Abstract
Mutations at the flügellos (fl) locus in Bombyx mori produce wingless pupae and moths because of the repressed response of wing discs to ecdysteroid. Four recessive fl alleles occurred spontaneously and were mapped at 13.0 of the silkworm genetic linkage group 10. By positional cloning, we confirmed that the gene responsible for fl is fringe (fng) encoding Fng glycosyltransferase, which is involved in regulating the Notch signaling pathway. In four different fl alleles, we detected a large deletion of the fng gene in flk and nonsense mutations in fl, flo, and fln. In the wild-type (WT) silkworm, fng is expressed actively in the wing discs, brain, and reproductive organs from the fourth to final instars but barely in the other tissues tested. In situ hybridization showed that fng mRNA is expressed in the dorsal layer of the WT wing discs. The wingless (wg) mRNA, a downstream marker of Fng-mediated Notch signaling, is localized at the dorsoventral boundary in the WT wing discs but repressed markedly in the fl wing discs. Although null mutants of Drosophila fng result in postembryonic lethality, loss of fng function in Bombyx affects only wing morphogenesis, suggesting different essential roles for fng in tissue differentiation among insects.
WING formation is a major morphological change during larval–pupal development in holometabolous insects. Wings develop from the wing imaginal discs, which differentiate in response to insect hormones, juvenile hormone, and pulses of ecdysteroid (Nardi and Willis 1979; Blais and Lafont 1980; Fujiwara and Hojyo 1997; Fujiwara and Ogai 2001; Truman and Riddiford 2007). The wing disc of Drosophila is a well-studied model system for identifying numerous genes involved in pattern formation and understanding the genetic regulation of morphological processes (Williams et al. 1993). However, whether the mechanism revealed in Drosophila is actually applicable to other insect species remains to be clarified. Furthermore, although many studies have shown the effects of ecdysteroid on the development of cultured wing discs in vitro (Nardi and Willis 1979; Blais and Lafont 1980; Fujiwara and Ogai 2001; Truman and Riddiford 2007), little is known about the molecular mechanisms of hormone-mediated wing morphogenesis and tissue differentiation during metamorphosis.
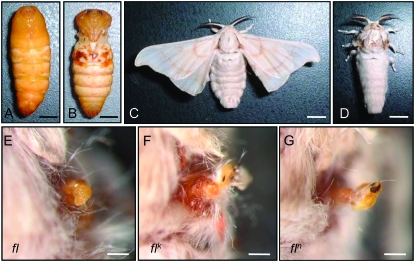
The silkworm Bombyx mori has three wing-deficient mutants: flügellos (fl), Vestigial (Vg), and rudimentary wing (rw) (Fujii et al. 1998). The recessive homozygote of the fl mutant has the most severe phenotype of the three, with almost undetectable wings in the pupal and adult stages (Figure 1). Four recessive fl mutants (fl, flk, fln, and flo) occurred spontaneously and independently (Katsuki 1935; Harizuka 1948; Ueda et al. 1959; Fujii et al. 1998) and were mapped at the same position (13.0) of the silkworm genetic linkage group 10 (LG10) (Figure 2A; Fujii et al. 1998). Despite the complete loss of wings, other larval and adult organs appear normal in the fl mutants. Histological studies revealed that, although the fl wing discs are slightly smaller than those of the wild type (WT), they develop normally until the fourth larval instar (Fujiwara and Hojyo 1997). In the fifth instar, however, developmental events such as wing epithelial invagination and tracheal migration into the lacunal space do not occur (Nagata 1962; Hojyo and Fujiwara 1997). Moreover, fl wing discs cultured in medium containing 20-hydroxyecdysone (20E) do not develop, whereas the WT wing discs differentiate normally under the same conditions (Fujiwara and Hojyo 1997). During the prepupal stage, the transcription of two ecdysteroid-induced genes, BHR3 (early-late gene; Bombyx homolog of DHR3) and Urbain (wing-specific late gene), is blocked in the flk wing disc in a tissue-specific manner (Matsuoka and Fujiwara 2000), and the expression of an ecdysteroid-induced 41-kDa protein is also reduced (Fujiwara and Hojyo 1997). These observations suggest that fl wing discs cannot respond to ecdysteroid during metamorphosis in the same way as WT wing discs do.
Figure 1.—
Bombyx wingless mutant, fl lacks four wings in the pupa and adult stages. (Top) The pupa (A and B) and adult (C and D) of wild type (A and C) and fl mutant (B and D). Bars, 5 mm. (Bottom) The vestigial wing of fl (E), flk (F), and fln (G). Bars, 0.5 mm.
Figure 2.—
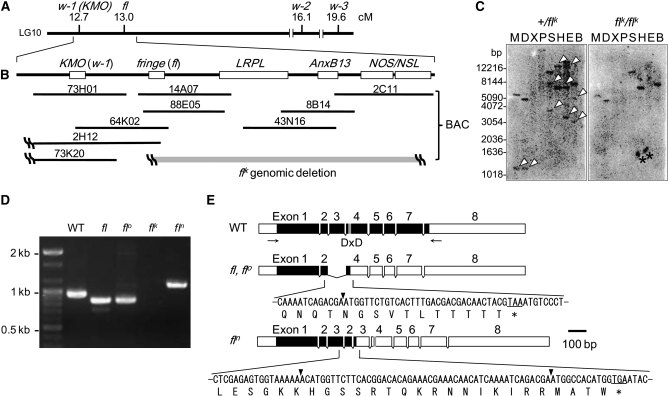
Map of fl locus and mutations in fng of each fl allele. (A) Genetic map of the silkworm LG10 near the fl locus. fl is mapped at 13.0 and distant 0.3 cM (∼100 kb) from w-1 (KMO). (B) Gene organization of fng (fl) and other genes across the fl locus. Shaded bar indicates the deletion region in the flk genome. (C) Genomic Southern analysis for +/flk and flk/flk genome with the fng probe. The cDNA of fng ORF region is used as the probe (see materials and methods). Arrowheads in +/flk indicate the bands that are lost in flk/flk. The DNA-size markers are shown to the left. Asterisks indicate nonspecific signals. M, MspI; D, DraI; X, XhoI; P, PstI; S, SacI; H, HindIII; E, EcoRI; B, BglII. (D) RT–PCR for fng mRNA from wing discs of fifth instar larvae of WT, fl, flo, flk, and fln strains, using a primer set indicated by arrows in Figure 2E. (E) Schematic representation of deletion of exon 3 of fng in fl, flo, and duplication of exon 2–3 of fng in fln. Solid and open boxes indicate ORF and untranslated region (UTR), respectively. The shaded box represents DxD (D, aspartic acid; x, any amino acid) motif. Arrowheads and asterisks indicate exon junction and stop codon, respectively.
The cause of wing deficiency in fl mutants is speculated to involve the functional loss of the “fl gene,” although the gene has not yet been identified. Previously, we identified genes expressed abnormally in the flk wing discs and found that the expression of Annexin b13 (Anxb13) mRNA is repressed completely (Matsunaga and Fujiwara 2002). By comparing the WT and flk genomic structures by Southern hybridization using the cDNA of Anxb13 as the probe, we found that the entire gene is lost from the flk genome (Matsunaga and Fujiwara 2002). Moreover, we have found that Anxb13 is located on the chromosome (LG10) that includes the fl locus (Matsunaga and Fujiwara 2002). These findings suggested that the fl gene might be located near Anxb13. We undertook to identify the fl gene by positional cloning, beginning at the Anxb13 locus, on the basis of bacterial artificial chromosome (BAC) library screening and shotgun sequencing. First, we compared the structures of the surrounding regions in the flk mutant with those of the WT. We detected a large chromosomal deletion in flk, which includes Anxb13 and another four genes. We compared the structures of these five genes in other fl mutants with the WT and found that all four fl mutants have deletion or nonsense mutations in only one gene, the fringe (fng) ortholog (Irvine and Wieschaus 1994). Fng is an O-fucose-specific β-1,3-N-acetylglucosaminyltransferase that modifies the receptor Notch (N) and thereby modulates its ligand sensitivities (Panin et al. 1997; Haines and Irvine 2003). Fng-mediated N signaling plays a crucial role in the developmental processes of various tissues in Drosophila melanogaster (Cho and Choi 1998; Domínguez and De Celis 1998; Papayannopoulos et al. 1998; Grammont and Irvine 2001; Rauskolb 2001; Thomas and Van Meyel 2007) and is especially well known for its participation in dorsoventral boundary formation in wing discs (Irvine and Wieschaus 1994; Haines and Irvine 2003). During wing development in Drosophila, fng is expressed specifically in the dorsal cells under the control of Apterous (encoded by ap), which is a LIM-homeodomain transcription factor that is required for dorsal cell identity (Irvine and Wieschaus 1994; Kim et al. 1995). Fng-mediated N signaling is activated along the border between dorsal and ventral cells, leading to the expression of wingless (wg), which results in the organization of wing growth and patterning (Couso et al. 1994; Panin et al. 1997; Klein and Martinez Arias 1998; Baonza and Garcia-Bellido 2000; Haines and Irvine 2003). In the fl wing discs, the expression of BmWnt-1, a Bombyx homolog of wg, is completely repressed, suggesting a functional deficiency of Fng in the fl mutants. These observations clarify that fng is the gene responsible for the fl mutant. This is the first report of the positional cloning of an important mutation locus in B. mori. It is interesting that null or nonsense mutations in fng cause postembryonic lethality in D. melanogaster (Irvine and Wieschaus 1994; Correia et al. 2003), whereas the functional deficiency of the fng mutants in B. mori affects only wing morphogenesis. Our findings demonstrate the crucial role of fng in Bombyx wing formation and the existence of a fng-independent signaling pathway for tissue differentiation in tissues other than wings.
MATERIALS AND METHODS
Silkworm strains:
The fl, fln, and flk mutant strains were provided from the silkworm stock center of Kyusyu University supported by the National Bio-Resource Project (NBRP), and the flo mutant strain was kindly provided by the National Institute of Agrobiological Sciences (NIAS, Kobuchizawa, Japan). The silkworms were reared with leaves of a mulberry or an artificial diet (Nihon Nosan Kogyo, Yokohama, Japan) under a 16-hr light:8-hr dark photoperiod at 25°. The newly molted fourth and fifth instar larvae were segregated immediately after the onset of the photophase and this day was designated as day 0. Under this condition, most larvae began wandering on day 8 and pupated on day 12 of the fifth instar.
BAC library screening and shotgun sequencing:
A Bombyx BAC library RPCI-96 was used for screening. We used BAC high-density replica (HDR) filter sets, which are available from BACPAC Resources, Children's Hospital Oakland Research Institute (Oakland, CA). The DNA probe for screening the BAC library containing the genomic sequence of 5′ upstream of LRPL was synthesized by PCR with a primer set, 5′-GCCGTCGCCTGTGGTACTGC-3′ and 5′-ACTCTTTAGACCAGCCTTGGTTGAC-3′. The hybridization of the BAC HDR filter with DNA probes was carried out using the ECL direct nucleic acid labeling and detection systems kit (GE Healthcare, Waukesha, WI), according to the manufacturer's instructions (Koike et al. 2003). Isolation and random shotgun sequencing analysis of a BAC DNA were carried out using the large-construction kit (QIAGEN, Valencia, CA) and TOPO shotgun subcloning kit (Invitrogen, Carlsbad, CA). Multiple sequence alignments were carried out using the VectorNTI software (Invitrogen). We predicted the protein-coding regions by the NCBI Blast search program, Silkbase (http://morus.ab.a.u-tokyo.ac.jp/cgi-bin/index.cgi), KAIKOBLAST (http://kaikoblast.dna.affrc.go.jp/) and KAIKOGAAS (http://kaikogaas.dna.affrc.go.jp/) (Mita et al. 2004; Xia et al. 2004).
Cloning of Bombyx fng cDNA:
Total RNA was isolated from several tissues by the TRI-reagent kit (Sigma, St. Louis) and reverse transcribed with random primer (N6) by the first-strand cDNA synthesis kit (GE Healthcare). The full-length cDNA was obtained by 5′ and 3′ rapid amplification of cDNA ends (RACE) technique using the Marathon cDNA amplification kit (Clontech, Mountain View, CA). We performed reverse transcription–polymerase chain reaction (RT–PCR) in the open reading frame (ORF) region of Bombyx fng of four fl alleles using the primer set 5′-ATGGGCGGACGAAGAATGCT-3′ and 5′-CTATCGCTGCACTGTTTCCTCTTCC-3′ in 35–40 cycles of 96° for 30 sec, 55° for 40 sec, and 72° for 3 min. The RT–PCR products were subcloned into pGEM-T easy Vector (Promega, Madison, WI) and sequenced by an ABI3130xl genetic analyzer (Applied Biosystems, Foster City, CA).
Sequence comparison and phylogenetic tree construction:
Sequence alignment was constructed using Clustal_X. The neighbor-joining method with the MEGA2 program was used for the construction of a phylogenetic tree for the fng sequences. The following fng sequences were used to create the diagram: BmFringe, Bombyx mori (this study, AB360592); PcFringe, Precis coenia (AAO38754); DmFringe, Drosophila melanogaster (AAF51658); SgFringe, Schistocerca gregaria (AAF17565); AmFringe, Apis mellifera (XP_623898); Bflo-Fringe, Branchiostoma floridae (CAD97418); Hsap-LF, -MF, and -RF, Homo sapiens (NP_002908, NP_002295, NP_002396); Mmus-LF, -MF, and -RF, Mus musculus (NP_033079, BAE31866, NP_032621); Rnor-LF, -MF, and -RF, Rattus norvegicus (AAH62031, NP_596884, AAH61801); Ggal-LF, -MF, and -RF, Gallus gallus (NP_990278, NP_990279, XP_416278); Xlae-LF and -RF, Xenopus laevis (AAB38363, NP_001017051); Nvir, Notophthalmus viridescens (AAD10827); Drer-LF, -MF, and -RF, Danio rerio (NP_001001830, NP_571046, AAT46070); LF, Lunatic fringe; MF, Manic fringe; RF, Radical fringe.
Semiquantitative RT–PCR for fng expression:
Total RNA was isolated from several tissues of WT larva at various stages as described above, using the TRI-reagent kit (Sigma) and reverse transcribed with random primer (N6) by a first-strand cDNA synthesis kit (GE Healthcare). We performed semiquantitative RT–PCR analysis for fng expression using the cDNAs and following primer sets: 5′-ATGGGCGGACGAAGAATGCT-3′ and 5′-CTATCGCTGCACTGTTTCCTCTTCC-3′ for fng, 5′-AGCACCCCGTCATGGGTCTA-3′ and 5′-TGCGTCCAAGCTCATCCTGC-3′ for ribosomal protein L3 (rpL3) as an internal standard. To quantify the transcripts of fng, we performed a quantitative RT–PCR of rpL3 as previously described (Matsuoka and Fujiwara 2000). PCR conditions were as follows: 20 cycles of 96° for 30 sec, 58° for 40 sec, and 72° for 1 min for rpL3; 20 cycles of 96° for 30 sec, 55° for 40 sec, and 72° for 2 min for fng. The fluorescence intensities with the ethidium bromide staining were determined using the ImageJ free software (National Institutes of Health, http://rsb.info.nih.gov/ij/).
Tissue dissection and in vitro tissue culture:
The wing discs were dissected from day 2 fifth instar larvae. After rinsing in phosphate-buffered saline (PBS), the discs were cultured in 1 ml of Grace's medium (Sigma). 20E (Sigma) was dissolved in 10% isopropyl alcohol and added to the culture medium to give the desired concentration. Incubation was performed in a 1.5-ml centrifuge tube rotating so that the discs were exposed to the air.
Northern and Southern hybridization:
Total RNA (10 μg) was separated on a formaldehyde–agarose (1%) gel and transferred to a Hybond-N nylon membrane (GE Healthcare). DNA probe for fng was synthesized by RT–PCR with the primer set, 5′-ATGGGCGGACGAAGAATGCT-3′ and 5′-CTATCGCTGCACTGTTTCCTCTTCC-3′, and labeled with [α-32P]dCTP using a BcaBEST labeling kit (Takara Bio, Japan). Hybridization was performed at 48° for 18 hr in 50% formaldehyde, 5× SSC (0.15 m sodium chloride and 0.15 m sodium citrate, pH 7.4), 10× Denhardt's solution (0.2% each of bovine serum albumin, Ficoll, and polyvinylryrrolidone), 25 μg/ml of sonicated salmon sperm DNA, 50 mm sodium phosphate (pH 7.0), and 32P-labeled DNA. The membranes were washed twice at room temperature for 20 min in 2× SSC containing 0.1% sodium dodecyl sulfate (SDS). Further washes were performed by 30 min at 65° successively in 2× SSC containing 0.1% SDS and in 0.2× SSC containing 0.1% SDS.
We performed genomic Southern hybridization for fng in the flk mutant. Genomic DNA isolated from posterior silk glands was digested with several restriction enzymes, separated on an agarose (1%) gel, and transferred to a Hybond-N nylon membrane (GE Healthcare). DNA probe synthesis and hybridization were performed with the same procedures for Northern hybridization, as mentioned above.
Whole-mount in situ hybridization:
Wing discs were dissected and then fixed immediately in 4% paraformaldehyde in PBS for 30 min at 4°. The peripodial membrane of wing discs was removed during fixation on ice. After washing in PBS containing 0.5% Tween 20 (PBST), the wing discs were treated with proteinase K (20 μg/ml; Merck, St. Louis) in PBS for 20 min at 37° and postfixed in 4% paraformaldehyde in PBS for 30 min. Hybridization was performed at 50° for 24 hr in the hybridization buffer (50% formamide, 5× SSC, 500 μg/ml yeast tRNA, 0.5% Tween 20 and 50 μg/ml heparin sodium) with a digoxigenin-labeled antisense RNA probe. RNA probes for Bombyx fng and BmWnt-1 were prepared using the Roche biochemicals kit and primers for fng described above in Northern and Southern hybridization and 5′-GTGAAGACTTGCTGGATGAGGC-3′ and 5′-CTAGGTATCCCCGGCACGCAC-3′ for BmWnt-1. After hybridization, the wing discs were washed at 50° for 10 min in hybridization buffer. The wing discs were then washed for 5 min successively in PBST. After replacing in PBST, the wing discs were incubated with alkaline phosphatase-labeled anti-DIG antibody (Roche, Indianapolis) diluted 1:1,200 in PBST for 2 hr at room temperature. Excess antibody was washed away with at least four changes of PBST. The sample was equilibrated with two changes of the staining buffer (100 mm Tris-HCl, pH 9.5, 100 mm NaCl, 50 mm MgCl2, and 0.1% Tween 20) and then reacted with the staining buffer containing 3.5 μg/ml 5-bromo-4-chloro-3-indolyl-phosphate, 4-toluidine (BCIP; Roche) and 4.5 μg/ml nitroblue tetrazolium chloride (NBT; Roche). After staining, wing discs were embedded in paraffin and sectioned at 7 μm and observed.
RESULTS
Comparison of genomic structures across the fl locus in flk and WT:
Previous studies have suggested that Anxb13 is deleted in the flk genome and is located near the fl locus (Matsunaga and Fujiwara 2002). We isolated Anxb13 from fl mutants, except the flk mutant, and compared its sequences with that of the WT but found no significant mutation (supplemental Figure 2). Therefore, we inferred that the Anxb13 gene is not the fl gene.
We deduced that there is a large chromosomal deletion in the flk genome that includes Anxb13 and the true fl gene. We undertook to define the deleted region by comparing the genomic structures of flk and WT. The white egg 1 (w-1) locus, which corresponds to the kynurenine 3-monooxigenase gene (KMO) (Quan et al. 2002), has been mapped nearest to the fl locus at 12.7 of LG10 (Figure 2A; Fujii et al. 1998). The 0.3 cM distance between fl and w-1 (KMO) on the classical genetic map is estimated to be ∼100–200 kb. The w-1 mutant is characterized morphologically by its white eggs and white adult eyes. The fact that the eggs and adult eyes are normally colored in the flk mutant indicates that KMO is not deleted in flk. Therefore, we presumed that KMO is located near the fl gene and that one of the ends of the chromosomal deletion lies between KMO and Anxb13 in the flk genome. Using BAC clones containing Anxb13 as the starting point, chromosomal walking was performed until both ends of the chromosomal deletion were found in the flk genome or the KMO (w-1) gene was reached (Figure 2, A and B). In the first round of chromosomal walking from Anxb13, we screened for BAC clones containing Anxb13 and analyzed these clones (43N16, 8B14, and 2C11 in Figure 2B) by random shotgun sequencing. We finally assembled these sequences and obtained contiguous stretches of genomic sequences. Analyzing these with Blast homology searches and gene prediction programs, we predicted that three genes were included in the neighborhood of Anxb13, those encoding low-density lipoprotein receptor-related protein-like protein (LRPL, GenBank accession no. AB360591), nitric oxide synthase (NOS) (Imamura et al. 2002), and NOS-like protein (NSL, GenBank accession no. AB360590) (Figure 2B). By comparing the WT and flk genomic structures by Southern hybridization using each BAC end sequence as the probe, we found that one BAC end of 2C11 (NOS/NSL side) is not deleted in the flk genome but that the other BAC end is deleted. In addition, both BAC ends of 43N16 and 8B14 are deleted. We inferred that the right end of the chromosomal deletion lies within the 2C11 region of the flk genome (shaded bar in Figure 2B). Another round of chromosomal walking from LRPL was performed until the left end of the chromosomal deletion in the flk genome was detected or KMO was reached. We screened the BAC library using the cDNA of KMO or the unique genome sequence upstream from LRPL (the left end of LRPL in Figure 2B; see materials and methods) as the probe and successfully juxtaposed the BAC clones between KMO and LRPL using each BAC end sequence and contiguous genomic sequences from a Bombyx genome database (Figure 2B). By comparing the genomic structures of the WT and flk by PCR analysis using primer sets designed to bind within each BAC end region, we found that one BAC end of 88E05 (KMO side) is not deleted in the flk genome but that the other end (LRPL side) is deleted. Therefore, we inferred that the left end of the chromosomal deletion lies within the 88E05 region of the flk genome (shaded bar in Figure 2B). We then analyzed one of these BAC clones (88E05) and obtained contiguous stretches of genomic sequences across the fl region. Using Blast homology searches and gene prediction programs to analyze the obtained sequences, we predicted that the fng homolog in Bombyx (GenBank accession no. AB360592) is included within this region (Figure 2B).
We further isolated the cDNAs of the genes fng, LRPL, NOS, and NSL from the fl, flo, and fln mutants and compared their sequences with those of the WT. The LRPL, NOS, and NSL genes contained no significant nucleotide changes causing severe amino acid mutations in the fl mutants (supplemental Figures 3–5). Therefore, we deduced that these genes are not the fl gene. However, we detected remarkable mutations in the fng genes of all fl mutants, as described below (Figure 2, C and D, and supplemental Figure 1B).
Identification and characterization of the fng ortholog in Bombyx:
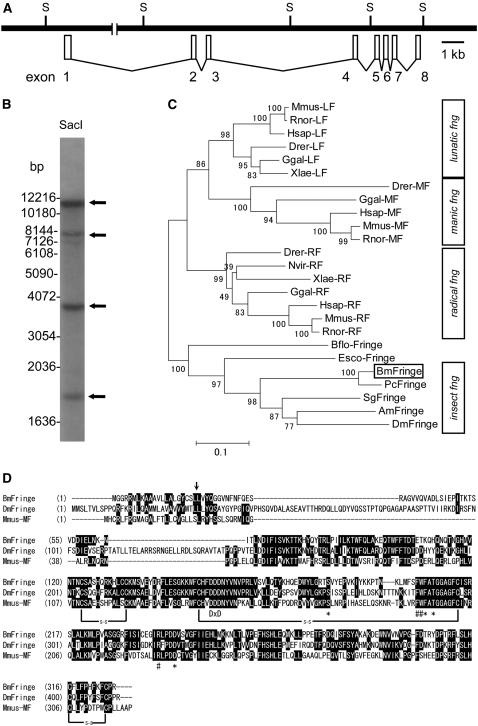
To clarify the structural deficiency of fng in fl mutants, we determined the complete fng cDNA sequence of the WT using RT–PCR and RACE techniques. The complete cDNA sequence (1781 bp) revealed that Bombyx fng has eight exons encoding 327 amino acids, constituting a 38-kDa protein (supplemental Figure 1A). On the basis of the shotgun sequence data, the genomic region of fng is predicted to be cut into at least four fragments by the SacI restriction enzyme (Figure 3A). Southern hybridization of the SacI-digested genomic DNA of the WT with the full-length fng cDNA as the probe showed four bands, which coincided with the predicted SacI sites (Figure 3B). We also performed a tBLASTn search of the Bombyx genome database using Bombyx Fng and Drosophila Fng proteins as the queries, to examine whether there is another fng-like gene in Bombyx. However, we did not detect any sequence apart from this fng gene (data not shown). Phylogenetic analysis of the amino acid sequences of Fng from Bombyx, Precis, and Drosophila together with other glycosyltransferases from Bombyx indicated that Bombyx Fng clusters with the other insect homologs (supplemental Figure 6). These results suggest that the Bombyx genome contains only one copy of the fng gene, although we cannot completely exclude the possibility that there is a gene that compensates for fng function in tissues other than the wing discs, despite its low sequence similarity to fng. Comparison of Bombyx Fng with that of other insects and vertebrates revealed that Bombyx Fng is most similar to the Fng homolog from another Lepidopteran species, P. coenia (Figure 3C and supplemental Figure 7). The amino acid sequence alignment of Bombyx Fng with Drosophila Fng and mouse Manic-fng (Mfng), the crystal structure of which has been revealed (Jinek et al. 2006), is shown in Figure 3D. Bombyx Fng is 44.2 and 37.7% identical to the homologous sequences of Drosophila Fng and mouse Mfng, respectively. A DxD (D, aspartic acid; x, any amino acid) motif, which is involved in sugar-transferase activities (Munro and Freeman 2000), is well conserved in all the Fng proteins. Furthermore, the amino acids required for UDP-GlcNAc binding and putative fucose binding are also highly conserved (Figure 3D) (Jinek et al. 2006). These observations confirm that the fng gene identified here is an authentic and exclusive ortholog of fng in B. mori.
Figure 3.—
Genomic and phylogenetic characterization of Bombyx fng. (A) The genomic structure of Bombyx fng, which consists of eight exons. S indicates SacI restriction site. (B) Genomic Southern analysis with the fng probe for WT genomic DNA restricted with SacI. The cDNA of fng ORF region is used as the probe (see materials and methods). Arrows indicate four positive bands. The DNA-size markers are shown to the left. (C) Phylogenetic tree of fng based on the amino acid sequences. The numbers at the tree edge represent the bootstrap values. The scale bar indicates the evolutionary distance between the groups. Bombyx fng (BmFringe) is boxed. Respective fng sequences are shown in materials and methods. (D) Amino acids alignment of Fng from Bombyx (BmFringe), Drosophila (DmFringe), and Manic-fng from mouse (Mmus-MF). Black shading with white letters indicates identical amino acid residues. Arrow shows the putative signal peptide cleavage site. DxD show conserved DxD motif. Asterisks and number signs (#) show the putative amino acid residues involved in UDP-GlcNAc binding and fucose-binding, respectively. s-s indicates disulfide bond.
Mutations of Bombyx fng in each fl mutant:
We determined the complete cDNA sequences of the fng genes amplified by RT–PCR from the fl, flo, and fln mutants. In the fng ORF of fl and flo, we found a 103-bp deletion, which corresponds to nearly all of exon 3. This deletion could cause a frameshift within exon 4 and a premature TAA stop codon before the completion of normal translation (Figure 2E, fl and flo). In the fng ORF of fln, we found a duplication of exons 2 and 3, which possibly cause a premature TGA stop codon in the second exon 3 (Figure 2E, fln). These results demonstrate that the three alleles, fl, flo, and fln, encode nonsense-mutated fng genes. The DxD motif, which is important for Fng function (Munro and Freeman 2000), is encoded in exon 4 of Bombyx fng (Figure 2E, WT). It is noteworthy that premature stop codons in the three nonsense mutants, fl, flo, and fln, appear before the DxD motif, which would cause the expression of incomplete Fng proteins without essential enzymatic activity (Figure 2E, fl, flo, and fln). We further characterized the deletion of the fng gene in the flk mutant by Southern hybridization in the whole genomes of the WT (+/flk heterozygote) and flk (flk/flk homozygote) using the full-length fng cDNA as the probe (Figure 2C). One to three bands appeared in each lane of the +/flk genome, and some of these were also present in the flk/flk genome, suggesting that the fng gene is not completely deleted from the flk genome. We designed primer sets to the genomic region of Bombyx fng and found that the genomic region containing exon 8 of fng was amplified in the flk genome by PCR but exons 1–3 of fng were not amplified (supplemental Figure 8). We inferred that the 5′ end of the fng gene has been removed from the flk genome. We also found that the complete Anxb13 and LRPL and most of NOS/NSL have been deleted from the flk genome (Figure 2B). From the average size of the BAC clones, we estimated that a region of >300 kb, from fng to NOS/NSL, is deleted (Figure 2B). This structural analysis of the genomes of the four fl mutant strains clearly shows that the functional Fng protein is not expressed from any of the fl alleles.
Expression profiles of Bombyx fng in wing discs and other larval tissues:
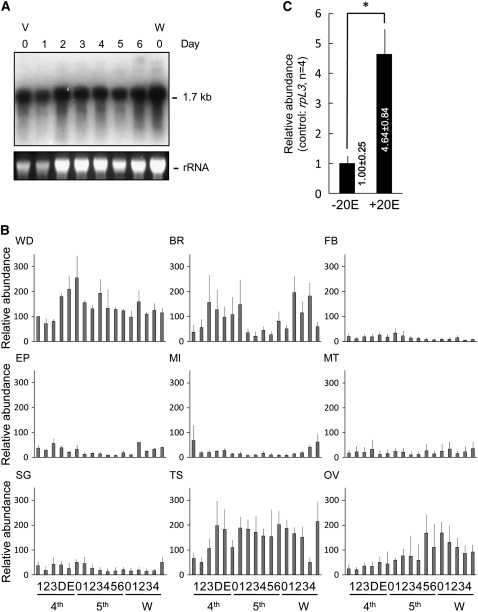
To identify the functional role of Bombyx Fng in wing morphogenesis, we first analyzed the expression of fng mRNA in the wing discs of WT fifth instar larva by Northern hybridization (Figure 4A). Bombyx fng is expressed continually as a single 1.7-kb transcript during the last larval instar. To compare fng expression semiquantitatively among larval organs, we used RT–PCR to amplify transcripts from various larval organs from the fourth instar to the prepupal stages (Figure 4B and supplemental Table S1). We observed significant expression of Bombyx fng in the wing discs (WD), brain (BR), testis (TS), and ovary (OV), but only a little in the other tissues, including the epidermis (EP), posterior silk gland (SG), midgut (MI), fat body (FB), and Malpighian tubule (MT). In Drosophila, the expression of fng transcripts has also been observed in the wing, eye, leg imaginal discs, ovarian follicle cells, and longitudinal glial cells of the central nervous system and is necessary for the correct development of those tissues (Irvine and Wieschaus 1994; Cho and Choi 1998; Domínguez and De Celis 1998; Papayannopoulos et al. 1998; Grammont and Irvine 2001; Rauskolb 2001; Thomas and Van Meyel 2007). It is intriguing that the major defect in the fl mutants, which have lost the Fng function, appears only in wing morphogenesis, even though Bombyx fng is expressed in several tissues other than the wing discs.
Figure 4.—
Expression patterns of Bombyx fng in WT larva. (A) Northern analysis of fng in wing discs during fifth larval instar. Ethidium bromide staining of rRNA is shown as a loading control. V0 to V6, days 0–6 of fifth instar larva; W0, day 0 of wandering stage. (B) Developmental profiles of fng expression in wing discs (WD), brain (BR), fat body (FB), epidermis (EP), midgut (MI), Malpighian tubule (MT), silk gland (SG), testis (TS), and ovary (OV), by semiquantitative RT–PCR. Relative abundance of fng mRNA during days 1–3 of fourth larval instar (4th), D and E of 4th molting stage (4th), days 0–6 of fifth larval instar (5th) and days 0–4 of prepupal (W, wandering stages) is shown by shaded columns. Vertical thin line indicates standard deviation (n = 3) (see supplemental Table S1). The rpL3 expression was used as an internal control. (C) Induction of mRNA of the fng gene by 20E in the wing discs of Bombyx. The discs were cultured for 12 hr with Grace's medium with 2 μg/ml 20E (+20E) or without (−20E), and the relative amount of mRNA was estimated by semiquantitative RT–PCR using rpL3 as an internal control. *P < 0.001.
The increase of the fng expression at the fourth molting stage (4th D and E) and after the wandering stage in several tissues, such as the WD, seems to be concurrent with the rising titer of ecdysteroid in those stages. To examine whether the expression of Bombyx fng is ecdysteroid inducible, we cultured the wing discs from the WT fifth instar larvae in Grace's medium with or without 20E and compared their responses to ecdysteroid by semiquantitative RT–PCR (Figure 4C). We observed an upregulation of fng mRNA expression in the wing discs cultured in the presence of 20E (Figure 4C). The general responses to ecdysteroid and the ecdysteroid-activated pathway have been studied extensively in Drosophila and Manduca sexta (Thummel 1995; Riddiford et al. 2003). Ecdysteroid exerts its effects by the transcriptional stimulation of its response genes, which define the functional and morphological properties of the tissue. This result suggests that Bombyx fng expression is induced by ecdysteroid and is involved in the ecdysteroid-activated pathway.
Expression patterns of Bombyx fng and BmWnt-1 mRNAs in wing discs:
In Drosophila wing discs, fng mRNA is especially detected in the dorsal region from the second to mid-third instars (Irvine and Wieschaus 1994). The expression pattern of Schistocerca fng has been investigated in segment morphogenesis and in the compound eye, legs, and ovary, but not in the wing (Dearden and Akam 2000). Among the fng orthologs, Precis fng (Correia et al. 2003) is most closely related to Bombyx fng (Figure 3C), but its expression pattern has not been established. To examine whether Bombyx fng localizes in the wing discs like Drosophila fng, whole-mount in situ hybridization was performed on the wing imaginal discs using the full-length ORF of Bombyx fng as the probe. In the wing imaginal discs of Bombyx, the fng transcripts were expressed in the dorsal apical region (Figure 5, A and B). This expression pattern of Bombyx fng is consistent with that of Drosophila fng. To our knowledge, this is the first report of the fng expression pattern in the wing discs of a Lepidopteran insect or of an insect other than Drosophila.
Figure 5.—
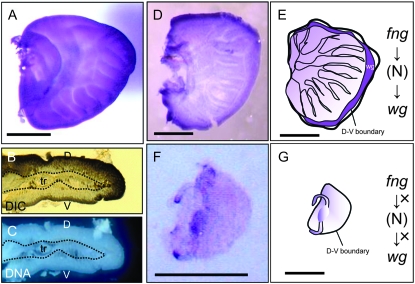
Whole mount in situ hybridization of Bombyx fng and BmWnt-1, Bombyx homolog of wg in wing discs. (A and B) Expression of Bombyx fng in wing discs (A, dorsal view; B, cross-section). The fng cDNA antisense probe was hybridized to the wing discs of last instar larvae. (C) DAPI staining in cross-section. D, dorsal; V, ventral; tr, trachea. (D–G) Expression of BmWnt-1 in wing discs. BmWnt-1 cDNA antisense probe was hybridized to the wing discs of the last instar larvae. D and F show the expression patterns in wild-type and fl wing discs, respectively. E and G show the schematic representation of the BmWnt-1 expression in wild-type and fl wing discs, respectively. Bars, 0.5 mm.
Shutting down the Fng function in fl wing discs was expected to severely affect the expression of genes in its downstream signaling pathway. The expression of wg is induced in the dorsoventral boundary region by the activation of Fng-mediated N signaling (Panin et al. 1997; Haines and Irvine 2003), and wg expression along the wing margin is important for wing morphogenesis in Drosophila (Couso et al. 1994; Klein and Martinez Arias 1998). Therefore, we first analyzed the expression pattern of BmWnt-1 mRNA in the wing discs of fifth instar WT larvae with whole-mount in situ hybridization using the full-length BmWnt-1 cDNA as the probe (Figure 5D). BmWnt-1 expression was observed along the entire dorsoventral boundary of the WT wing disc, which occupies the margin of the wing disc in the Lepidoptera (Figure 5, D and E). Next, we examined BmWnt-1 expression in the fl wing disc of fifth instar larvae, which is smaller than the WT disc and does not show any morphological changes, such as tracheal invasion. In the fl wing disc, we detected no distinct signals for BmWnt-1 mRNA at the wing margin or in any other wing area (Figure 5F). These results suggest that the Fng-mediated N signaling pathway and wg expression are conserved and are essential for the process of wing morphogenesis in Bombyx, as in Drosophila and other organisms. These data also suggest that the loss of Fng function inhibits the activation of the N signaling pathway, which results in the wing-deficient phenotype observed in the fl mutants of the silkworm (Figure 5G).
DISCUSSION
In this study, we elucidated the genome structure across the fl locus and identified that the responsible gene for fl is fng. To our knowledge, this is the first report to use positional cloning of the responsible gene for an intriguing allele, such as the wing-deficient mutant in the silkworm, B. mori. We initially thought that the wing-deficient mutant of the silkworm, fl, may be caused by the functional loss of some known genes identified in Drosophila, such as ap or vestigial, mutations of which cause a wingless phenotype similar to that of fl. On this point, the finding that fng is the fl gene is an unexpected result, because the severe loss-of-function mutants of fng are the postembryonic lethal in Drosophila (Irvine and Wieschaus 1994; Correia et al. 2003).
fng is the gene responsible for fl:
In the flk mutant, we identified a large chromosomal deletion that includes five putative genes: fng, Anxb13, LRPL, NOS, and NSL (Figure 2, B and C). In the other three fl alleles, we detected nonsense mutations in the fng gene (Figure 2, D and E), but only synonymous or insignificant amino acid changes in the other four genes (supplemental Figures 2–5), strongly suggesting that fng is the gene responsible for fl. In the flk homozygote, all five genes are deleted from the chromosomes, but the loss of Anxb13, LRPL, NOS, and NSL seems to have no vital effect on flk development, because there is no remarkable difference in the phenotypes of flk and the other fl mutants. We also estimated the distance between fng and the nearest known gene, KMO, to be <200 kb by calculating the average size of the BAC clones. This value coincides well with the 0.3-cM distance between them on the classical genetic map (Figure 2A).
We found no fng homolog other than Bombyx fng in the silkworm genome or expressed sequence tag (EST) databases (data not shown and supplemental Figure 6). Southern hybridization also demonstrated that the silkworm genome contains a single copy of the fng homolog. These results suggest that fng at the fl locus is the only functional fng ortholog in B. mori, although we cannot completely exclude the possibility that there is a gene that compensates for fng function in the tissues other than the wing discs, despite its low sequence similarity to fng. Bombyx Fng has the amino acids that are required for UDP-GlcNAc binding and putative fucose binding, as in mouse Mfng and Drosophila Fng. On the basis of a phylogenetic tree constructed from the amino acid sequences of Fng from a wide variety of organisms, Bombyx Fng (BmFringe) on silkworm LG10 is most closely related to butterfly Fng (PcFringe) (Figure 3C). Furthermore, Lepidopteran Fng proteins (BmFringe and PcFringe) are more distantly related to Drosophila or Apis Fng than to Schistocerca Fng. Several unique amino acid residues are observed, which are either Lepidopteran type (conserved in Lepidopteran Fng) or non-Lepidopteran type (conserved in Fng of other insects) (supplemental Figure 7). Although the biological significance of these differences is unknown, we presume that these amino acid residues are responsible for the phylogenetic relationships of the Lepidopteran Fng proteins and other insect Fng proteins.
Tissue- and temporal-specific fng expression:
Males and females of the recessive fl homozygote, which lack wings, can be crossed with each other to produce an F1 generation that also develops normally, except for wing morphogenesis, suggesting that these homozygotes are fertile and their sexual behavior is normal. As shown in Figure 4B, RT–PCR analysis of various tissues from the fourth instar to the prepupal stages showed that fng mRNA is expressed abundantly in wing discs, brain, testis, and ovary but only to a small extent in other tissues. Bombyx Fng in the wing discs must be involved in wing morphogenesis because the expression of a downstream marker of Fng-mediated N signaling pathway, wg, is repressed in the fl wing discs (Figure 5, F and G). However, we cannot explain the exact functional role of Bombyx fng in the brain or reproductive organs because it seems that the reproductive ability and behavior of the fl mutants are not affected. As a trivial effect, we usually observed that the third thoracic legs of the fl mutants move more slowly than the corresponding WT legs, suggesting that the neurogenesis of the fl mutants is affected through the loss of fng function in the brain and nervous systems.
The fng expression in the wing discs and brain increased in the fourth molting stage and after the wandering stage (Figure 4B). Previous morphological studies have shown that several defects in fl wing development become evident immediately after the fourth larval ecdysis, suggesting that the fl gene product is required around the fourth molting stage. The titer of ecdysteroid, which regulates various morphological events, peaks at each molting stage and at the prepupal stages. The correlation between the fng expression pattern and the ecdysteroid titer implies that fng mRNA is induced by ecdysteroid. We demonstrated that fng is an ecdysteroid-inducible gene in Bombyx (Figure 4C). These results suggest that Bombyx fng is part of the ecdysteroid-activated pathway and plays a wing-specific role.
We have shown previously that fl wing discs lose their responsiveness to ecdysteroid in a wing-disc specific manner (Fujiwara and Hojyo 1997; Hojyo and Fujiwara 1997; Matsuoka and Fujiwara 2000). The expressions of a 20E-inducible 41-kDa protein, BHR3, and Urbain were also reduced in fl wing discs. These results imply that the loss of fng function specifically affects the downstream pathway of ecdysteroid signaling and leads to a deficiency in wing formation. Although we do not have direct evidence at present, the fl wing disc is a good model in which to study the unknown crosstalk between the ecdysteroid and Fng-mediated N signaling pathways in insect development.
Crucial role of fng in Bombyx wing morphogenesis:
Here, we detected fng expression in the dorsal-specific region of Bombyx wing discs, which is similar to that in Drosophila (Figure 5, A and B). Previously, the analysis of Precis homologs of Drosophila appendage patterning genes revealed that butterfly ap and wg are expressed exclusively in dorsal cells and in cells along the future wing margin of the Precis wing discs, respectively (Carroll et al. 1994; Weatherbee et al. 1999). This fact implies that Butterfly fng is also expressed in the dorsal region of the wing discs, under the control of ap, as in Drosophila. Furthermore, from our results, wg is expressed along the border between the dorsal and ventral cells in the WT but not in fng null mutants of B. mori (Figure 5, D–G). These results suggest that the wing fields of Drosophila and Lepidopteran insects are organized and regulated in a similar manner and that the function of fng is conserved in Bombyx and Drosophila, although the mode of action of Bombyx fng remains speculative at present.
The Lepidopteran wing margin contains a special structure called the “bordering lacuna” (BL) (Fujiwara and Ogai 2001). In the early pupal stage of wing development, ecdysteroid induces cell proliferation and differentiation proximal to the BL and programmed cell death distal to the BL (Fujiwara and Ogai 2001). The outline of the adult wings of moths and butterflies emerges as a result of the disappearance of the peripheral region during pupal stages. These phenomena are specific to the Lepidoptera and are widely observed in most butterflies and moths. We recently found that two ecdysteroid receptor isoforms, EcR-A and EcR-B1, are expressed precisely in the cell death and cell proliferation areas in the wings, respectively (Lobbia et al. 2007). It is noteworthy that the EcR-A expression region corresponds to the region of the wg expression region during wing morphogenesis. Fng-mediated N signaling induces wg expression, which is involved in the formation of the wing margin in Drosophila and probably also in Bombyx. Therefore, it is possible that Bombyx fng regulates the expression of wg, EcR-A, or both, to produce the BL structure in Lepidoptera, although further studies are required to support this hypothesis.
Different developmental effects of the fng defect on Bombyx and Drosophila:
In Drosophila, fng null mutants or fng mutants with an abnormal DxD motif are lethal during early larval stages (Irvine and Wieschaus 1994; Correia et al. 2003). Drosophila Fng plays essential roles in the morphogenesis of the leg and compound eye imaginal discs and in several other tissues, as well as in the wing imaginal discs (Papayannopoulos et al. 1998; Grammont and Irvine 2001; Rauskolb 2001; Thomas and Van Meyel 2007). In contrast, null mutants of Bombyx fng are viable and fertile, and Bombyx fng seems to be required for wing development in Bombyx, unlike in Drosophila. We are interested in understanding the difference in fng functions between Bombyx and Drosophila. It has been suggested that the developmental processes of organogenesis differ between the Lepidoptera and Drosophila (Kim 1959; Tanaka and Truman 2005; Allee et al. 2006; Franco et al. 2007). Drosophila imaginal organs develop from each imaginal disc in a concentric region. However, in Bombyx and other Lepidoptera, most adult organs are derived from the immature epithelial region called the “primordium,” except for the wings, which develop from the wing imaginal discs. For example, Lepidoptera legs do not develop from imaginal discs but from the cylindrical epidermis of the caterpillar's legs (Kim 1959; Tanaka and Truman 2005). In the developing leg imaginal discs of Drosophila, Fng-mediated N signaling functions to establish segmentation and the promotion of leg growth (Rauskolb 2001). Our results suggest that Bombyx legs and adult organs, other than the wings, develop and grow independently of fng function. Thus, the critical difference in the developmental systems of Drosophila and the Lepidoptera may relate to the different functional roles of fng. It will be interesting to investigate in more detail the mechanism underlying fng function in the development of the adult organs of Lepidoptera and to determine the developmental systems in the Lepidoptera.
Acknowledgments
This work was supported by grants from the Ministry of Education, Science, and Culture (MEXT) of Japan, National Bio-resource Project (NBRP), the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN), and a research fellowship of the Japan Society for the Promotion of Science for Young Scientists to T.M.M. The sequencing of BAC clones was partly supported by funds from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
References
- Allee, J. P., C. L. Pelletier, E. K. Fergusson and D. T. Champlin, 2006. Early events in adult eye development of the moth, Manduca sexta. J. Insect Physiol. 52 450–460. [DOI] [PubMed] [Google Scholar]
- Baonza, A., and A. Garcia-Bellido, 2000. Notch signaling directly controls cell proliferation in the Drosophila wing disc. Proc. Natl. Acad. Sci. USA 97 2609–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais, C. B., and R. Lafont, 1980. In vitro differentiation of Pieris brassicae imaginal wing discs: effects and metabolism of ecdysone and ecdysterone. Wilhelm Roux' Arch. Entwicklungsmech. Org. 188 27–36. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., J. Gates, D. N. Keys, S. W. Paddock, G. E. Panganiban et al., 1994. Pattern formation and eyespot determination in butterfly wings. Science 265 109–114. [DOI] [PubMed] [Google Scholar]
- Cho, K. O., and K. W. Choi, 1998. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature 396 272–276. [DOI] [PubMed] [Google Scholar]
- Correia, T., V. Papayannopoulos, V. Panin, P. Woronoff, J. Jiang et al., 2003. Molecular genetic analysis of the glycosyltransferase Fringe in Drosophila. Proc. Natl. Acad. Sci. USA 100 6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso, J. P., S. A. Bishop and A. Martinez Arias, 1994. The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development 120 621–636. [DOI] [PubMed] [Google Scholar]
- Dearden, P., and M. Akam, 2000. A role for Fringe in segment morphogenesis but not segment formation in the grasshopper, Schistocerca gregaria. Dev. Genes Evol. 210 329–336. [DOI] [PubMed] [Google Scholar]
- Domínguez, M., and J. F. De Celis, 1998. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature 396 276–278. [DOI] [PubMed] [Google Scholar]
- Franco, M. D., J. Bohbot, K. Fernandez, J. Hanna, J. Poppy et al., 2007. Sensory cell proliferation within the olfactory epithelium of developing adult Manduca sexta (Lepidoptera). PLoS ONE 2 e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H., Y. Banno, H. Doira, H. Kihara and Y. Kawaguchi, 1998. Genetical Stocks and Mutations of Bombyx mori: Important Genetic Resources, Ed. 2. Institute of Genetic Resources, Kyushu University, Fukuoka, Japan.
- Fujiwara, H., and T. Hojyo, 1997. Developmental profiles of wing imaginal discs of flügellos (fl), a wingless mutant of the silkworm, Bombyx mori. Dev. Genes Evol. 207 12–18. [DOI] [PubMed] [Google Scholar]
- Fujiwara, H., and S. Ogai, 2001. Ecdysteroid-induced programmed cell death and cell proliferation during pupal wing development of the silkworm, Bombyx mori. Dev. Genes Evol. 211 118–123. [DOI] [PubMed] [Google Scholar]
- Grammont, M., and K. D. Irvine, 2001. fringe and Notch specify polar cell fate during Drosophila oogenesis. Development 128 2243–2253. [DOI] [PubMed] [Google Scholar]
- Haines, N., and K. D. Irvine, 2003. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 4 786–797. [DOI] [PubMed] [Google Scholar]
- Harizuka, M., 1948. The linkage between the genes for flügellos (fl) and the white egg 2 (w-2). J. Seric. Sci. Jpn. 17 6–8 (in Japanese). [Google Scholar]
- Hojyo, T., and H. Fujiwara, 1997. Reciprocal transplantation of wing discs between a wing deficient mutant (fl) and wild type of the silkworm, Bombyx mori. Dev. Growth Differ. 39 599–606. [DOI] [PubMed] [Google Scholar]
- Imamura, M., J. Yang and M. Yamakawa, 2002. cDNA cloning, characterization and gene expression of nitric oxide synthase from the silkworm, Bombyx mori. Insect Mol. Biol. 11 257–265. [DOI] [PubMed] [Google Scholar]
- Irvine, K. D., and E. Wieschaus, 1994. fringe, a boundary-specific signaling molecule, mediates interaction between dorsal and ventral cells during Drosophila wing development. Cell 79 595–606. [DOI] [PubMed] [Google Scholar]
- Jinek, M., Y. W. Chen, H. Clausen, S. M. Cohen and E. Conti, 2006. Structural insights into the Notch-modifying glycosyltransferase Fringe. Nat. Struct. Mol. Biol. 13 945–946. [DOI] [PubMed] [Google Scholar]
- Katsuki, K., 1935. Weitere vershche uber erbliche mosaikbildung und gynandromorphismus bei Bombyx mori L. Biol. Zentralbl. 55 361–383. [Google Scholar]
- Kim, C. W., 1959. The differentiation centre inducing the development from larval to adult leg in Pieris brassicae (Lepidoptera). J. Embryol. Exp. Morphol. 7 572–582. [PubMed] [Google Scholar]
- Kim, J., K. D. Irvine and S. B. Carroll, 1995. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell 82 795–802. [DOI] [PubMed] [Google Scholar]
- Klein, T., and A. Martinez Arias, 1998. Interactions among Delta, Serrate and Fringe modulate Notch activity during Drosophila wing development. Development 125 2951–2962. [DOI] [PubMed] [Google Scholar]
- Koike, Y., K. Mita, M. G. Suzuki, S. Maeda, H. Abe et al., 2003. Genomic sequence of a 320-kb segment of the Z chromosome of Bombyx mori containing a kettin ortholog. Mol. Genet. Genomics 269 137–149. [DOI] [PubMed] [Google Scholar]
- Lobbia, S., R. Futahashi and H. Fujiwara, 2007. Modulation of the ecdysteroid-induced cell death by juvenile hormone during pupal wing development of Lepidoptera. Arch. Insect Biochem. Physiol. 65 152–163. [DOI] [PubMed] [Google Scholar]
- Matsunaga, T. M., and H. Fujiwara, 2002. Identification and characterization of genes abnormally expressed in wing-deficient mutant (flügellos) of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 32 691–699. [DOI] [PubMed] [Google Scholar]
- Matsuoka, T., and H. Fujiwara, 2000. Expression of ecdysteroid-regulated genes is reduced specifically in the wing discs of the wing-deficient mutant (fl) of Bombyx mori. Dev. Genes Evol. 210 120–128. [DOI] [PubMed] [Google Scholar]
- Mita, K., M. Kasahara, S. Sasaki, Y. Nagayasu, T. Yamada et al., 2004. The genome sequence of silkworm, Bombyx mori. DNA Res. 11 27–35. [DOI] [PubMed] [Google Scholar]
- Munro, S., and M. Freeman, 2000. The Notch signalling regulator Fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr. Biol. 10 813–820. [DOI] [PubMed] [Google Scholar]
- Nagata, T., 1962. Development of the wings in normal type and wingless mutant of the silkworm, Bombyx mori L. Bull. Fac. Text Sci. 3 341–371 (in Japanese). [Google Scholar]
- Nardi, J. H., and J. H. Willis, 1979. Control of cuticle formation by imaginal discs in vitro. Dev. Biol. 68 381–395. [DOI] [PubMed] [Google Scholar]
- Panin, V. M., V. Papayannopoulos, R. Wilson and K. D. Irvine, 1997. Fringe modulates Notch-ligand interactions. Nature 387 908–912. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos, V., A. Tomlinson, V. M. Panin, C. Rauskolb and K. D. Irvine, 1998. Dorsal-ventral signaling in the Drosophila eye. Science 281 2031–2034. [DOI] [PubMed] [Google Scholar]
- Quan, G.X., I. Kim, N. Komoto, H. Sezutsu, M. Ote et al., 2002. Characterization of the kynurenine 3-monooxygenase gene corresponding to the white egg 1 mutant in the silkworm Bombyx mori. Mol. Genet. Genomics 267 1–9. [DOI] [PubMed] [Google Scholar]
- Rauskolb, C., 2001. The establishment of segmentation in the Drosophila leg. Development 128 4511–4521. [DOI] [PubMed] [Google Scholar]
- Riddiford, L. M., K. Hiruma, X. Zhou and C. A. Nelson, 2003. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol. 33 1327–1338. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., and J. W. Truman, 2005. Development of the adult leg epidermis in Manduca sexta: contribution of different larval cell populations. Dev. Genes Evol. 215 78–89. [DOI] [PubMed] [Google Scholar]
- Thomas, G. B., and D. J. Van Meyel, 2007. The glycosyltransferase Fringe promotes Delta-Notch signaling between neurons and glia, and is required for subtype-specific glial gene expression. Development 134 591–600. [DOI] [PubMed] [Google Scholar]
- Thummel, C. S., 1995. From embryogenesis to metamorphosis: the regulation and function of Drosophila nuclear receptor superfamily members. Cell 83 871–877. [DOI] [PubMed] [Google Scholar]
- Truman, J. W., and L. M. Riddiford, 2007. The morphostatic actions of juvenile hormone. Insect Biochem. Mol. Biol. 37 761–770. [DOI] [PubMed] [Google Scholar]
- Ueda, S., T. Hirao and M. Harizuka, 1959. The crossing-over rate between the genes for flügellos (fl) and white egg 1 (W-1) of the silkworm, Bombyx mori. Sansi-Kenkyu 28 6–8 (in Japanese). [Google Scholar]
- Weatherbee, S. D., H. F. Nijhout, L. W. Grunert, G. Halder, R. Galant et al., 1999. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr. Biol. 9 109–115. [DOI] [PubMed] [Google Scholar]
- Williams, J. A., S. W. Paddock and S. B. Carroll, 1993. Pattern formation in a secondary field: a hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete subregions. Development 117 571–584. [DOI] [PubMed] [Google Scholar]
- Xia, Q., Z. Zhou, C. Lu, D. Cheng, F. Dai et al., 2004. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306 1937–1940. [DOI] [PubMed] [Google Scholar]