrosine (Cell Signaling Technology, 1:1000); anti-PSTAIRE (Santa Cruz SC-53, 1:500). Chemiluminescent signal was captured using Bio-Rad ChemiDoc XRS 170-8070 system and QuantityOne Analysis software after treatment with SuperSignal Elisa Femto maximum-sensitivity substrate (Pierce). For examination of Sld2 phosphorylation, protein extracts were prepared by trichloro-acetic acid (TCA) precipitation as described (Foiani et al. 1995).
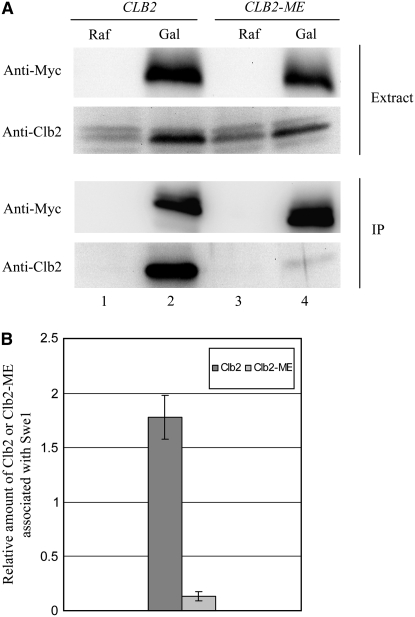
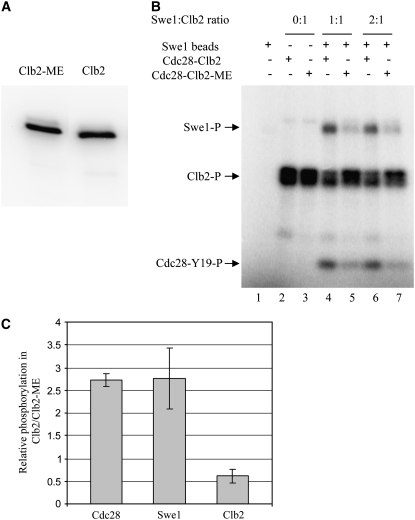
Co-immunoprecipitation (Figure 5) of Clb2-Ha3-Cdc28 and Swe1-Myc6 from whole-cell extracts was carried out as described in (Mortensen et al. 2002; Harvey et al. 2005) with some modifications. Cells were broken by bead beating in lysis buffer containing protease inhibitor cocktail (Roche). Four milligrams of total protein was incubated with 9E10 (1:100) in 500 μl volume for 1 hr at 4°; 30 μl of protein G-sepharose beads (50:50 slurry) was added and incubated with gentle rotation for 1 hr at 4°. The beads were washed and suspended in 45 μl of protein sample buffer and heated at 95° for 5 min. Fifteen microliters of supernatant was loaded onto gels. One percent of the whole-cell extract was loaded in the “extract” lanes. Cdc28-Clb2-Ha3 was immunoprecipitated with anti-Ha antibody (16B12, 1:250) for examination of Cdc28-Y19 phosphorylation.
Figure 5.—
Clb2 requires N and K for interaction with Swe1. Unsynchronized GAL-SWE1-MYC6 cells expressing CLB2 (YGy72) or CLB2-ME (YGy69) from the endogenous CLB2 promoter were incubated in YEP/2% raffinose (Raf) or YEP/2% galactose (Gal) for 2 hr at 30°. (A) Equal amounts of soluble proteins (Extract) were subjected to immunoprecipitation (IP) with anti-Myc antibody, followed by immunoblot analysis of the precipitated proteins with anti-Myc and anti-Clb2 antibodies. (B) Quantification of the amounts of Clb2 and Clb2-ME relative to the amounts of precipitated Swe1. The values are based on chemiluminescent signal intensities and are dimensionless; error bars indicate standard deviation (n = 2).
Immunoaffinity isolation of Clb2-Ha3-Cdc28 was carried out as described with some modifications (Harvey et al. 2005). Five hundred milliliters of cells expressing Clb2-Ha3 or Clb2-N260M, K270E-Ha3 from the GAL1 promoter were harvested after 3 hr of growth in YEP + 2% galactose at 30°. The cells were washed and resuspended in 5 ml lysis buffer containing phosphatase inhibitors (25 mm β-glycerophosphate, 50 mm NaF, 100 μm sodium orthovanadate) and protease inhibitors [1 mm PMSF, 50 μg/ml TPCK and protease inhibitor cocktail (Roche)] at 4°. The cell suspension was distributed into 10 2-ml tubes for cell lysis by bead beating in a Fastprep instrument (MP Biomedicals). The extracts were pooled (∼5 ml) and incubated with anti-Ha antibody mixture (16B12, 1:500 and 12CA5, 1:250) overnight at 4°, followed by 1 hr incubation with 300 μl of protein G-sepharose beads with gentle rotation. The beads were washed and incubated at 23° for 15 min with 300 μl of elution buffer containing 0.5 mg/ml Ha dipeptide (YPYDVPDYASLYPYDVPDYA, University of Southern California Norris Cancer Center) with occasional agitation. The beads were precipitated by centrifugation and the eluate was collected. This step was repeated once at 23° and three additional times at 30°. The eluates were pooled and concentrated to ∼120 μl using an Amicon Ultra-4 (Millipore).
Kinase reactions:
Swe1-Myc6 immunoprecipitate beads were prepared freshly for each experiment as described for isolation of Clb2-Ha3-Cdc28 above. Swe1-Myc6 beads were suspended in 400 μl of kinase buffer (50 mm Hepes-KOH, pH 7.6, 2 mm MgCl2, 10% glycerol, 1 mm DTT, 0.05% Tween-20). Three microliters of purified Clb2-Ha3-Cdc28 or Clb2-ME-Ha3-Cdc28 was incubated with 0, 3, or 6 μl of freshly prepared Swe1-Myc6 immunoprecipitate beads on ice. Eleven microliters of kinase buffer containing 10 μm ATP and 0.5 μl [γ-32P]ATP (6000 Ci/mmol) was added to each reaction. Samples were incubated for 20 min at 30° with occasional agitation and reactions were terminated by the addition of 4 μl of 6× SDS–PAGE sample buffer. The samples were boiled for 3 min and separated by SDS–PAGE. The gels were exposed to the imaging screen K-HD (Bio-Rad) for 2–4 hr; the radioactive signal of 32P incorporation was captured on an FX Scanner (Bio-Rad) and analyzed by QuantityOne Analysis software.
RESULTS
Identification of Clb2 residues required for its regulation by Swe1:
To examine the structural basis for specificity in Swe1 inhibition of different Clb-Cdc28 complexes, we aligned the sequences of Clb1 to -6 encompassing the conserved region implicated in Clb5 interactions with substrates and in cyclin A interaction with p27 inhibitor and substrates (Russo et al. 1996; Schulman et al. 1998; Cross and Jacobson 2000). This region contains the HP motif (consisting of M197, L201, and W204 in Clb5) and two additional residues (E207 and Q241 in Clb5) that make contacts in the cyclin A-p27 structure and have also been implicated in Clb5–substrate interactions (Figure 1) (Russo et al. 1996; Cross and Jacobson 2000). Among these five residues, positions 2, 3, and 5 are invariant among the six Clbs, with a single exception (Figure 1, amino acids in boldface type). However, positions 1 and 4 exhibit significant variability, with the respective residues being N and K in Clb1 and Clb2, F and Q in Clb3 and Clb4, and M and E in Clb5 and Clb6. Interestingly, these amino acid differences correspond to these cyclin pairs' relative susceptibilities to inhibition by Swe1 (Hu and Aparicio 2005), leading to the hypothesis that N and K at HP-region positions 1 and 4 mediate Swe1 interaction, while M and E at these positions do not.
Figure 1.—
Sequence alignment of Clb1-Clb6. Numbers to the left of the sequences refer to amino acid positions. Amino acid positions indicated in boldface type are numbered 1–5 for description in the text.
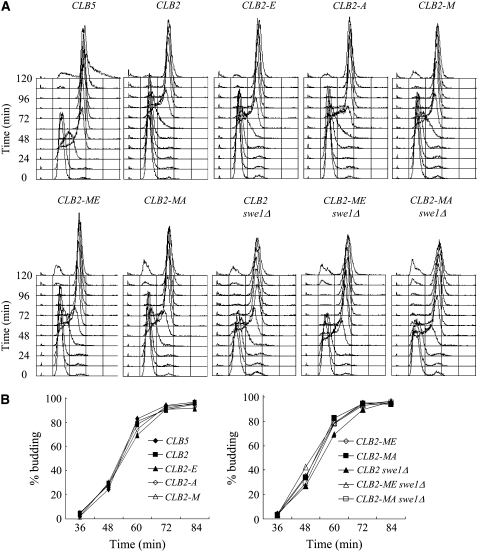
Expression of wild-type CLB2 from the CLB5 promoter does not stimulate replication initiation on schedule due to inhibition of early expressed Clb2 by Swe1 (Figure 2A) (Hu and Aparicio 2005). Hence, we reasoned that mutation of Clb2 residues that are required for regulation by Swe1 should allow Clb2 to trigger earlier S-phase entry in the presence of Swe1. To determine the importance of the N and K residues of Clb2 for regulation of Clb2-Cdc28 by Swe1, we mutated these residues to the corresponding residues of Clb5 (M and E, respectively), which does not appear to be subject to Swe1 regulation in vivo (Hu and Aparicio 2005). We also constructed CLB2 alleles with K mutated to A instead of E. The resulting single-mutant alleles, CLB2-N260M, CLB2-K270E, and CLB2-K270A, and double-mutant alleles, CLB2-N260M, K270E and CLB2-N260M, K270A, are referred to as CLB2-M, CLB2-E, CLB2-A, CLB2-ME, and CLB2-MA, respectively (the presence of a letter designating the allele or protein always indicating the mutation at position 1 and/or 4). A wild-type or mutated CLB2 allele was introduced into clb5Δ clb6Δ yeast cells under control of the CLB5 promoter at the endogenous locus, resulting in CLB2 expression in late G1 and early S phase. We monitored S-phase kinetics of these cells by DNA content analysis after release from G1 arrest with α-factor.
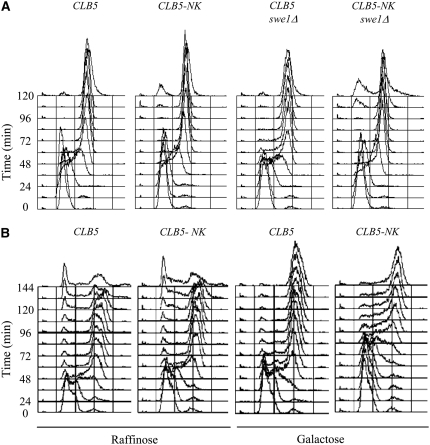
Figure 2.—
Clb2 HP region residues N and K mediate regulation by Swe1. Each strain expresses the indicated CLB5 or CLB2 allele from the endogenous CLB5 promoter and lacks CLB6. Strains DGy221 (CLB5), FHy116 (CLB2), FHy285 (CLB2-E), FHy284 (CLB2-A), FHy363 (CLB2-M), FHy302 (CLB2-ME), FHy364 (CLB2-MA), FHy134 (CLB2 swe1Δ), FHy303 (CLB2-ME swe1Δ), and FHy366 (CLB2-MA swe1Δ) were synchronized in G1 with α-factor and released at 23°. Samples were collected every 12 min for DNA content analysis (A) and budding analysis (B).
Analysis of the timing of S-phase entry shows that the M and E mutations of Clb2 enhance its ability to stimulate S phase in the presence of Swe1. Clb5-expressing cells enter S phase 12–24 min after α-factor release, whereas Clb2-expressing cells enter S phase at least 36 min later (Figure 2A). Expression of Clb2-M, Clb2-E, or Clb2-A advances S-phase entry by at least 12 min compared with Clb2; however, S-phase entry remains ∼24 min behind Clb5-expressing cells. The effect of each mutation is additive, as the double mutants Clb2-ME and Clb2-MA advance S phase by an additional 12 min, entering S phase with almost identical timing as Clb2-expressing cells lacking Swe1. These results suggest that the ME and MA mutations disrupt the interaction between the early expressed Clb2 protein and Swe1. The similar effects of the E and A mutations (singly or in combination with the M mutation) suggest that the positive charge of K at position 4 contributes to Swe1 interaction.
Consistent with the idea that the ME and MA mutations disrupt Clb2 interaction with Swe1, SWE1 deletion only slightly advances S-phase entry (<12 min) of Clb2-ME- and Clb2-MA-expressing cells (Figure 2A). Furthermore, swe1Δ cells expressing Clb2-ME or Clb2-MA enter S phase ∼12 min earlier than swe1Δ cells expressing wild-type Clb2, although still delayed compared with Clb5-expressing cells. This result suggests that Clb2-ME and Clb2-MA have enhanced ability to target replication substrates than Clb2, although still weaker than Clb5. The differential effects on the timing of DNA replication of these CLB2 mutations are not due to indirect effects on progression through G1 or release from pheromone arrest as all strains initiate budding with similar timing (Figure 2B).
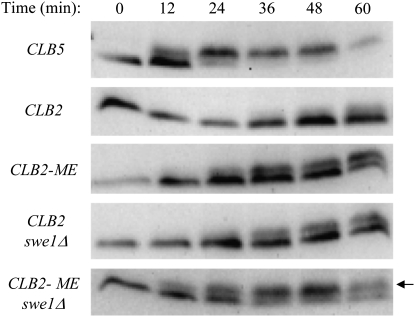
To examine directly CDK activity associated with Clb5, Clb2, and Clb2-ME, we analyzed phosphorylation of Sld2, a normal Clb5-Cdc28 substrate, phosphorylation of which is essential for replication initiation and can be monitored by its reduced gel mobility (Masumoto et al. 2002). The timing of Sld2 phosphorylation closely corresponds with the timing of S-phase entry in these strains (Figure 3, compare with Figure 2A). For example, expression of Clb5 results in partial Sld2 phosphorylation by 12 min and virtually complete phosphorylation by 24 min, corresponding to early and mid-S phase, respectively. In contrast, Clb2 expression results in partial Sld2 phosphorylation at 60 min, which corresponds to early S phase in these cells. As shown previously, deletion of SWE1 in Clb2-expressing cells significantly advances the timing and level of Sld2 phosphorylation (Figure 3), whereas deletion of SWE1 has no effect on timing of Sld2 phosphorylation (or S phase) in Clb5-expressing cells (data not shown; Hu and Aparicio 2005). In accordance with the effect of the Clb2 mutations on the timing of S-phase entry, expression of Clb2-ME significantly advances the timing of Sld2 phosphorylation compared with Clb2 in cells expressing SWE1. Furthermore, the effect of Clb2-ME is comparable to deletion of SWE1 in the Clb2-expressing cells, consistent with the idea that the Clb2 mutations disrupt interaction with Swe1.
Figure 3.—
Clb2-ME alters Sld2 phosphorylation kinetics. Each strain expresses the indicated CLB5 or CLB2 allele from the endogenous CLB5 promoter, lacks CLB6, and expresses SLD2-MYC9. Strains FHy336 (CLB5), FHy340 (CLB2), YGy74 (CLB2-ME), FHy341 (CLB2 swe1Δ), and YGy73 (CLB2-ME swe1Δ) were synchronized in G1 with α-factor and released at 23°; samples were collected every 12 min. Protein extracts were prepared by TCA precipitation and examined by immunoblotting with anti-Myc antibody. The arrow indicates phosphorylated Sld2.
Analysis of Sld2 phosphorylation also provides evidence that Clb2-ME has enhanced ability to interact with Sld2. In cells lacking Swe1, Clb2-ME expression leads to slightly earlier Sld2 phosphorylation than wild-type Clb2 expression (Figure 3). Furthermore, the proportion of phosphorylated Sld2 is greater in cells expressing Clb2-ME than Clb2. Nevertheless, Sld2 phosphorylation is not as rapid or complete as in Clb5-expressing cells. These results indicate that the M and E mutations enhance Clb2's ability to target replication substrates, but Clb2-ME still does not match Clb5's targeting ability. Taken together, these findings show that Clb2-ME largely escapes regulation by Swe1 while also interacting more avidly with replication substrates than Clb2.
Clb2-ME diminishes inhibitory Y19 phosphorylation of Cdc28 in vivo:
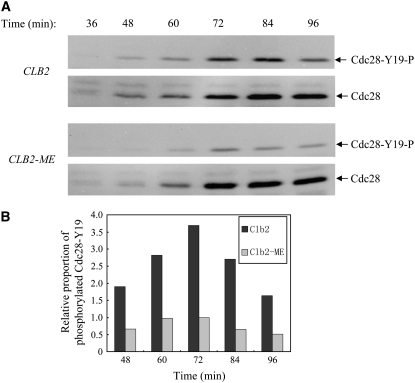
Reduced interaction between Swe1 and Clb2-ME should reduce the level of phosphorylation of Cdc28-Y19. We examined phosphorylation of Clb2-associated Cdc28-Y19 by immunoprecipitation of Clb2-Ha3 or Clb2-ME-Ha3 from cell extracts after G1 block and release. These strains expressed CLB2-HA3 or CLB2-ME-HA3 from the CLB2 locus. Total coprecipitated Cdc28 was analyzed by immunoblotting with anti-PSTAIRE antibody, while Y19-phosphorylated Cdc28 was detected with Cdc2-phosphotyrosine-specific antibody. Phosphorylated Cdc28-Y19 is first detected ∼48 min after release from G1 and peaked at ∼72–84 min in wild-type cells (Figure 4A). Cells expressing Clb2-ME show similar timing of phosphorylation of Cdc28-Y19; however, the level is markedly reduced. We quantified the level of phosphotyrosine-Cdc28 relative to the total Clb2-associated Cdc28 by quantitative chemiluminescence (Figure 4B). The quantified data show three- to fourfold decrease of phosphorylated Cdc28-Y19 associated with Clb2-ME. These results indicate that the HP region N and K residues are required for inhibitory phosphorylation of Clb2-associated Cdc28 by Swe1, and suggest that these Clb2 residues mediate physical interaction with Swe1.
Figure 4.—
Clb2-ME diminishes Cdc28-Y19 phosphorylation in vivo. Strains expressing CLB2-HA3 (FHy259) or CLB2-ME-HA3(YGy78) from the endogenous CLB2 promoter were synchronized in G1 with α-factor and released at 23°. Soluble protein extracts were prepared at the indicated intervals and immunoprecipitated with anti-Ha antibody. (A) Precipitates were analyzed by immunoblotting with anti-PSTAIRE antibody to detect Cdc28 (Cdc28), and anti-phosphotyrosine-Cdc2 antibody to detect specifically Y19-phosphorylated Cdc28 (Cdc28-Y19-P). CLB2 and CLB2-ME samples were run on the same blots for accurate quantification. (B) Quantification of the chemiluminescent signal of Cdc28-Y19-P relative to Cdc28. The values are dimensionless and a value of 1 was assigned for the maximum proportion achieved in the CLB2-ME mutant cells.
Swe1 interaction with Clb2-Cdc28 requires HP region N and K residues:
To provide direct evidence that Swe1 interaction with Clb2-Cdc28 depends on the HP region N and K residues of Clb2, we examined the physical interaction between Swe1 and Clb2 in whole-cell extracts. Swe1-Myc6 was overexpressed from the GAL1 promoter in logarithmically growing cells expressing Clb2 or Clb2-ME from the endogenous CLB2 promoter. Whole-cell extracts were prepared and subject to immunoprecipitation with anti-Myc antibody; coprecipitation of Clb2 was determined by immunoblotting with anti-Clb2 antibody. Wild-type Clb2 coprecipitates robustly with Swe1-Myc6, whereas Clb2-ME coprecipitates much less efficiently (Figure 5A, compare lanes 2 and 4). Quantification of two independent experiments shows an ∼12-fold reduction in the amount of Clb2-ME associated with Swe1 relative to wild-type Clb2 (Figure 5B). This result demonstrates that the HP region N and K residues of Clb2 are required for stable association with Swe1 and is consistent with the reduced phosphorylation of Clb2-ME-associated Cdc28-Y19 (Figure 4) and the enhanced ability of early expressed Clb2-ME to drive S-phase entry in the presence of Swe1 (Figure 2A).
Clb2-ME disrupts mutual phosphorylations between Swe1 and Cdc28 in vitro:
Recent studies have elucidated additional details of the interaction between Swe1 and Clb2-Cdc28, showing that Clb2-Cdc28 first phosphorylates Swe1, thereby promoting a more stable association and activating Swe1 tyrosine kinase activity toward Cdc28-Y19, which ultimately results in inhibition of Clb2-Cdc28 (Asano et al. 2005; Harvey et al. 2005). Thus, the defective regulation of Clb2-ME-Cdc28 by Swe1 may reflect initial failure of Clb2-ME-Cdc28 to phosphorylate Swe1 due to weakened physical interaction between the two. Considering that the HP region has been implicated in substrate targeting, it seems likely that N and K mediate phosphorylation of Swe1 by Clb2-Cdc28 and that Clb2-ME is defective in this activity.
To determine the effects of the Clb2 mutations on phosphorylation of Swe1, we immunoaffinity-isolated epitope-tagged Clb2-Cdc28 and Clb2-ME-Cdc28 from swe1Δ cells (Figure 6A), which ensured that the associated Cdc28-Y19 was unphosphorylated. Separately, we overexpressed and isolated Swe1-Myc6 on immunoaffinity beads. We incubated the isolated proteins alone or in combination, in the presence of [γ-32P]ATP, and detected the products by gel autoradiography. Swe1 beads alone show no significant protein kinase activity, while Clb2-Cdc28 alone and Clb2-ME-Cdc28 alone show robust autophosphorylation of Clb2 and Clb2-ME, respectively (Figure 6B). Clb2-Cdc28 and Clb2-ME-Cdc28 also show indistinguishable levels of kinase activity toward histone H1 (data not shown). The similar levels of autophosphorylation and phosphorylation of H1 indicate that the Clb2 mutations have little or no effect on Clb2's ability to target itself or on the inherent kinase activity of the associated Cdc28.
Figure 6.—
Clb2-ME disrupts mutual phosphorylations between Swe1 and Clb2-Cdc28 in vitro. Clb2-Ha3 and Clb2-ME-Ha3 were isolated from strains YGy65 (GAL-CLB2-HA3 swe1Δ) and YGy54 (GAL-CLB2-ME-HA3 swe1Δ), respectively, by immunoprecipitation and peptide elution (see materials and methods). (A) Eluates were analyzed by immunoblotting with anti-Clb2 antibody. (B) Swe1-Myc6 was isolated by immunoprecipitation from strain YGy29 (GAL-SWE1-MYC6) and incubated alone or with Clb2-Ha3 or Clb2-ME-Ha3, as indicated, all in the presence of [γ-32P]ATP. Samples were resolved by SDS–PAGE and phosphorylated Swe1, Clb2, and Cdc28 were detected by phosphorimaging. (C) Cdc28, Swe1, and Clb2 (or Clb2-ME) phosphorylation levels were quantified by standardization against the amount of Clb2 (or Clb2-ME) input (lanes 2 and 3) and plotted as the relative level of phosphorylation of each protein in the Clb2- vs. Clb2-ME-containing reactions. Error bars indicate standard deviation (n = 4).
Incubation of Swe1 beads with Clb2-Cdc28 results in phosphorylation of Swe1 (Figure 6B). However, phosphorylation of Swe1 is reduced about threefold when incubated with Clb2-ME-Cdc28 (Figure 6, B and C). These data are consistent with Clb2-Cdc28 targeting Swe1 as a substrate and strongly suggest that N and K mediate this reaction. Further examination of the reaction products shows that phosphorylation of Cdc28 results from incubation of Clb2-Cdc28 with Swe1 and is reduced about threefold in the incubation of Clb2-ME-Cdc28 with Swe1 (Figure 6, B and C). This reduced phosphorylation of Cdc28 likely stems from the reduced phosphorylation of Swe1 by Clb2-ME-Cdc28, which is required to activate Swe1 and stabilize its association with Clb2-Cdc28, both of which contribute to inhibition of Cdc28 (Booher et al. 1993; McMillan et al. 1999; Harvey et al. 2005). Finally, inhibition of Clb2-Cdc28 is indicated by the reduced autophosphorylation of Clb2, while similar inhibition of Clb2-ME autophosphorylation is not observed (Figure 6, B and C). Together with the increased S-phase activity of Clb2-ME (Figures 2 and 3), and its reduced interaction with Swe1 (Figures 4 and 5), these findings support the conclusion that residues N and K are essential for functional interaction between Clb2-Cdc28 and Swe1.
Clb5-NK is subject to regulation by Swe1:
Mutating N and K of Clb2 to the corresponding residues of Clb5, M and E, disrupts interaction of Clb2 with Swe1. We wondered whether introduction of N and K at positions 1 and 4 of Clb5 would place Clb5 under regulation by Swe1. To test this, we created CLB5-M197N, E207K (CLB5-NK) and expressed it from its native promoter. We analyzed the time of S-phase entry in clb6Δ and clb6Δ swe1Δ cells expressing Clb5 or Clb5-NK with the expectation that S phase would be delayed in a SWE1-dependent manner if the CLB5 mutations result in novel regulation by Swe1. However, S-phase entry is not significantly delayed in SWE1 cells expressing Clb5-NK compared with Clb5 (Figure 7A, compare CLB5 with CLB5-NK). This result suggests that these mutations of Clb5 are insufficient to confer interaction with Swe1.
Figure 7.—
Clb5-NK is subject to regulation by Swe1. (A) clb6Δ strains expressing CLB5 (DGy221) or CLB5-NK (FHy374) and swe1Δ derivatives (FHy136 and YGy24, respectively) were synchronized in G1 with α-factor and released at 23°. DNA content was measured at 12-min intervals. (B) GAL-SWE1 clb6Δ strains expressing CLB5 (YGy81) or CLB5-NK (YGy82) were grown in YEP/2% raffinose (noninducing) medium and synchronized in G1 with α-factor. Each culture was split and half was incubated in YEP/2% raffinose plus α-factor, while the other half of each was incubated in YEP/2% galactose plus α-factor. After 1 hr, the cultures were released from α-factor arrest into fresh raffinose or galactose medium (the same as in the previous incubation) at 25°. DNA content was analyzed every 12 min.
We considered the possibility that Swe1 abundance in the early cell cycle is insufficient to inhibit Clb5-NK. CLB5 and SWE1 are expressed with similar timing in late G1 (Mendenhall and Hodge 1998). Clearly, Swe1 abundance is sufficient to inhibit Clb2 expressed from the CLB5 promoter (Figure 2A). However, as Clb5 stimulates S phase earlier than Clb2 (even in the absence of Swe1) (Figure 2A), Clb5 may act before Swe1 has sufficiently accumulated. Therefore, we overexpressed Swe1 in G1-arrested Clb5- or Clb5-NK-expressing cells prior to their release into S phase. In Clb5-expressing cells, induction of Swe1 expression with galactose does not alter the timing of S phase (Figure 7B, compare galactose with raffinose). However, in Clb5-NK-expressing cells, Swe1 overexpression delays S-phase entry by at least 24 min (Figure 7B). Thus, N and K at positions 1 and 4 are sufficient to establish a functional interaction between Clb5 and Swe1, albeit dependent on Swe1 overexpression. These results further support the conclusion that these residues are key determinants of Swe1 regulation of different Clb-Cdc28 complexes.
DISCUSSION
HP-associated residues determine specificity of Swe1 regulation:
We have identified HP-associated residues N260 and K270 of Clb2 as critical to its regulation by Swe1. Mutation of these residues largely abrogates Swe1 inhibition of early expressed Clb2 in stimulation of DNA replication, reduces inhibitory phosphorylation of Clb2-associated Cdc28-Y19 in vivo, and disrupts stable association of Clb2-Cdc28 with Swe1 in extracts. In vitro, the mutual phosphorylations between Clb2-Cdc28 and Swe1 also depend on N and K. Although our analysis does not allow us to determine the order of these phosphorylation events, it seems most likely, and consistent with our data, that activating phosphorylation of Swe1 by Clb2-Cdc28 precedes inhibitory tyrosine phosphorylation of Cdc28 by Swe1 (Harvey et al. 2005). In the converse relationship, initial phosphorylation of Cdc28-Y19 should be associated with reduced phosphorylation of Swe1, which is not observed (Figure 6, B and C). It seems likely that N and K mediate both of the mutual phosphorylation events between Swe1 and Clb2-Cdc28. However, given the available knowledge, the defect in phosphorylation of Swe1 by Clb2-ME-Cdc28 may be sufficient to account for the loss of inhibitory regulation by Swe1.
Analogous to the function of N and K in mediating the physical interaction and phosphorylations between Clb2-Cdc28 and Swe1, and according to the model that Swe1 activation depends on its phosphorylation by Cdc28, we infer that introduction of N and K residues at positions 1 and 4 of Clb5 allows Clb5-NK-Cdc28 to interact with and phosphorylate Swe1, which triggers Swe1 phosphorylation of Clb5-NK-associated Cdc28 on Y19. The need to overexpress Swe1 to inhibit Clb5-NK likely reflects relatively weak interaction between Swe1 and Clb5-NK-Cdc28, suggesting that the unique feature(s) of Clb2 in addition to N and K contributes to its robust interaction with Swe1. Although overexpression of Swe1 inhibits Clb5-NK- but not Clb5-associated Cdc28, we have been unable to detect a differential interaction(s) between Swe1 and Clb5 or Clb5-NK. Instead, we found that overexpression of Swe1 results in phosphorylation of Clb5- and Clb5-NK-associated Cdc28-Y19 (data not shown). We speculate that more avid interaction between Clb5-NK and Swe1 results in Clb5-NK targeting overexpressed Swe1 preferentially over its normal replication substrates. Another possibility we discuss below is that interaction of Clb5 with Sic1 is altered in Clb5-NK, facilitating its inhibition by Swe1.
Interestingly, the N and K mutations in Clb5 do not significantly delay S-phase entry in the absence of SWE1, suggesting that the native M and E residues are not essential for effective interaction of Clb5 with essential replication targets such as Sld2 (Figure 7A, compare CLB5 swe1Δ with CLB5-NK swe1Δ). Previous studies have shown that mutation of all three HP residues (Figure 1, positions 1–3) to alanines significantly reduces Clb5 interaction with specific substrates and its ability to complement the lethality of clb3-6Δ cells (Cross and Jacobson 2000). Thus, the remaining two, highly conserved HP residues (positions 2 and 3, L and W) appear to be sufficient to maintain Clb5 interaction with critical substrates. Similarly, mutation of all three HP residues to alanines reduces Clb2's biological activity and ability to complement the lethality of clb(1, 3, 4)Δ, clb2ts cells (Cross and Jacobson 2000). However, Clb2-ME complements clb(1, 3, 4)Δ, clb2ts cells at the nonpermissive temperature (data not shown), indicating that the remaining two HP residues are sufficient to maintain Clb2's interactions with essential substrates. Together, these findings support the idea that N and K of Clb2 and M and E of Clb5 are crucial for their differential interaction with Swe1, while having a more limited role in targeting essential Clb2- or Clb5-specific substrates.
Role of the HP in CDK regulation:
The role of the HP region in mediating interactions with CDK substrates and CDK inhibitors, such as Swe1, potentially provides for strict control over CDK activation and deactivation, as well as substrate phosphorylations. Thus, in addition to inhibition of Clb2-Cdc28 function by tyrosine phosphorylation, Swe1 binding the HP may act as a competitive inhibitor of Clb2-Cdc28 activity on mitotic substrates. These controls might act redundantly to ensure tight control over Clb2-Cdc28. This is consistent with the finding that Swe1 partially inhibits Cdc28-Y19F, which cannot be phosphorylated (McMillan et al. 1999).
The HP region also may mediate CDK inhibition by Sic1, which is functionally and structurally related to mammalian p27 (Barberis et al. 2005); p27 binds the HP of cyclin A, and when expressed in S. cerevisiae can bind and inhibit Clb5-Cdc28 (Cross and Jacobson 2000). Sic1 overexpression prevents Swe1 phosphorylation of Clb5-associated Cdc28-Y19, suggesting that Sic1 and Swe1 bind overlapping sites on Clb5-Cdc28 (Keaton et al. 2007). However, the triple alanine mutation of the HP only slightly reduces the ability of Sic1 to bind and inhibit Clb5-Cdc28, indicating that other determinants are involved (Cross and Jacobson 2000). This previous study did not determine the significance of E at position 4 in the interaction between Sic1 and Clb5-Cdc28. However, we note that Clb5-NK-expressing cells show no evidence of loss of Sic1 inhibition, such as precocious S-phase entry (Figure 7). This result is consistent with the ability of Sic1 to inhibit both Clb5- and Clb2-Cdc28 (Schwob et al. 1994). Therefore, with regard to Sic1 inhibition, these data suggest that the HP region does not discriminate Clb5- from Clb2-Cdc28, although we cannot rule out that the HP region contributes nonspecifically to Sic1 interactions with Clb-Cdc28 complexes. Indeed, it is possible that the ability of Swe1 overexpression to inhibit Clb5-NK reflects decreased interaction with Sic1, which may normally sequester Clb5 from Swe1.
Evolutionary conservation of the HP region from yeast to humans has been attributed to its role in mediating CDK interactions with substrates as well as inhibitors of the p27 family (Archambault et al. 2005). Given the evolutionary conservation of Wee1 regulation and the HP regions of many cyclins, it would appear likely that Wee1 tyrosine kinases in other organisms bind their cognate CDKs through the cyclin HP region as well. Consistent with this notion, SWE1 and human WEE1 complement an Schizosaccharomyces pombe wee1 mutant (Booher et al. 1993; Watanabe et al. 1995). However, it is notable that the Clb5 HP region shares significantly greater similarity with the HP region of human cyclins A, E, and B and S. pombe cyclins Cdc13 and Cig2 than does the Clb2 HP, suggesting that Clb2 has diverged (data not shown) (Archambault et al. 2005). Interaction with Swe1 probably contributed to divergence between Clb5 and Clb2 in their respective HP regions and, indirectly, to the evolution of distinct sets of substrates. This is because the acquisition (or loss) of Swe1 interaction through mutation of specific HP region residues would immediately create a distinctly regulated CDK, thereby providing a foundation for the selection of CDK-specific substrates. Swe1 itself may have diverged to gain interaction with Clb2 (or lose interaction with Clb5), as Swe1 shares little sequence homology with human Wee1 and Myt1, which are more closely related to S. pombe Mik1, which S. cerevisiae lacks (Watanabe et al. 1995). Because S. cerevisiae expresses only a single relevant Cdk (Cdc28), Swe1 must exploit differences between cyclins to regulate CDKs differentially. However, the presence of multiple Cdk subunits in multicellular organisms provides an alternative (or additional) basis for CDK regulation by Wee1 and Myt1. Thus, it remains to be determined whether Wee1 in other organisms will bind their CDKs via the HP region, as suggested by the functional conservation across species, and whether the HP will impart specificity to Wee1–CDK interactions in other organisms.
Acknowledgments
We thank S. Al-Bassam, Y. Chowdhury, A. Han, and L. Holden for strain and plasmid constructions, and J. Aparicio for critical reading of the manuscript. National Institutes of Health grant GM-65494 (to O.M.A.) supported this study.
References
- Amon, A., U. Surana, I. Muroff and K. Nasmyth, 1992. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature 355 368–371. [DOI] [PubMed] [Google Scholar]
- Aparicio, J. G., C. J. Viggiani, D. G. Gibson and O. M. Aparicio, 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24 4769–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault, V., E. J. Chang, B. J. Drapkin, F. R. Cross, B. T. Chait et al., 2004. Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 14 699–711. [DOI] [PubMed] [Google Scholar]
- Archambault, V., N. E. Buchler, G. M. Wilmes, M. D. Jacobson and F. R. Cross, 2005. Two-faced cyclins with eyes on the targets. Cell Cycle 4 125–130. [DOI] [PubMed] [Google Scholar]
- Asano, S., J. E. Park, K. Sakchaisri, L. R. Yu, S. Song et al., 2005. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 24 2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis, M., L. De Gioia, M. Ruzzene, S. Sarno, P. Coccetti et al., 2005. The yeast cyclin-dependent kinase inhibitor Sic1 and mammalian p27Kip1 are functional homologues with a structurally conserved inhibitory domain. Biochem. J. 387 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher, R. N., R. J. Deshaies and M. W. Kirschner, 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12 3417–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher, R. N., P. S. Holman and A. Fattaey, 1997. Human Myt1 is a cell cycle-regulated kinase that inhibits Cdc2 but not Cdk2 activity. J. Biol. Chem. 272 22300–22306. [DOI] [PubMed] [Google Scholar]
- Chow, J. P., W. Y. Siu, H. T. Ho, K. H. Ma, C. C. Ho et al., 2003. Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. J. Biol. Chem. 278 40815–40828. [DOI] [PubMed] [Google Scholar]
- Cross, F. R., and M. D. Jacobson, 2000. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol. Cell. Biol. 20 4782–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., M. Yuste-Rojas, S. Gray and M. D. Jacobson, 1999. Specialization and targeting of B-type cyclins. Mol. Cell 4 11–19. [DOI] [PubMed] [Google Scholar]
- Donaldson, A. D., 2000. The yeast mitotic cyclin Clb2 cannot substitute for S phase cyclins in replication origin firing. EMBO Rep. 1 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani, M., G. Liberi, G. Lucchini and P. Plevani, 1995. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol. Cell. Biol. 15 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D. G., J. G. Aparicio, F. Hu and O. M. Aparicio, 2004. Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 24 10208–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, K. L., and P. Nurse, 1989. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342 39–45. [DOI] [PubMed] [Google Scholar]
- Gu, Y., J. Rosenblatt and D. O. Morgan, 1992. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 11 3995–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, S. L., and D. R. Kellogg, 2003. Conservation of mechanisms controlling entry into mitosis: budding yeast wee1 delays entry into mitosis and is required for cell size control. Curr. Biol. 13 264–275. [DOI] [PubMed] [Google Scholar]
- Harvey, S. L., A. Charlet, W. Haas, S. P. Gygi and D. R. Kellogg, 2005. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell 122 407–420. [DOI] [PubMed] [Google Scholar]
- Hu, F., and O. M. Aparicio, 2005. Swe1 regulation and transcriptional control restrict the activity of mitotic cyclins toward replication proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, P., Y. Gu and D. O. Morgan, 1996. Role of inhibitory CDC2 phosphorylation in radiation-induced G2 arrest in human cells. J. Cell Biol. 134 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton, M. A., E. S. Bardes, A. R. Marquitz, C. D. Freel, T. R. Zyla et al., 2007. Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr. Biol. 17 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, D. J., and S. I. Reed, 1995. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 129 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., J. J. Stanton, Z. Wu and H. Piwnica-Worms, 1997. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol. Cell. Biol. 17 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., and Y. Wang, 2006. The function and regulation of budding yeast Swe1 in response to interrupted DNA synthesis. Mol. Biol. Cell 17 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Loog, M., and D. O. Morgan, 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434 104–108. [DOI] [PubMed] [Google Scholar]
- Lundgren, K., N. Walworth, R. Booher, M. Dembski, M. Kirschner et al., 1991. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64 1111–1122. [DOI] [PubMed] [Google Scholar]
- Masumoto, H., S. Muramatsu, Y. Kamimura and H. Araki, 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415 651–655. [DOI] [PubMed] [Google Scholar]
- McMillan, J. N., R. A. Sia, E. S. Bardes and D. J. Lew, 1999. Phosphorylation-independent inhibition of Cdc28p by the tyrosine kinase Swe1p in the morphogenesis checkpoint. Mol. Cell. Biol. 19 5981–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty, J. J., and D. J. Lew, 2005. Swe1p responds to cytoskeletal perturbation, not bud size, in S. cerevisiae. Curr. Biol. 15 2190–2198. [DOI] [PubMed] [Google Scholar]
- Mendenhall, M. D., and A. E. Hodge, 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62 1191–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. E., and F. R. Cross, 2001. Cyclin specificity: How many wheels do you need on a unicycle? J. Cell Sci. 114 1811–1820. [DOI] [PubMed] [Google Scholar]
- Morgan, D. O., 1996. The dynamics of cyclin dependent kinase structure. Curr. Opin. Cell Biol. 8 767–772. [DOI] [PubMed] [Google Scholar]
- Morgan, D. O., 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13 261–291. [DOI] [PubMed] [Google Scholar]
- Mortensen, E. M., H. McDonald, J. Yates, III and D. R. Kellogg, 2002. Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell 13 2091–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, P. R., T. R. Coleman, A. Kumagai and W. G. Dunphy, 1995. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science 270 86–90. [DOI] [PubMed] [Google Scholar]
- Murray, A. W., 2004. Recycling the cell cycle: cyclins revisited. Cell 116 221–234. [DOI] [PubMed] [Google Scholar]
- Rhind, N., and P. Russell, 1998. Tyrosine phosphorylation of cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell Biol. 18 3782–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind, N., and P. Russell, 2001. Roles of the mitotic inhibitors Wee1 and Mik1 in the G(2) DNA damage and replication checkpoints. Mol. Cell Biol. 21 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, P., and P. Nurse, 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45 145–153. [DOI] [PubMed] [Google Scholar]
- Russell, P., S. Moreno and S. I. Reed, 1989. Conservation of mitotic controls in fission and budding yeasts. Cell 57 295–303. [DOI] [PubMed] [Google Scholar]
- Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massague and N. P. Pavletich, 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382 325–331. [DOI] [PubMed] [Google Scholar]
- Schulman, B. A., D. L. Lindstrom and E. Harlow, 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95 10453–10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob, E., T. Bohm, M. D. Mendenhall and K. Nasmyth, 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79 233–244. [DOI] [PubMed] [Google Scholar]
- Sorger, P. K., and A. W. Murray, 1992. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature 355 365–368. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., M. Broome and T. Hunter, 1995. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 14 1878–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes, G. M., V. Archambault, R. J. Austin, M. D. Jacobson, S. P. Bell et al., 2004. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 18 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]