Abstract
We report on the construction of sex-specific high-density linkage maps and identification of sex-linked markers for the black tiger shrimp (Penaeus monodon). Overall, we identified 44 male and 43 female linkage groups (2n = 88) from the analysis of 2306 AFLP markers segregating in three full-sib families, covering 2378 and 2362 cM, respectively. Twenty-one putatively homologous linkage groups, including the sex-linkage groups, were identified between the female and male linkage maps. Six sex-linked AFLP marker alleles were inherited from female parents in the three families, suggesting that the P. monodon adopts a WZ–ZZ sex-determining system. Two sex-linked AFLP markers, one of which we converted into an allele-specific assay, confirmed their association with sex in a panel of 52 genetically unrelated animals.
SHRIMP represent one of the most valuable aquaculture crops (Food and Agriculture Organization of the United Nations 2005). Several species are cultivated and are in varying stages of domestication (Argue and Alcivar-Warren 2000; Cuzon et al. 2004). The process is most advanced for the Pacific white shrimp [Penaeus (litopenaeus) vannamei] where the bulk of production is from domesticated stocks and for which there are a number of specific genetic improvement programs (e.g., Gitterle et al. 2005). For most species, including the other dominant farmed species P. monodon, only a small proportion of production, if any, is from domesticated sources, and specific genetic improvement programs are few and largely experimental (Argue and Alcivar-Warren 2000).
Despite the early stage of domestication of shrimp, some molecular markers and associated tools such as genetic maps are available. Molecular markers have been used for the analysis of the population structure of wild shrimp resources (for review, see Benzie 2000; Xu et al. 2001), strain and species identification (Khamnamtong et al. 2005), and parentage analysis (e.g., Jerry et al. 2006). First-stage linkage maps have been generated for P. monodon (Wilson et al. 2002; Maneeruttanarungroj et al. 2006), P. vannamei (Perez et al. 2004; Zhang et al. 2006), P. chinensis (Z. Li et al. 2006), and P. japonicus (Moore et al. 1999; Li et al. 2003). In the case of P. japonicus, a QTL for growth has been identified on the male map (Y. Li et al. 2006). Nevertheless, the majority of these tools are preliminary and some quite fundamental aspects of shrimp biology important for further genetic work remain unclear, such as the mechanism of sex determination in shrimp.
In the case of sex determination, circumstantial evidence on interspecies hybrids between P. monodon and P. esculentus indicates that the female is the heterogametic sex, as the sex ratio of the surviving progeny was skewed to males (Benzie et al. 2001). According to Haldane's rule, a higher mortality in such crosses is experienced by the heterogametic sex. In P. japonicus and in P. vannamei, sex was found to map to a linkage group on the female map (Li et al. 2003; Zhang et al. 2006), suggesting that sex in these penaeids is determined by a WZ–ZZ chromosomal system. However, none of the markers reported was actually closely linked to the sex locus. Only more extensive mapping will clarify this situation.
Most of the maps for shrimp were generated using AFLP technology (Vos et al. 1995), in part because other markers such as simple sequence repeats proved difficult to develop (Li et al. 2003). AFLP markers offer several advantages. Foremost among these is that the AFLP technology requires no prior sequence information and, hence, has a relatively low start-up cost. In addition, the AFLP technique is amenable to automation and is highly multiplexed. This offers the potential to improve the efficiency and throughput of marker data production in organisms that lack the genomics platform necessary for the development of genotyping arrays (Vuylsteke et al. 2007a), as in most aquaculture species. Finally, the ability to score AFLP markers codominantly on the basis of quantitative measurements of the band intensities using AFLP-QuantarPro software (http://www.keygene-products.com) allows extraction of more genetic information from AFLP fingerprints than dominant (presence/absence) scoring, and, hence, speeds up the mapping process (Vuylsteke et al. 2007a).
Here, we report on AFLP-based linkage maps of P. monodon with a marker density and genome coverage that exceeds the currently available linkage maps of any penaeid species. We also present sex-linked markers, of which one is converted into an allele-specific assay, that disclose linkage groups corresponding to both the W and the Z chromosomes and female heterogamety in P. monodon.
MATERIALS AND METHODS
Production of families:
Three full-sib (FS) families, containing ∼115 individual F1 progeny each (Table 1), were produced from controlled crosses under specific pathogen-free conditions at Moana Technologies in Hawaii. All four parental animals originated from breeding stock of Moana Technologies. Two FS families, named FAM1 and FAM2, had “IC67” as a female parent in common, while FAM2 and FAM3 had “AL91” as a male parent in common. As a result, we had two female and two male half-sib (HS) families with one family in common. At post-larval stage 120 (i.e., 120 days after metamorphosis from mysis stage), the phenotypic sex of each individual progeny was recorded and tail/muscle samples were flash frozen in a mixture of alcohol and dry ice and stored at −70°. Muscle samples were also taken from the parental animals.
TABLE 1.
Parents and progeny (genotyped) for each family
| Family | Dam | Sire | Total no. of progeny | Female progeny genotyped | Male progeny genotyped |
|---|---|---|---|---|---|
| FAM1 | IC67 | AL99 | 120 | 47 | 46 |
| FAM2 | IC67 | AL91 | 113 | 53 | 40 |
| FAM3 | IC100 | AL91 | 111 | 51 | 42 |
DNA extraction:
The frozen tail samples were pulverized with mortar and pestle and DNA was extracted with a modified CTAB method for shrimp tissue. Briefly, 150 μl of CTAB buffer (2% w/v cetyl trimethyl-ammonium bromide, 1.4 m NaCl, 20 mm EDTA, 100 mm Tris–HCl, pH 7.5, and 0.25% v/v 2-mercaptoethanol) was added to 25 mg of sample powder and gently homogenized. After complete homogenization, an extra 750 μl of CTAB buffer was added. The tissue slurry was incubated for 30 min at 25°. After transferring the supernatant to a clean tube, 600 μl PCA solution (25 vol phenol, 24 vol chloroform, 1 vol isoamylalcohol) was added and the mixture was mixed vigorously for 20 sec. Phases were separated by centrifugation (10,000 × g for 5 min), and 800 μl of the upper aqueous phase was transferred to a clean tube and mixed vigorously for 20 sec with 600 μl of CA solution (24 vol chloroform, 1 vol isoamylalcohol). Next, 700 μl of the upper aqueous phase was removed to a clean tube and gently mixed with 630 μl (i.e., 0.9 vol) isopropanol, followed by incubation for 1 hr at −70°. Genomic DNA was precipitated by centrifugation (12,000 × g for 15 min), washed with 70% ethanol, air dried, and resuspended in 50 μl of water.
Bulked segregant analysis:
We performed a large-scale bulked segregant analysis (BSA; Michelmore et al. 1994) using AFLP to detect sex-linked AFLP markers for P. monodon. Two bulks containing either five male or female segregants were generated for each family. AFLP analysis was performed as described by Vuylsteke et al. (2007a) using the enzyme combination EcoRI/MseI. AFLP adapter and primer sequences are as described by Vuylsteke et al. (2007a). AFLP images were generated with LI-COR-automated DNA sequencers (NEN Global IR2 system; LI-COR Biosciences, Lincoln, NE) and infrared dye (IRD) detection technology.
Segregation and linkage analysis:
From each family, 93 segregants were analyzed with 125 EcoRI + 3/MseI + 3 AFLP primer combinations (PCs). AFLP PCs used and the number of polymorphic fragments observed for each PC in the different mapping families are listed in supplemental Table 1. AFLP markers were scored on the basis of relative fragment intensities, using the specific image analysis software AFLP-QuantarPro (http://www.keygene-products.com). Each AFLP marker was identified by (1) a code referring to the corresponding PC (encoded according to supplemental Table 1) followed by the estimated molecular size of the fragment in nucleotides as estimated by AFLP-QuantarPro. AFLP markers heterozygous in one parent and homozygous in the other, and expected to segregate 1:1 in the F1 generation, were termed “female” or “male,” depending on the sex of the heterozygous parent. AFLP markers heterozygous in both parents, and expected to segregate 1:2:1 in the F1 generation were termed “biparental” markers. Whenever feasible, biparental markers were scored codominantly (i.e., following a 1:2:1 segregation pattern), but dominantly (i.e., conforming to a 3:1 segregation pattern) when the heterozygotes could not reliably be discriminated from the individuals homozygous for the “band present” allele.
Linkage analysis and segregation distortion tests were performed using the software package JoinMap 3.0 (Van Ooijen and Voorrips 2001). The appropriate mapping population type was set to option CP [a population resulting from a cross between two heterogeneously heterozygous and homozygous diploid parents, linkage phases (possibly) originally unknown]. As for population type CP, the segregation type (SEG) might vary across the loci, a code indicating the segregation type has to be given. The SEG of the female and male markers was set to <lm × ll> and <nn × np>, respectively, and biparental markers to the <hk × hk> SEG type. The two characters left and right of the “×” in these codes correspond to the two AFLP marker alleles of the first and second parent, respectively; each distinct AFLP marker allele is represented by a different character. We first ran through a fairly wide range of LOD thresholds, from 2.0 to 10.0, to obtain a proper view of what might be the best grouping. The LOD scores used by JoinMap are based on χ2 tests for independence of segregation, which are somewhat different from the usual LOD scores used in linkage analysis. In general, we decided to use the grouping obtained with a LOD score of 6.0. In a few cases, the grouping obtained at a LOD threshold of 7.0 was used. Only linkage groups containing at least three markers were considered for map construction.
When a linkage group contained both female and male markers, separate female and male linkage maps were constructed. Biparental markers were included in both the female and the male maps. Maps were constructed in three rounds, each producing a linkage map. In this map-building procedure, each map was calculated by using the pairwise data of loci present on the map, with default settings (recombination frequency (REC) < 0.4; LOD threshold > 1). Once the well-fitting markers (causing a change in goodness of fit smaller than the threshold: 5) were positioned on the map (after two rounds), the remaining markers were forced onto the map by annulling the jump threshold. When the markers in the third map caused a jump in goodness of fit larger than an arbitrary threshold of 10, the second map was selected as the final map, and the third map otherwise. Single markers with a segregation ratio in discordance with the flanking markers (i.e., markers showing heavy segregation distortion flanked by a number of nondistorted markers) were discarded, and the map construction was repeated. A marker order was not forced on any linkage group during map construction. Recombination frequencies were converted to Kosambi centimorgans prior to the map estimation.
Sex-specific map integration:
The genotype data from FAM1 and FAM2 were combined to calculate an integrated female map. Likewise, marker groups from FAM2 and FAM3 that relate to the same linkage group were combined to calculate an integrated male map. Corresponding linkage groups were identified on the basis of the presence of identical AFLP markers. Integrated female and male maps were calculated on the basis of the mean recombination frequencies and combined LOD scores (Van Ooijen and Voorrips 2001) using JoinMap.
Testing genetic association of sex-linked AFLP marker alleles to sex on a population scale:
The genetic association between the six sex-specific AFLP marker alleles and sex was tested in a panel of 52 unrelated female and male animals from Moana Technologies breeding stock (i.e., 48 unrelated animals and the four parents from the mapping families). DNA was extracted from frozen tails as described for the families.
Conversion of a sex-linked AFLP marker into a PCR-based allele-specific assay:
Purifying 33P-labeled AFLP fragments from polyacrylamide gels followed by PCR amplification and subsequent sequencing was done as described in Vuylsteke et al. (2007b). To detect internal DNA sequence variation among the male and female E06M45M347.0 marker alleles, locus-specific primers ATT/GCA-1 and ATT/GCA-2 (Figure 3) were designed on the basis of the sequence obtained and used to amplify male and female genomic DNA. PCR reactions were done in 1×GeneAmp PCR buffer II (Applied Biosystems, Foster City, CA) supplemented with 1.5 ng/μl of each primer, 0.2 mm of each dNTP, 2.5 mm of MgCl2, 0.025 units of Taq polymerase, and 100 ng of DNA in a total volume of 20 μl. An initial denaturation step of 4 min at 95° was followed by 35 cycles of 30 sec denaturation at 95°, 30 sec annealing at 52°, 30 sec elongation at 72°, and a final elongation step of 2 min at 72°. To detect internal DNA sequence variation upstream of the primer ATT/GCA-1, primer ATT/GCA-2 and the corresponding EcoRI-specific AFLP primer (EcoRI + ATT) were used. PCR reactions were done in a total volume of 50 μl with 5 μl of a 20-fold diluted preamplification reaction product as template. PCR reactions were as follows: 35 cycles of 30 sec denaturation at 95°, 30 sec annealing at 53°, 30 sec elongation at 72°, and a final elongation step of 2 min at 72°.
Figure 3.—
ClustalW alignment (http://www.ebi.ac.uk/clustalw) showing polymorphisms between the female and male allelic sequences of the sex-specific AFLP marker E06M45M347.0 (solid background). The EcoRI and MseI sites are shaded; primers ATT/GCA-1 and ATT/GCA-2, used for amplifying both the female and male fragments from genomic DNA, are underlined; primers indel-4 and indel-5, flanking the 6-bp indel polymorphism, are boxed.
To convert the 6-bp indel into a PCR-based allele-specific assay, primers “indel-4” and “indel-5,” flanking the 6-bp indel (Figure 3), were designed. To allow amplification product detection with conventional autoradiography, the indel-4 primer was radiolabeled by phosphorylating the 5′-end of the primer with [γ-33P]ATP. The PCR reaction was performed in 1×GeneAmp PCR buffer II (Applied Biosystems) supplemented with 0.3 ng/μl of each primer, 0.2 mm of each dNTP, 2.5 mm of MgCl2, 0.025 units of Taq polymerase, and 50 ng of genomic DNA. The PCR program was as described above for the detection of internal DNA sequence variation. After electrophoresis of the amplification product on standard denaturing polyacrylamide gel, fingerprints were visualized with phosphorimager technology.
RESULTS
Bulked segregant analysis:
Initially, BSA was utilized to identify sex-linked AFLP markers. Two bulked samples containing either five male or five female segregants from FAM3 were screened for ∼70,400 AFLP fragments amplified by 1408 AFLP PCs. Totals of 114 and 42 AFLP fragments were present only in the female and male bulks, respectively.
Segregation analysis:
A total of 125 AFLP PCs, generating on average 10.6 segregating markers (supplemental Table 1) per family, resulted in a total of 2306 polymorphic AFLP fragments. The number of segregating AFLP markers common to the three families are illustrated with a Venn diagram (supplemental Figure 1). The two female HS families FAM1 and FAM2 and the two male HS families FAM2 and FAM3 share 693 and 898 markers, respectively, with 382 markers shared by the three families.
We discarded from the analysis 59, 95, and 90 marker loci for which 23 or more individuals (≥25%) could not be reliably genotyped in FAM1, FAM2, and FAM3, respectively. This resulted in 1141, 1207, and 1361 AFLP markers, most of which are female or male markers, available for linkage mapping for FAM1, FAM2, and FAM3, respectively (Table 2). Of the marker loci, 19–25% displayed significant distorted segregation ratios at the α = 0.05 level. As segregation distortion is a normal phenomenon in wide crosses, these loci were not a priori excluded, but evaluated after map construction.
TABLE 2.
Markers available for linkage mapping for each family
| Family
|
||||
|---|---|---|---|---|
| Marker type | FAM1 | FAM2 | FAM3 | Average |
| Female | 439 (92) | 431 (89) | 564 (121) | 478 |
| Male | 571 (155) | 624 (124) | 596 (113) | 597 |
| Biparental | ||||
| 1:2:1 | 42 (0) | 66 (0) | 98 (0) | 69 |
| 3:1 | 89 (37) | 86 (32) | 103 (28) | 93 |
| Total | 1141 (284) | 1207 (245) | 1361 (262) | 1236 |
Only markers with <25% missing genotypes are included. Numbers in parentheses represent the number of markers showing significant segregation distortion (P < 0.05).
Linkage mapping:
Linkage analysis was carried out for each of the three families separately and sex-specific maps were created for each family. The sex-specific genetic map data for each family are summarized in Table 3. Strikingly, in each family, more markers could be mapped in the male map than in the female map. However, there was a strong correlation between the number of markers available and those that were mapped (Pearson r = 0.926, P < 0.01). The female linkage maps generated from FAM1 and FAM2 are similar in length and number of mapped markers, but smaller than the FAM3 female linkage map in number of markers and in total map length (Table 3). In contrast, the three male maps are comparable to each other in number of markers and in total map length, irrespective their origin.
TABLE 3.
Statistics for the female and male maps for each family
| FAM1
|
FAM2
|
FAM3
|
||||
|---|---|---|---|---|---|---|
| Female | Male | Female | Male | Female | Male | |
| No. of markers | 378 | 552 | 377 | 602 | 499 | 598 |
| Map size (cM) | 1814 | 2286 | 1822 | 2300 | 2474 | 2283 |
| No. of linkage groups | 43 | 43 | 43 | 46 | 53 | 43 |
Construction of integrated sex-specific maps:
Linkage analysis was carried out on the combined genotype data of 39 corresponding linkage groups to calculate a common female linkage map across FAM1 and FAM2. The 39 corresponding linkage groups shared at least two markers. Two linkage groups were specific for the FAM1 and FAM2 female map each and were added to the common female map as such. As a result, a common female map containing 494 AFLP markers grouped in 43 linkage groups, spanning a total of 2362 cM, was constructed (Table 4 and supplemental Figure 2). Likewise, a common male linkage map containing 757 markers and having 44 linkage groups covering a genetic distance of 2378 cM (Table 4 and supplemental Figure 3) was calculated from genotype data of 42 corresponding linkage groups, with at least two markers in common. One linkage group was specific to FAM2 and FAM3. Despite some large genomic regions without markers [e.g., 49.4 cM on the linkage group IC67_14 of the female map (supplemental Figure 2) and 21.6 cM on the linkage group AL91_21 of the male map (supplemental Figure 3)], the median intermarker distance was relatively low (2.8 and 2.1 cM for the female and male map, respectively) (Table 4).
TABLE 4.
Statistics for the integrated female and male map
| Female (IC67) | Male (AL91) | |
|---|---|---|
| No. of linkage groups | 43 | 44 |
| No. of markers mapped | ||
| Min | 4 | 3 |
| Max | 28 | 52 |
| Median | 10 | 16.5 |
| Mean | 11 | 17.2 |
| Total | 494 | 757 |
| Size of linkage groups (cM) | ||
| Min | 20.9 | 10.7 |
| Max | 132.7 | 111.3 |
| Median | 54.9 | 53.9 |
| Mean | 54.9 | 54.0 |
| Total | 2362 | 2378 |
| Intermarker distance (cM) | ||
| Min | 0 | 0 |
| Max | 49.4 | 21.6 |
| Median | 2.8 | 2.1 |
| Mean | 5.2 | 3.3 |
The female and the male linkage maps share a total of 70 common AFLP markers. Twenty linkage groups of the two sex-specific linkage maps have at least two markers in common and hence can be considered as putatively homologous linkage groups (supplemental Figure 4). Another 13 linkage groups of the two sex-specific linkage maps share only one identical marker, indicative for potential homology (supplemental Figure 4). One of these 13 potential homologous linkage group pairs (i.e., IC67_1 and AL91_1) was found to represent the sex chromosomes (see below).
On the basis of the inclusion of 70 shared markers in the female and male maps, we identified 37 marker pairs for which the intermarker distance (or marker interval) on the female and male maps, expressed in centimorgans, could be compared. A significant correlation was found (Pearson r = 0.446; P < 0.01) between the marker intervals in the male and female map. In addition, a paired t-test indicates that the 37 marker intervals were not significantly different between the male and female maps (t = 0.287; P = 0.78), suggesting that the recombination frequencies in the female and the male do not differ significantly in P. monodon.
Sex locus:
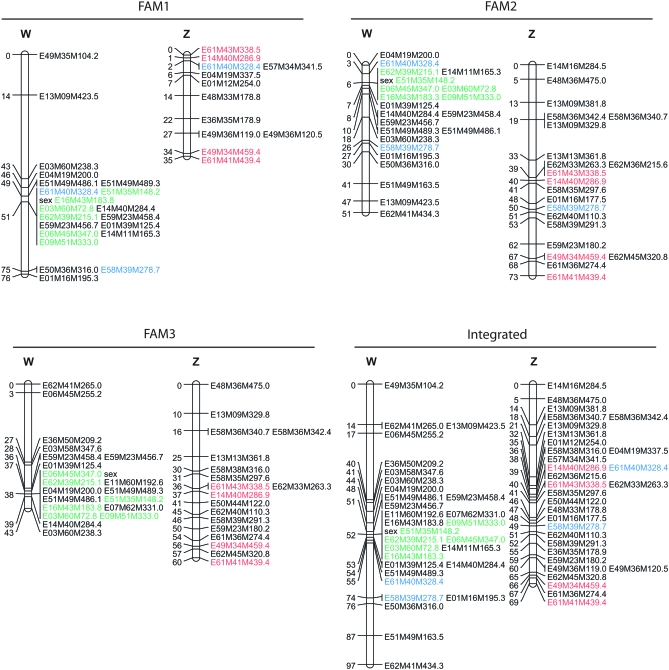
Assuming female heterogamety, 6 of the 156 markers identified previously by BSA as putatively sex linked in FAM3 showed complete linkage to phenotypic sex in each of the three families (markers E16M43M183.8, E51M35M148.2, E06M45M347.0, E03M60M72.8, E62M39M215.1, and E09M51M333.0; LOD scores ranging from 25.3 to 28 in each of the three families) (Figure 1). These markers map to the IC67_1 linkage group of the integrated female map, covered by 24 AFLP markers and spanning 89 cM (supplemental Figure 2). Hence, the six AFLP marker alleles were inherited from female parents in the three families, strongly suggesting female heterogamety in P. monodon (i.e., an WZ–ZZ system).
Figure 1.—
Separate and integrated linkage maps of the sex chromosomes from FAM1, FAM2, and FAM3. Distances are given in Kosambi centimorgans. Markers in blue were used for the identification of the Z-linkage group, markers in red on the Z chromosome segregate in each family, and markers in green are completely linked to the sex locus in each family. The integrated linkage groups, constructed with linkage data from FAM1, FAM2, and FAM3, are slightly different in marker order and intermarker distance from the linkage groups IC67_1 and AL91_1 that were constructed with linkage data from FAM2 and FAM3 and FAM1 and FAM2, respectively.
Alternatively, assuming male heterogamety (i.e., an XX–XY system), linkage is then found only with a single biparental marker in FAM1 (E61M40M328.4). This marker was also found to be linked to sex under the previous assumption of a WZ–ZZ system. This observation is not surprising because a biparental marker will show linkage to any other non-biparental marker, irrespective of the latter being encoded as a female or a male marker. In FAM2, the sex locus and four male markers, which do not map together, were moderately linked. In FAM3, no linkage was found. Therefore, data from linkage analysis favor a female over male heterogamety.
A more specific test of the sex-determination system can be made by comparing the observed segregation patterns to those expected for sex-linked AFLP markers under the assumption of a WZ–ZZ system or a XX–XY system (Table 5). The segregation patterns that can unequivocally differentiate the sex-determination system are patterns 1, 2, and 3, expected only under the assumption of female heterogamety, and patterns 6, 7, and 8, expected only under the assumption of male heterogamety. While a number of markers segregating according to patterns 1 and 2 (but not 3) were observed, none segregated according to patterns 6, 7, or 8 (Table 5). These segregation patterns, together with the linkage data, firmly establish female heterogamety in P. monodon.
TABLE 5.
Expected and observed segregation patterns for sex-linked AFLP markers under the assumption of either a WZ–ZZ or a XX–XY sex-determination system
| No. of observations
|
|||||||
|---|---|---|---|---|---|---|---|
| Patterna | Dam genotype | Sire genotype | Genotype of female offspring | Genotype of male offspring | FAM1 | FAM2 | FAM3 |
| WZ–ZZ | |||||||
| 1 | Aa | aa | Aa | aa | 7 | 7 | 10 |
| 2 | aA | aa | aa | Aa | 1 | 0 | 3 |
| 3 | aA | AA | aA | AA | 0 | 0 | 0 |
| 4 | aA | aA | Aa or aa | AA or Aa | NI | NI | NI |
| 5 | aa | Aa | Aa or aa | Aa or aa | NI | NI | NI |
| XX–XY | |||||||
| 6 | aa | aA | aa | Aa | 0 | 0 | 0 |
| 7 | aa | Aa | Aa | aa | 0 | 0 | 0 |
| 8 | AA | Aa | AA | Aa | 0 | 0 | 0 |
| 9 | Aa | Aa | AA or Aa | Aa or aa | NI | NI | NI |
| 10 | Aa | aa | Aa or aa | Aa or aa | NI | NI | NI |
WZ–ZZ, patterns 1–5; XX–XY, patterns 6–10. The A allele represents the “AFLP band present” allele. NI, not informative.
Offspring genotype classes as observed for segregation patterns 4 and 9 cannot unambiguously discriminate between the two sex-determination mechanisms. Offspring genotype classes observed for patterns 5 and 10 are identical to those expected for non-sex-linked markers. Hence, observations of these four patterns are not informative in characterizing the sex-determination system. Segregation patterns unique for the two sex-determination mechanisms are in italics.
Having evidenced that linkage group IC67_1 corresponds to the W chromosome, biparental markers allowed the identification of the homologous linkage group in the male map, i.e., the representative of the Z chromosome. Marker E61M40M328.4 and E58M39M278.7 identify the FAM1 and FAM2 male linkage group, respectively (Figure 1). Notably, E61M40M328.4 and E58M39M278.7 segregate as female markers in FAM2 and FAM1, respectively. In both cases, the linkage to the sex locus is confirmed. Six markers are shared between the two identified male linkage groups, supporting the belief that the identified male linkage groups represent not only the same chromosome, but also the homologous chromosome for the W chromosome, i.e., the Z chromosome. Four of the six markers led to the identification of the Z linkage group in FAM3 (Figure 1). Together, these data allow the unambiguous identification and integration of the homologous sex linkage groups (Figure 1).
Genetic association between AFLP marker alleles and sex on a population scale:
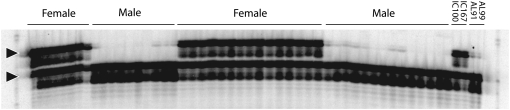
To assess the strength of the six marker–sex linkages, we tested the genetic association of the AFLP marker alleles to sex in a panel of 52 genetically unrelated animals. Two AFLP marker alleles, E03M60M72.8 and EO6M45M347.0, confirmed the association to sex. Only for marker EO6M45M347.0 did sequencing of the AFLP band result in a DNA sequence of a length sufficient to optimally design primers for a locus-specific assay (Figure 2). Amplification, sequencing, and careful examination of the male and female allelic sequences at the EO6M45M347.0 locus revealed 10 sex-specific polymorphisms (P1–P10; Figure 3): seven single nucleotide polymorphisms, two 1-bp indel markers, and one 6-bp indel marker. A 1-bp indel (P8; Figure 3) created an additional MseI restriction site in the male allele, most probably causing the absence of the E06M45M347.0 fragment in the male individuals. Taking advantage of the flanking DNA sequence of the 6-bp indel (P3; Figure 3), we designed primers indel-4 and indel-5 (Figure 3) to amplify a female-specific fragment of 82 bp and a sex-nonspecific fragment of 76 bp, converting the EO6M45M347.0 marker into a PCR-based allele-specific assay. These allelic amplicons were readily distinguishable when analyzed on a denaturing polyacrylamide gel. In a panel of 52 broodstock animals, all females were heterozygous while all males showed homozygosity for the 76-bp amplicon (Figure 4), confirming the strong linkage between marker and sex.
Figure 2.—
Sequences of the sex-linked AFLP fragments E03M60M72.8 and E06M45M347.0. EcoRI and MseI restriction sites are shaded and selective nucleotides are underlined.
Figure 4.—
PCR-based genotyping of the set of 52 unrelated broodstock animals for the sex-specific indel polymorphism. The 82-bp female-specific allele and the 76-bp allele are indicated by arrowheads.
DISCUSSION
We present sex-specific high-density AFLP linkage maps of P. monodon. For this purpose, we exploited the high multiplex ratio of the AFLP technique, combined with the high level of polymorphism in P. monodon to generate a large number of markers with relative ease. We analyzed a total of 125 AFLP PCs, resulting in an average of 10.6 AFLP markers/PC, which is comparable to previously reported frequencies for penaeid species, namely 7.3 in P. chinensis (Z. Li et al. 2006), 10 in P. monodon (Wilson et al. 2002), and 10 in P. japonicus (Li et al. 2003). Compared to mapping efforts in other penaeid species, our study was novel in that segregation data were generated in two female and two male HS families with one family in common. This greatly improved marker density, genome coverage, map alignment, and identification of homologous linkage groups between the female and male linkage map. The female and male linkage maps, incorporating 494 and 757 markers, respectively, have a median intermarker distance of 2.8 and 2.1 cM, respectively, outnumbering by far all published linkage maps for a penaid species in terms of marker density. The map consisted of 43 female and 44 male linkage groups, respectively, approximating well the expected number of linkage groups in penaeids (2n = 88 for P. monodon; Fanjun and Dong 1993). This supports the assumption that the representation of all chromosomes, and hence the complete coverage of the P. monodon genome, is asymptotically approached as the number of mapped markers increases.
In total, 21 homologous linkage groups, including the linkage groups representing the sex chromosomes, were identified between the two sex-specific linkage maps. Another 12 female linkage groups shared one common marker with a male linkage group, indicative for potential homology. The identification of these homologous linkage groups was greatly enhanced by codominant scoring of AFLP markers segregating in a 1:2:1 mode in the mapping populations, using the specific software AFLP-QuantarPro.
In all penaeid maps published to date (Moore et al. 1999; Wilson et al. 2002; Li et al. 2003; Z. Li et al. 2006; Maneeruttanarungroj et al. 2006), except for P. vannamei (Perez et al. 2004; Zhang et al. 2006), more markers could be mapped on the male map than on the female map. The intermarker distance was also slightly lower in the male map and the estimated genome size was larger in the female than in the male map. On the basis of this evidence, all authors have suggested that recombination frequency in the male was lower than in the female. We observed the same bias toward a higher marker density in male maps. However, because we identified biparental markers that were mapped on the male as well as the female map, we were able to directly compare marker distances between the two sexes, which other studies could not. This limited analysis indicates that recombination frequency does not differ between males and females.
The marked sexual dimorphism in growth observed between male and female penaeid shrimp (Primavera et al. 1998) has led to suggestions that the efficiency of production of culture systems could be improved by setting up a mono-sex production of the faster-growing sex (females in the case of penaeid shrimp) (Moss et al. 2002). To establish mono-sex production in P. monodon, knowledge of the genetic sex-determining mechanism needs to be well understood. Interspecies crosses between P. monodon and P. esculentis suggested that the female sex is most likely to be heterogametic (Benzie et al. 2001), and the mapping of phenotypic sex to a linkage group on the female map in P. japonicus (Li et al. 2003) and in P. vannamei (Zhang et al. 2006) was also consistent with this suggestion. However, none of the markers reported in these studies was completely linked to the sex locus. So while these data were consistent with the assumption of a WZ–ZZ system, they were not conclusive evidence for the system. Here, we characterized the genetic basis of sex determination in P. monodon. Combining BSA with genomewide linkage analysis, we identified six sex-linked AFLP marker alleles that were all inherited from the female parents in each of the three families, evidencing female heterogamety in P. monondon. Two sex-linked AFLP markers confirmed their association with sex in a panel of 52 genetically unrelated animals, suggesting tight linkage between the two markers and sex. Conversion of the sex-linked AFLP marker EO6M45M347.0 into a PCR-based allele-specific assay formally demonstrated that females are the heterogametic sex in P. monodon. The identification of a sex-linked marker in a general population of tiger shrimp is vital for the further development of mono-sex culture in shrimp, i.e., by allowing the identification of homogametic females that, upon crossing with a homogametic male, would give completely uniform heterogametic offsprings.
Acknowledgments
The authors thank J. Brock and the technical staff at Moana Techonologies in Hawaii for expert technical support and INVE Technologies, Dendermonde, Belgium, for their assistance. Moana Technologies and the VIB are willing to allow access to genotype information for all markers and the samples of the mapping families for others to add markers. The sex-linked AFLP markers are covered by a patent owned by Moana Technologies and VIB. Information concerning licenses to use the sex-linked AFLP markers for commercial purposes can be obtained from VIB. This work was funded in part by a grant from the Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (IWT 040263).
References
- Argue, B. J., and A. Alcivar-Warren, 2000. Genetics and breeding applied to the Penaeid shrimp farming industry, pp. 29–54 in Controlled and Biosecure Production Systems: Evolution and Integration of Chicken and Shrimp Models (Proceedings of a Special Session on Integration of Shrimp and Chicken Models, Sydney, April 27–30, 1999), edited by R. A. Bullis and G. D. Pruder. The Oceanic Institute, Waimanalo, HI.
- Benzie, J. A. H., 2000. Population genetic structure in penaeid prawns. Aquac. Res. 31 95–119. [Google Scholar]
- Benzie, J. A. H., M. Kenway and E. Ballment, 2001. Growth of Penaeus monodon × Penaeus esculentus tiger prawn hybrids relative to the parental species. Aquaculture 193 227–237. [Google Scholar]
- Cuzon, G., L. Arena, J. Goguenheim and E. Goyard, 2004. Is it possible to raise offspring of the 25th generation of Litopenaeus vannamei (Boone) and 18th generation Litopenaeus stylirostris (Stimpson) in clear water to 40g? Aquac. Res. 35 1244–1252. [Google Scholar]
- Fanjun, K., and Z. Dong, 1993. The karyotype of tiger shrimp Penaeus monodon. J. Fish. China 17 83–84. [Google Scholar]
- Food and Agriculture Organization of the United Nations, 2005. Fisheries Technical Paper no. 476. Food and Agriculture Organization of the United Nations, Rome.
- Gitterle, T., M. Rye, R. Salte, J. Cock, H. Johansen et al., 2005. Genetic (co)variation in harvest body weight and survival in Penaeus (Litopenaeus) vannamei under standard commercial conditions. Aquaculture 243 83–92. [Google Scholar]
- Jerry, D. R., B. S. Evans, M. Kenway and K. Wilson, 2006. Development of a microsatellite DNA parentage marker suite for black tiger shrimp Penaeus monodon. Aquaculture 255 542–547. [Google Scholar]
- Khamnamtong, B., S. Klinbunga and P. Menasveta, 2005. Species identification of five penaeid shrimps using PCR-RFLP and SSCP analyses of 16S ribosomal DNA. J. Biochem. Mol. Biol. 38 491–499. [DOI] [PubMed] [Google Scholar]
- Li, Y., K. Byrne, E. Miggiano, V. Whan, S. Moore et al., 2003. Genetic mapping of the kuruma prawn Penaeus japonicus using AFLP markers. Aquaculture 219 143–156. [Google Scholar]
- Li, Y., L. Dierens, K. Byrne, E. Miggiano, S. Lehnert et al., 2006. QTL detection of production traits for the Kuruma prawn Penaeus japonicus (Bate) using AFLP markers. Aquaculture 258 198–210. [Google Scholar]
- Li, Z., J. Li, Q. Wang, Y. He and P. Liu, 2006. AFLP-based genetic linkage map of marine shrimp Penaeus (Fenneropenaeus) chinensis. Aquaculture 261 463–472. [Google Scholar]
- Maneeruttanarungroj, C., S. Pongsomboon, S. Wuthisuthimethavee, S. Klinbunga, K. J. Wilson et al., 2006. Development of polymorphic expressed sequence tag-derived microsatellites for the extension of the genetic linkage map of the black tiger shrimp (Penaeus monodon). Anim. Genet. 37 363–368. [DOI] [PubMed] [Google Scholar]
- Michelmore, R. W., I. Paran and V. Kesseli, 1994. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, S. S., V. Whan, G. P. Davis, K. Byrne, D. J. S. Hetzel et al., 1999. The development and application of genetic markers for the Kuruma prawn Penaeus japonicus. Aquaculture 173 19–32. [Google Scholar]
- Moss, D. R., O. L. Hennig and S. M. Moss, 2002. Sexual growth dimorphism in penaeid shrimp. Potential for all female culture? Global Aquac. Advocate 5 60–61. [Google Scholar]
- Perez, F., C. Erazo, M. Zhinaula, F. Volckaert and J. Calderon, 2004. A sex-specific linkage map of the white shrimp Penaeus (Litopenaeus) vannamei based on AFLP markers. Aquaculture 242 105–118. [Google Scholar]
- Primavera, J. H., F. D. Parado-Estepa and J. L. Lebata, 1998. Morphometric relationship of length and weight of giant prawn Penaeus monodon according to life stage, sex, and source. Aquaculture 164 67–75. [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap 3.0, Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuylsteke, M., J. Peleman and M. J. van Eijk, 2007. a AFLP technology for DNA fingerprinting. Nat. Protoc. 2 1387–1398. [DOI] [PubMed] [Google Scholar]
- Vuylsteke, M., J. Peleman and M. J. van Eijk, 2007. b AFLP-based transcript profiling (cDNA-AFLP) for genome-wide expression analysis. Nat. Protoc. 2 1399–1413. [DOI] [PubMed] [Google Scholar]
- Wilson, K., Y. Li, V. Whan, S. Lehnert, K. Byrne et al., 2002. Genetic mapping of the black tiger shrimp Penaeus monodon with amplified fragment length polymorphism. Aquaculture 204 297–309. [Google Scholar]
- Xu, Z., J. H. Primavera, L. D. de la Pena, P. Pettit, J. Belak et al., 2001. Genetic diversity of wild and cultured black tiger shrimp (Penaeus monodon) in the Philippines using microsatellites. Aquaculture 199 13–40. [Google Scholar]
- Zhang, L., C. Yang, Y. Zhang, L. Li, X. Zhang et al., 2006. A genetic linkage map of Pacific white shrimp (Litopenaeus vannamei): sex-linked microsatellite markers and high recombination rates. Genetica 131 37–49. [DOI] [PubMed] [Google Scholar]