Abstract
In zebrafish, mutations in the gap junction gene connexin43 lead to short bony fin ray segments that give rise to the short fin phenotype. The sofb123 mutant exhibits fins that are half the length of wild-type fins and have reduced levels of cx43 mRNA. We find that sofb123 regenerating fins exhibit reduced levels of cell proliferation. Interestingly, the number of dividing cells per unit length of fin growth is similar between wild-type and mutant fins, suggesting that the number of cells that enter the cell cycle is specifically affected in sofb123. Expression of cx43 is identified in mitotic cells, which further suggests that Cx43 may contribute to establishing or maintaining the population of dividing cells. Indeed, missense alleles exhibiting high or low levels of gap junctional communication reveal a correlation between defects in direct cell-cell communication, cell proliferation, and segment length. Finally, targeted gene knockdown of cx43 in adult regenerating fins recapitulates the sofb123phenotype, revealing that the loss of Cx43 is sufficient to reduce both cell proliferation and segment length. We hypothesize that the level of gap junctional intercellular communication among dividing cells regulates the level of cell proliferation and ultimately regulates bone growth.
Keywords: Bone growth, short fin, Regeneration, Zebrafish, Cell proliferation, Cx43, GJIC
Introduction
The underlying mechanisms regulating the size and shape of bony structures are largely unknown. We utilize growth of the zebrafish fin to reveal such mechanisms. The fin grows throughout the lifetime of the fish, providing extended opportunities to monitor growth. The zebrafish fin is comprised of skeletal elements called fin rays (or leipidotrichia) which grow by the addition of new bony segments to their distal ends (Goss and Stagg, 1957; Haas, 1962). Each ray is made of two concave hemirays surrounding undifferentiated mesenchymal cells, vasculature, and nerves (Santamaria et al., 1992). Growth of the bony segments occurs in the absence of a cartilaginous precursor, by the process of intramembraneous ossification (Landis and Geraudie, 1990).
Fins have the capacity for regeneration, enabling more precisely timed experiments and evaluation of a more rapid growth process. Following wound healing (12–24 hours post-amputation, hpa), a specialized structure called a blastema forms in the distal mesenchyme (Poss et al., 2000). The blastema is required for outgrowth and replacement of lost tissue. Organization of the blastema into two morphologically indistinct compartments occurs by 48–72 hpa (Nechiporuk and Keating, 2002). The distal-most blastema (distal most 10–50 μm) contains non-proliferative, msxb positive cells that may provide directional growth information. Rapidly dividing cells are located in the proximal blastema or proliferation zone, 100–200 μm proximal to the distal-most blastema. During outgrowth (72 hpa and beyond), cells migrate proximally and laterally (Nechiporuk and Keating, 2002; Poleo et al., 2001) prior to differentiation outside of the proliferation zone.
Mechanisms regulating the growth and size of fin ray segments remain unknown. Fin length mutants with defects in segment length will provide insights into the molecular and cellular requirements for bone growth. To date, the short finb123 (sofb123) mutant is the only fin mutant affecting segment length (Iovine and Johnson, 2000). The sofb123phenotype was recently found to be caused by mutations in the connexin43 (cx43) gene (Iovine et al., 2005). Three non-complementing ENU-induced missense mutations were identified in the coding region of cx43 (sofj7e1 codes for Cx43-F30V, sofj7e2 codes for Cx43-P191S, sofj7e3 codes for Cx43-F209I). The original allele, sofb123, lacks mutations in the coding sequence, although it exhibits reduced cx43 mRNA levels. All four alleles are recessive adult viable mutations, and appear to be hypomorphs (Iovine et al., 2005). The Cx43-F30V and Cx43-P191S alleles behave similarly to sofb123, causing segment length and fin length to be the most severely affected. In contrast, the Cx43-F209I allele is relatively mild, resulting in segment length and fin length more similar to wild-type animals (but measurably smaller, Iovine et al., 2005).
Connexins are the subunits of gap junctions, proteinaceous channels required for direct cell-cell communication among adjacent cells. A single connexin is a four-pass transmembrane protein containing two extracellular domains and one intracellular loop. Six connexin proteins make a connexon and two connexons (one from each neighboring cell) form a gap junction channel. Gap junctional intracellular communication (GJIC) occurs by the exchange of small molecules (≤1000 Da) via these channels. GJIC is important for development, homeostasis, and tissue function as evidenced by the association of mutations in connexin genes with human disease phenotypes (reviewed in Laird, 2006). For example, (primarily) missense mutations in human CX43 cause oculodentodigital dysplasia (ODDD), a syndrome characterized by malformations of the craniofacial and distal limb skeleton among other pleiotropic phenotypes (Paznekas et al., 2003). It is not immediately clear how mutations in connexin genes result in defects in bone growth and morphology. Correlations between particular missense mutations and disease phenotypes are not apparent (Paznekas et al., 2003), nor is there a correlation between channel activity of missense alleles and the severity or type of symptoms (Roscoe et al., 2005; Seki et al., 2004; Shibayama et al., 2005). The zebrafish sof mutant may provide opportunities to evaluate Cx43 function with respect to a single, relatively simple structure.
The correlation between the zebrafish cx43 mutant fin phenotypes in vivo and the defects of gap junctional coupling in vitro suggests the possibility that the level of GJIC via Cx43 gap junctions contributes to bone growth (Hoptak-Solga et al., 2007). The goal of this study is to identify the underlying cellular defect in the sof mutants. Previous studies revealed that cx43 is expressed both in the distal mesenchymal compartment of growing fin rays (i.e. in the same compartment as dividing cells) and more proximally in differentiated osteoblasts flanking joints (Iovine et al., 2005). Here we examine cell proliferation in wild-type and sof regenerating fins. We find that the number of dividing cells is reduced in all four sof alleles and that cx43 is expressed in mitotic cells, suggesting a cell autonomous role for cx43 in proliferating cells. Furthermore, we find that targeted gene knockdown of cx43 recapitulates both the segment length and cell proliferation defects of sof mutants, indicating that reduced Cx43 function is sufficient to cause both phenotypes. We suggest that GJIC via Cx43 gap junctions coordinates cell proliferation with joint formation to regulate normal segment growth.
Materials and Methods
Fish maintenance
Both wild-type and sofb123 stocks were from the C32 strain (Rawls et al., 2003) and were kept at a constant 25°C. Fish were exposed to a 14 light:10 dark photoperiod (Westerfield, 1993).
Immunoblots and Cx43 antibody
The final 16 amino acids of zebrafish Cx43 was chosen as the antigen in conjunction with Quality Controlled Biochemicals (www.qcb.com). QCB completed peptide synthesis, immunization, rabbit maintenance, and affinity purification of bleeds.
E. coli lysates expressing the GST-Cx43CT fusion protein were grown to confluency before adding 0.3mm IPTG to induce protein expression. Lysates were prepared using 50 mg/ml lysozyme in lysis buffer (50 mM glucose, 20 mM Tris-Cl pH 8.0, 10 mM EDTA pH 8.0). In addition, 2.2N NaOH and 8%BME was added to the mixture. Total protein was precipitated using TCA and pellets were resuspended in SDS buffer. Samples were diluted 1:100 in SDS buffer and increasing volumes were loaded (0.6μl to 1.6μl, all in a total volume of 10μl). For the antibody competition identical gels were prepared. The anti-Cx43 antibody was either used directly (1:2000) or following pre-incubation with the peptide made as the Cx43 antigen. Band densities were determined using Scion imaging software (Scion Corp., Frederick, MD) and the percent reduction was calculated.
Wild-type and sof fin (5 dpa regenerates) and whole body (21 days post fertilization) tissue was dried and homogenized using a mortar and pestle stored on dry ice and resuspended in homogenization buffer (20 mM Tris-HCl pH 7.4, 10 mM MgCl2, 0.6 mM CaCl2, 0.5mM EGTA, 0.005% Triton-X 100, 0.1 mM PEFABLOC SC). Protein concentration was determined using a spectrophotometer (OD 280). Samples were prepared in SDS sample buffer. Protein was first separated using 12% SDS-PAGE at 26mA and then transferred onto nitrocellulose membranes. Following transfer, blots were rinsed in 40% isopropanol, rinsed in dH2O and blocked in 5% milk in SuperTBST for 30 minutes at room temperature. Blots were then incubated with either anti-Cx43 (1:2000) or anti-α-tubulin (Sigma, clone B512, 1:1000) for one hour at room temperature, rinsed for one hour in TBST, and incubated with peroxidase-conjugated Goat anti-rabbit IgG or Goat anti-mouse IgG (1:250,000, pre-absorbed with fin tissue, Pierce, Rockford, IL) for one hour at room temperature. Following incubation, blots were rinsed in TBST for one hour at room temperature. Using ECL chemiluminescent reagents (SuperSignal® West Femto Maximum Sensitivity Substrate, Pierce, Rockford, IL), blots were developed and exposed to X-ray film (Xposure™ film, Pierce, Rockford, IL). Band densities were determined as above.
Whole mount Cx43 staining
Fins (5 dpa) were harvested and fixed in 2% PFA in 0.1 mM phosphate buffer (PB) for 30 min at room temperature. Fins were washed (3 × 10 min) in 25 mM PB, incubated in Trypsin/EDTA (Gibco) for 10 min on ice and washed in 25 mM PB+10% sucrose for 1 hour at room temperature. Fins were blocked (1M Tris-HCl pH 7.4, 5M NaCl, 0.3 % Triton-X 100, 6 % goat serum) for 30 minutes at room temperature and incubated with Cx43 antibody (1:200) overnight at 4°C. After incubation, fins were washed in block (3x5min) and incubated in Goat anti-rabbit Alexa 546 (1:200, Molecular Probes) antibody for 2 hours at room temperature followed by 1xPBS washes (3 × 10 min).
For double labeling, fins (3 dpa) were processed for H3P as described (see below, beginning after the rehydration series). Fins were mounted on slides in 50 % glycerol for whole mount imaging. Fins were cryosectioned to identify doubly-stained cells in the mesenchymal compartment. Sixty-two sections (intact epithelium and mesenchymal compartments, and containing at least one H3P positive cell) were sampled from five different wild-type fins.
Cryosectioning
Fins were rinsed in 1xPBS (3 × 10 min) and embedded in 1.5% agarose/5% sucrose blocks, and submerged in 30% sucrose overnight at 4 °C. Blocks were frozen on dry ice and mounted using O.C.T. Compound (Tissue Tek®, Sakura, the Netherlands), and 20μm sections were cut using a cryostat (Leica 2800 Frigocut E; Cambridge Instruments, Germany). Sections were collected on Superfrost Plus slides.
Morpholino injections and electroporation
Injection and electroporation experiments were performed as described (Thummel et al., 2006). Two targeting and two control morpholinos were used in this study, and all four were modified with fluorescein to provide a charge and for detection. cx43-MO (Iovine et al., 2005) was designed against the translational start site of cx43. As a control, a related morpholino containing 5 mis-matches was used, 5mmcx43-MO (5′-CCT CTT ACC TCA GTT ACA ATT TAT A 3′). cx43-MO2 (5′-GTT CTA GCT GGA AAG AAG TAA AGA G 3′) was designed against the 5′UTR of cx43 with its mismatch control (5mmcx43-MO2; 5′-GTT GTA GGT GGA AAC AAC TAA ACA G 3′). Morpholinos were purchased from Gene Tools, LLC and were diluted to 1.2 mM in dH2O.
Adult fish were first anesthetized using Tricane-S. Fin amputation was performed under a dissecting microscope using a scalpel and ruler to precisely amputate 50% of the caudal fin. At three days post amputation, morpholino was injected into the blastema of the three longest fin rays in either lobe using a Narishige IM 300 Microinjector. Approximately 50 nl of morpholino was injected per ray. Immediately following injection, both dorsal and ventral halves were electroporated using a CUY21 Square Wave Electroporator (Protech International, Inc.). The following parameters were used: 10 50-msec pulses of 15V with a 1 second pause between pulses. At 24 hpe (hours post electroporation), success was evaluated by monitoring fluorescein uptake. Fish were returned to their tanks for either H3P analyses (1 dpe) or segment length analyses (4 dpe). In all experiments, 3–4 fins were tested per data point. Cell counts or segment length measurements were completed and standard deviation was calculated. Student’s t-tests were completed to determine statistical significance.
In situ hybridization
In situ hybridization (ISH) as well as riboprobe synthesis (using digoxigenein-labeled UTP) was performed as described by Poss et al. (2000) using a probe against full length cx43 (Iovine et al., 2005).
Detection of proliferating cells using H3P
Fins were amputated to 50% and permitted to regenerate for 1,3,7, or 9 days (3–4 fins per time point per strain) before harvesting and fixation in 4% paraformaldehyde in PBS overnight at 4 °C. Fins were dehydrated in 100% methanol overnight at −20 °C and re-hydrated in a methanol/PBS series. The fins were treated with 1 mg/ml collagenase in PBS for 45 min at room temperature and blocked using a 0.5% BSA in PBS solution with 0.1% Triton-X. A rabbit antibody against anti-phosphohistone H3 (H3P, Upstate Biotechnology) was diluted 1:100 in block and fins were incubated overnight at 4 °C. Following a series of washes in block, fins were incubated with an anti-rabbit antibody conjugated to Alexa-546 (Molecular Probes) (diluted 1:200 in block) for 2.5 hours at room temperature. Washes were performed in block before mounting in Vectashield. H3P positive cells were visualized using a Nikon Eclipse E80 compound microscope and photographed using ImagePro software. Labeled cells were counted within the distal-most 250μm. In the day 1 fins (which do not have 250 μm of tissue in the regenerate) only positive cells in the regenerate were counted. Standard deviation was calculated and student’s t-tests were performed.
Measurements of segment length
At 4 dpe, the fins were amputated asymmetrically to maintain dorsal and ventral axes, harvested and fixed in 4% paraformaldehyde at 4 °C overnight. After washing in PBS, fins were mounted on slides in 50 % glycerol and photos were taken under a Nikon SMZ800 stereomicroscope. Measurements were completed using ImagePro Plus software. Standard deviation was calculated and student’s t-tests were performed.
Results
Cx43 function is required in dividing cells during fin regeneration
During ontogeny, the number of dividing cells was shown to be reduced in sofb123 mutants (Iovine et al., 2005), providing evidence that the level of cell proliferation is affected. Here we followed the growth rate and the number of mitotic cells during the course of fin regeneration, from 1 day post amputation (dpa) to 9 dpa. Throughout this time, the length of the sofb123 regenerate is half as long as the wild-type regenerate, and the rate of growth is also half that of wild-type fins (Figure 1 A,B). Fins were examined concurrently for the number of dividing cells by labeling with the mitosis marker histone-3-phosphate (H3P, Wei et al., 1999). Since this marker detects cells only during mitosis, relatively few dividing cells are identified (see Lee et al., 2005 for comparison of BrdU labeling and H3P co-staining), making it possible to count all H3P positive cells reliably. H3P positive cells were counted from the distal-most 250 μm of the regenerates since this is the area where mesenchymal proliferating cells contribute to fin outgrowth (Goldsmith et al., 2003). The number of mitotic cells in the sofb123 mutant is approximately half the number present in wild-type (Figure 1C). Interestingly, when the number of H3P positive cells is divided by the length of growth (from Figure 1A), wild-type and sofb123 fins behave similarly (Figure 1D). Therefore, the number of mitotic cells per unit length of fin growth is the same for both wild-type and sofb123 mutants. Note that at 9 dpa it appears that sofb123 fins actually exhibit a greater number of H3P positive cells/length of growth. However, this is likely a reflection of the greater variability observed in wild-type fins at this late time point. Thus, for a given number of dividing cells, sofb123 mutants exhibit the same amount of tissue growth as wild-type regenerating fins, suggesting that events including and following the cell division cycle are not affected. Rather, the primary defect in sofb123 fins may be that fewer cells enter the cell cycle.
Figure 1.
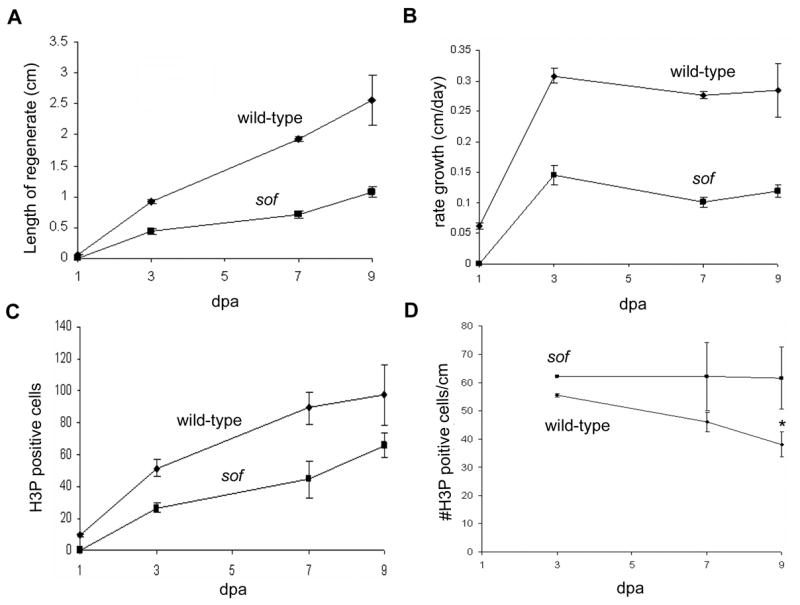
Growth rate and the number of mitotic cells are reduced in sofb123 mutants during regeneration. Measurements of regenerates in wild-type and sofb123 fins were conducted over a period of nine days. (A) The length of the sofb123 regenerate is half as long as the wild-type regenerate. Standard deviation is shown. (B) The rate of regeneration is slower in sofb123 mutants. Standard deviation is shown. (C) The number of mitotic (H3P positive) cells is reduced in sofb123 mutants compared to wild-type. Standard deviation was calculated and the student’s t-test reveals all data points between wild-type and sofb123 are statistically different (p = 0.0001; 0.0008; 0.0042; 0.0454 for 1,3,7,9 dpa). (D) The number of mitotic cells per cm is the same between wild-type and sofb123 fins. Standard deviation was calculated and the student’s t-test reveals data points through 7 dpa are not statistically different (p > 0.06). The data points at 9 dpa are statistically different, p = 0.0001 (*).
To determine if cx43 is expressed autonomously or non-autonomously in this dividing cell population, 3 dpa regenerating fins were double stained for cx43 mRNA and H3P. Mitotic cells are observed throughout the fin in the overlying epithelium (Figure 2B). However, only H3P positive cells in the distal-most 250 μm of the mesenchymal compartment were examined for co-localization with cx43. Fins were sectioned to facilitate the identification of doubly-labeled cells in the distal mesenchyme. We find that the majority of H3P positive cells in the germinal compartment were also cx43 positive (89 % or 108/121 cells), revealing that cx43 is expressed in the majority of dividing cells (Figure 2).
Figure 2.
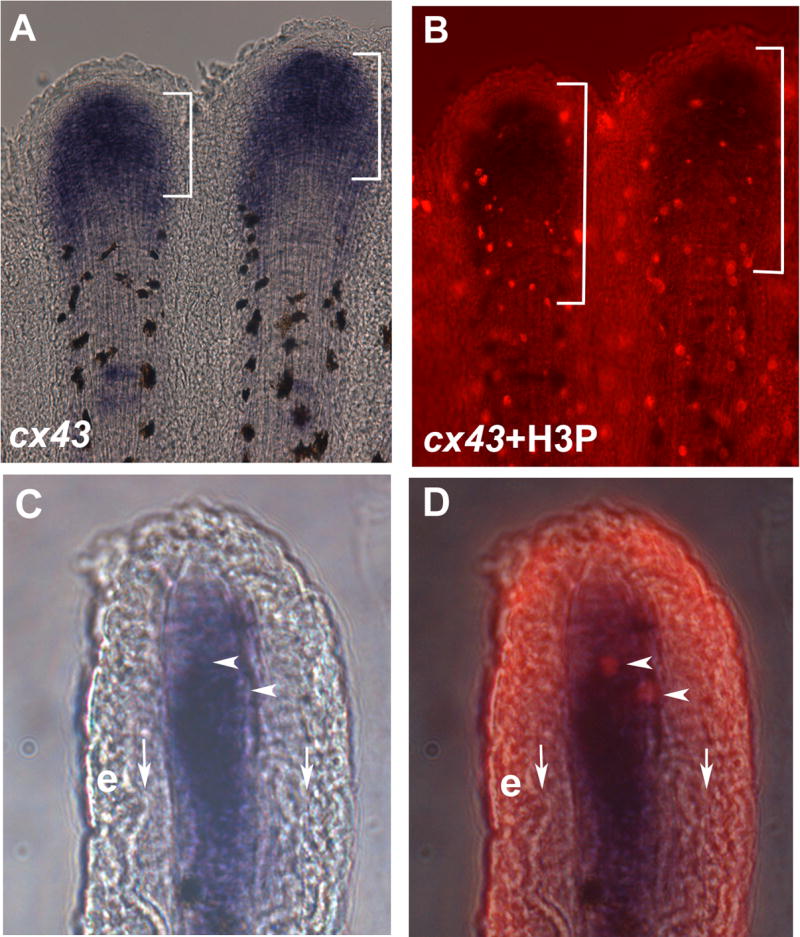
cx43 is expressed in mitotic cells. Regenerating fins were labeled for both cx43 mRNA and H3P. (A) Whole mount in situ hybridization shows cx43 expression (brackets). The cx43 positive compartment measured from 70–140 μm from the distal end of the fin. (B) Double-staining for H3P and cx43 in situ hybridization. A distance of 250 μm from the distal end of the fin is shown in brackets. (C) Brightfield image of a single cryosection showing cx43 expression in the distal mesenchymal compartment. (D) Brightfield plus fluorescence reveals that the two H3P positive cells in this section are coincident with cx43 positive cells. Vertical arrows point to bone matrix; e, epithelium; arrowheads point to doubly-labeled cells.
Since the sofb123 allele exhibits reduced expression of an otherwise wild-type cx43 mRNA, it is not clear if the defect in cell proliferation is due to defects in GJIC or in another possible function of the connexin proteins (Wei et al., 2004). Therefore, we next determined the number of dividing cells in each of our missense alleles of Cx43. Two of these alleles (Cx43-F30V and Cx43-P191S) exhibit significant defects in ionic coupling assays completed in Xenopus oocytes, and the third (Cx43-F209I) functions similar to wild-type Cx43 (Hoptak-Solga et al., 2007). Fins were harvested at a single time point (3 dpa) and processed for the detection of H3P positive cells. A significant decrease in the number of H3P positive cells was found between wild-type and the more severe alleles, Cx43-F30V and Cx43-P191S (Figure 3). The weak allele, Cx43-F209I, exhibited levels of cell proliferation not statistically different from wild-type (Figure 3). Together with our previous studies this demonstrates that the severity of the reduction of cell proliferation correlates with the in vivo phenotype of each allele (i.e. segment length) and with the severity of GJIC defects.
Figure 3.
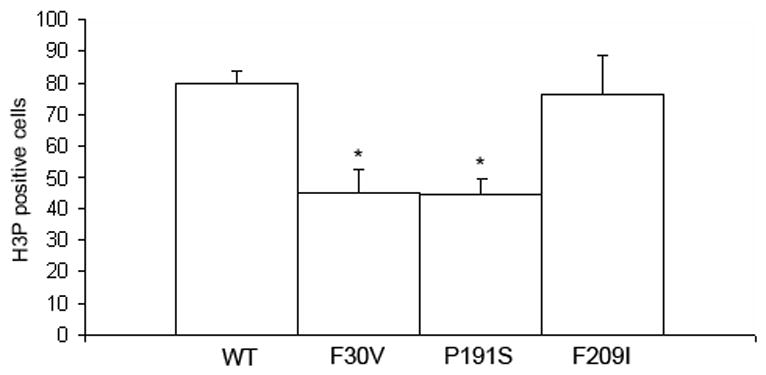
Cell proliferation is reduced in missense alleles of cx43. Fins were harvested at 3 dpa and stained for H3P detection. A statistically significant reduction in the number of H3P cells was observed for both the Cx43-F30V (p = 0.0001) and the Cx43-P191S (p = 0.0002) alleles in comparison to wild-type. The weaker Cx43-F209I allele was not statistically different from wild-type (p = 0.638). Standard deviation is shown for all alleles.
Morpholino-mediated gene knockdown of Cx43 recapitulates the sof phenotype
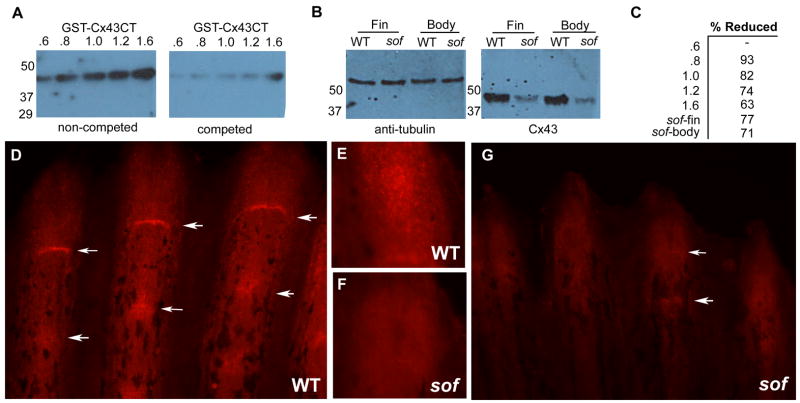
Recently, a method of targeted gene knockdown was developed for adult regenerating fins (Thummel et al., 2006). This method relies on the electroporation of charged antisense morpholino-modified oligonucleotides into cells of regenerating fins. Since morpholinos function by steric hindrance of ribosome binding and translation (Nasevicius and Ekker, 2000), a Cx43 antibody was generated to evaluate knockdown of the Cx43 gene product. The antibody was targeted against the final 16 amino acids of zebrafish Cx43. We utilized a GST fusion protein containing the carboxy-terminus (GST-Cx43CT) to evaluate the antisera and specificity. We loaded increasing amounts of bacterial lysates expressing GST-Cx43CT to demonstrate that the antibody recognizes its target. Further, pretreatment of the antibody with its target sequence resulted in a linear competition for GST-Cx43CT (Fig 4A,C). Next we tested the antibody in endogenous tissue. The antibody recognizes a single 43 kD band from whole animal and 5 dpa regenerating fins (Fig 4B), and from mouse heart (not shown). As a further test of specificity, we show that levels of Cx43 protein are reduced approximately 70 % in sofb123 fins and whole animal (Fig 4B,C).
Figure 4.
Cx43 protein expression is reduced in sofb123 mutants. (A) An antibody raised against zebrafish Cx43 detects a GST fusion protein with the carboxy tail of Cx43 (GST-Cx43CT). Increasing amounts of E. coli lysates expressing GST-Cx43CT were loaded and test for antibody detection (left). When the antibody was pre-incubated with the Cx43 target sequence, binding to the immunoblot was competed. (B) Protein levels of Cx43 in sofb123 regenerating fins and whole body are reduced by greater than 70 %. Tubulin was used as a loading control and to normalize the loading of fin or whole body lysates. (C) Percent reductions of anti-Cx43 antibody binding for the antibody competition (A) and sofb123 Cx43 levels (B). (D) Cx43 expression in wild-type fins. Distal tip staining in wild-type (E) and sofb123 fins (F). (G) Cx43 expression is substantially reduced in sofb123 mutant fins. Arrows point to Cx43 staining at the joints.
Whole-mount immunofluorescence in wild-type fins reveals an expression pattern of Cx43 protein similar to cx43 mRNA expression, which has been found in the distal mesenchymal cells and in cells surrounding newly formed joints (Iovine et al., 2005). Cx43 protein is detected similarly, both along the fin rays and in cells surrounding the joints (Figure 4 D,E). Interestingly, it appears that the Cx43 protein is found along the length of the fin ray while the cx43 mRNA is detected only in the distal-most area. This likely reflects an up-regulation of cx43 transcription during the rapid growth of regeneration, while the protein expression represents steady-state levels. Immunoflourescence in sofb123 fins also demonstrates that Cx43protein is reduced in sofb123 mutant fins as expected (Figure 4 F,G).
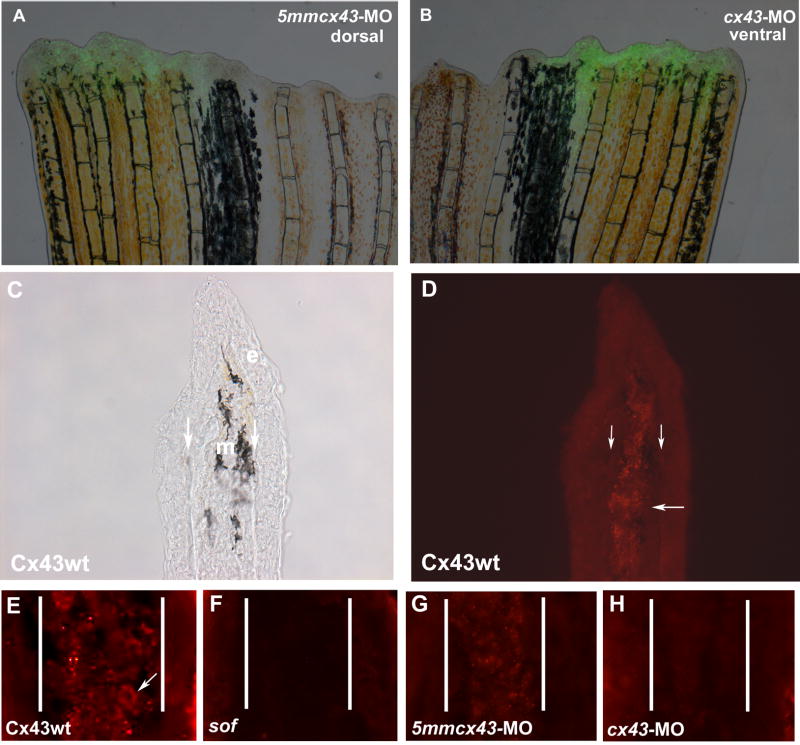
Targeted gene knockdown of cx43 was completed using either of two targeting morpholinos (cx43-MO or cx43-MO2, with corresponding mismatch control morpholinos, 5mm) modified with the charged fluorescent molecule fluorescein. Fins were harvested at 1 day post electroporation (dpe) and evaluated for morpholino uptake (incorporation of fluorescein, Figure 5 A,B) and Cx43 levels (Figure 5 G,H). To more carefully evaluate the staining pattern of Cx43, fins were cryosectioned to visualize staining of mesenchymal cells. Punctate Cx43 expression is detected at the plasma membrane of mesenchymal cells in wild-type fin tissue, consistent with the predicted location of connexin protein (Figure 5 C–E). Mesenchymal Cx43 expression is significantly reduced in sofb123 fin tissue (Figure 5F). Fins electroporated with either of the mismatch control MOs exhibit a pattern of similar intensity to that observed in wild-type fins (Figure 5 G, 5mmcx43-MO is shown). Expression of Cx43 protein was reduced in fins electroporated with either of the targeting morpholinos (Figure 5H, cx43-MO is shown). Therefore, morpholinos targeted against zebrafish cx43 are taken up by adult fin cells and are effective at reducing Cx43 protein levels.
Figure 5.
Injection and electroporation of cx43 morpholinos confirms targeted knockdown of Cx43. (A,B) Fins were injected with either 1.2 mM 5mmcx43-MO or 1.2 mM cx43-MO 1.2 and immediately electroporated. Cellular uptake is shown by fluorescein fluorescence. (C,D) Cryosections through fins stained with Cx43 antibody show mesenchymal staining in wild-type fins. Brightfield and fluorescence are shown. Vertical arrows indicate bone matrix, horizontal arrow points to one area of punctate staining. (E) Closer examination of Cx43 staining reveals punctate plasma membrane staining. Arrow points to one cell. (F) sofb123 regenerates exhibit a reduction in Cx43 protein in the mesenchyme (G) Fins injected/electroporated with the 5mmcx43-MO show similar expression of Cx43 as wild-type fins. (H) Fins injected/electroporated with the targeting morpholino cx43-MO exhibit a reduction in Cx43 expression. Vertical lines in E-H distinguish epithelium and mesenchymal compartments.
To determine the effect of cx43 gene knockdown on regenerating fins, wild-type fins were injected/electroporated with either of the targeting morpholinos (or the corresponding mismatch control morpholinos) and segment length was measured at 4 dpe. In this experiment, the dorsal side of each fin was injected/electroporated and the ventral side of the fin was electroporated only. The first three complete segments (formed following injection) were measured from the longest fin ray in both lobes. The injected side was compared to the non-injected side. In fins injected with either cx43-MO or cx43-MO2, a significant difference in the length of the first three newly formed segments was evident when compared to the un-injected side (Figure 6 A–C and G–I). In contrast, a significant difference in segment length was not observed in fins injected with either of the 5mm control morpholinos (Figure 6 D–F, 5mmcx43-MO shown). Therefore, knockdown of Cx43 in fins recapitulates the short segment (and reduced fin length) phenotype of sof mutants.
Figure 6.
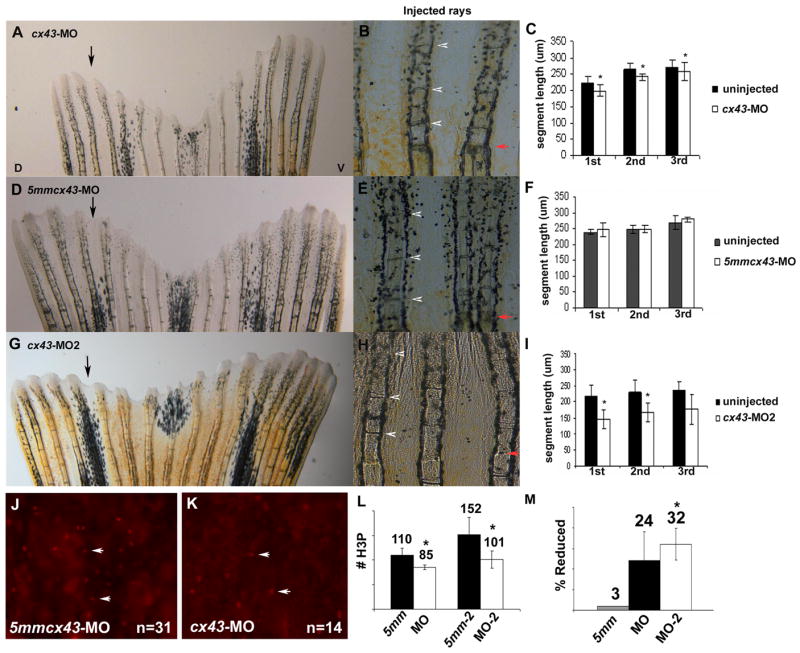
Knockdown of Cx43 recapitulates the sof phenotype. Wild-type fins were injected/electroporated in dorsal fin rays with either targeting morpholino (cx43-MO, cx43-MO2) and the respective control morpholino. The ventral side of the fin was electroporated only. At 4 dpe, segment length was measured for the first three newly formed segments for each fin ray. (A) Fin electroporated with 1.2 mM cx43-MO. (B) Segment size was measured for both injected and uninjected fin rays. (C) Graph comparing segment length (μm) for the cx43-MO and the uninjected rays. There is a significant difference in segment length for all three segments (p < .01). (D) Fin electroporated with 1.2 mM 5mmcx43-MO. (E) Segment size was measured for both injected and uninjected fin rays. (F) Graph comparing segment length (μm) for the 5mmcx43-MO and the uninjected rays. There is no significant different in segment length (p = 0.25, p = 0.94, p = 0.08, respectively). (G) Fin electroporated with 1.2 mM cx43-MO2. (H) Segment size was measured for both injected and uninjected fin rays. (I) Graph comparing segment length (μm) for the cx43-MO2 and the uninjected rays. There is a significant difference in segment length for all three segments (p < 0.01). Black arrows indicate the injected lobe, red arrows indicate the amputation plane, arrowheads designate newly formed joints. (J–L) Wild-type fins were injected/electroporated with either targeting or control morpholinos, harvested at 1 dpe and processed for H3P detection. Representative images of H3P positive cells are shown in J,K. (J) Dorsal rays were injected/electroporated with 5mmcx43-MO. (K) Ventral fin rays were injected/electroporated cx43-MO. (L) A significant reduction in H3P number was observed for either targeting morpholino (cx43-MO, p < 0.01 and cx43-MO2, p = 0.01) when compared to their corresponding controls. (M) The cx43-MO2 appears more effective at cx43 gene knockdown than cx43-MO since the percentage of H3P positive cells was more greatly affected when the cx43-MO2 was used (p < 0.05 when comparing the percent reduction of H3P positive cells of cx43-MO and cx43-MO2).
Next, knockdown fins were examined for cell proliferation defects by evaluating the number of H3P positive cells. Here, the dorsal side was injected with either 5mmcx43-MO or 5mmcx43-MO2, and the ventral side was injected with either cx43-MO or cx43-MO2. The number of mitotic cells was determined by counting the number of H3P positive cells in the distal-most 250μm of the injected/electroporated rays (a total of 54 fin rays) from both lobes (Figure 6 J,K for representative images). The number of H3P positive cells was significantly reduced in rays injected/electroporated with either cx43-MO or cx43-MO2 (Figure 6L). We noted a more significant reduction in H3P number in those fins injected with cx43-MO2 (p < 0.05), suggesting that cx43-MO2 may be more effective at Cx43 knockdown (Figure 6M). These results indicate that knockdown of Cx43 recapitulates not only the segment length phenotype but also the newly established cell proliferation phenotype of sofb123 mutants.
Discussion
Mutations in cx43 cause the short segment (and reduced fin length) phenotypes of the sofb123 mutant. Here we show that Cx43 function is related to the number of dividing cells during fin regeneration, and further, that cx43 is expressed in dividing cells. When the function of Cx43 is reduced either by lower expression levels of cx43 (as in sofb123) or by reduced GJIC in missense alleles, a reduced total number of mitotic cells contribute to fin growth. The activity of these cells does not appear to be affected, however, since the amount of growth is similar between wild-type and sofb123 when the same number of cells divide. Furthermore, manipulation of Cx43 function via targeted-gene knockdown recapitulates effects on cell proliferation and segment length, revealing that the loss of Cx43 is sufficient to cause both phenotypes. Collectively, these data suggest that GJIC via Cx43-gap junctions regulates the proliferation state of the population of dividing cells, which in turn may regulate growth and segment length.
Reduced cell proliferation is not sufficient to explain the observed short segment length phenotype. For instance, abrogation of either Shh function or Fgfr1 signaling in regenerating fins each results in reduced cell proliferation, but neither affects segment length (Quint et al., 2002; Lee et al., 2005). Loss of Cx43 function by reduced expression, missense mutations or gene knockdown causes defects in both processes, although is not clear if these phenotypes are linked (i.e. if all cx43 mutations will affect both proliferation and length) or separable. Further, it is not known if the cx43-positive cells surrounding the joint are derived from the distal cx43-expressing cells, or if these represent independent populations. However, since targeted-gene knockdown of cx43 in blastemal cells causes short segments, it appears that both the cell proliferation defect and the segment length defect are caused by related populations of cells.
We propose that Cx43 function coordinates signals regulating cell division and joint formation. This hypothesis is supported by the positive correlation between GJIC defects in the missense mutations, reduced number of dividing cells in the missense alleles, and segment length defects in the missense alleles. Cx43 positive cells in the germinal compartment may respond to a small molecule signal required either to initiate cell division in a direct manner, or to provide a “competence” factor that identifies Cx43 positive cells as a proliferating population. Candidate locations for the source(s) of the signal(s) include the distal-most blastema, the basal epithelium surrounding the germinal compartment, and the stump tissue. A graded signal from the stump tissue in particular may provide a mechanism to measure the length of the growing tissue and regulate segment length. For example, a sub-population of the distal cx43-expressing cells may initiate a joint when the stump signal is sufficiently “diluted” by growth (Iovine et al., 2005). Reduced Cx43 function would prevent adequate distribution of the stump signal to more distal tissue, and therefore premature joint formation.
Cx43 may mediate its function(s) directly or indirectly. For example, Cx43 is correlated with proliferating cells in normal skin and during wound healing in the mouse and may play a role in the maintenance of this tissue (Coutinho et al., 2003). Alternatively, GJIC via Cx43 gap junctions could establish a gradient of signaling along the proximal-distal axis that regulates the expression of other genes. Reciprocal signaling may be required to maintain or establish the population of proliferating cells. Candidates for potential Cx43 target genes include genes in the Wnt-βcat pathway (Stoick-Cooper et al., 2007), Fgfr1 (Lee et al., 2005), Shh (Quint et al., 2002), Bmp (Smith et al., 2006), and msxb (Thummel et al., 2006) since abrogation of each results in reduced cell proliferation and outgrowth. Indeed, knockdown of cx43 during limb growth in the chick has been shown to down-regulate genes in the Shh pathway, leading to digit truncation (Law et al., 2002). Either a candidate gene approach utilizing quantitative RT-PCR or a whole genome approach comparing gene expression in wild-type and sofb123 fins will reveal the putative Cx43 target genes regulating the population of dividing cells during fin regeneration.
The role of GJIC in proliferating cells is not well established. A recent report on gap junctions in planaria suggested that GJIC is required for the maintenance of the stem cell/neoblast population during animal homeostasis and regeneration (Oviedo and Levin, 2007). This report provides additional support for the hypothesis that GJIC regulates the proliferative state of certain populations of cells, and identifies Cx43-gap junctions as a key regulator in zebrafish fins. Future studies will elucidate the molecular mechanisms underlying communication among proliferating cells, and between proliferating cells and their surrounding cells, permitting a coordinated response to injury and essentially unlimited growth and regeneration.
Acknowledgments
The authors thank Jake Fugazzotto for care and maintenance of the fish colony. We thank Julia Hafter for suggestions for Cx43 whole mount fin staining protocols and Ken Poss for critical comments regarding the manuscript. This work was funded by the NIDCR (HD047737 to MKI) and the NEI (R21-EY017134 to DRH). R.T. was partially funded by a Fight for Sight Postdoctoral Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Coutinho P, et al. Dynamic changes in connexin expression correlate with key events in the wound healing process. Cell Biol Int. 2003;27:525–41. doi: 10.1016/s1065-6995(03)00077-5. [DOI] [PubMed] [Google Scholar]
- Goldsmith MI, et al. Saltatory control of isometric growth in the zebrafish caudal fin is disrupted in long fin and rapunzel mutants. Dev Biol. 2003;259:303–17. doi: 10.1016/s0012-1606(03)00186-6. [DOI] [PubMed] [Google Scholar]
- Goss RJ, Stagg MW. The regeneration of fins and fin rays in Fundulus heteroclitus. J Exp Zool. 1957;136:487–507. doi: 10.1002/jez.1401360306. [DOI] [PubMed] [Google Scholar]
- Haas HJ. Studies on mechanisms of joint and bone formation in the skeleton rays of fish fins. Dev Biol. 1962;5:1–34. doi: 10.1016/0012-1606(62)90002-7. [DOI] [PubMed] [Google Scholar]
- Hoptak-Solga AD, et al. Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Lett. 2007;581:3297–302. doi: 10.1016/j.febslet.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, et al. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol. 2005;278:208–19. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Johnson SL. Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics. 2000;155:1321–9. doi: 10.1093/genetics/155.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird DW. Life cycle of connexins in health and disease. Biochem J. 2006;394:527–43. doi: 10.1042/BJ20051922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis WJ, Geraudie J. Organization and development of the mineral phase during early ontogenesis of the bony fin rays of the trout Oncorhynchus mykiss. Anat Rec. 1990;228:383–91. doi: 10.1002/ar.1092280404. [DOI] [PubMed] [Google Scholar]
- Law LY, et al. Knockdown of connexin43-mediated regulation of the zone of polarizing activity in the developing chick limb leads to digit truncation. Dev Growth Differ. 2002;44:537–47. doi: 10.1046/j.1440-169x.2002.00666.x. [DOI] [PubMed] [Google Scholar]
- Lee Y, et al. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–83. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene, knockdown’ in zebrafish. Nat Genet. 2000;26:216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–17. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Levin M. smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development. 2007;134:3121–31. doi: 10.1242/dev.006635. [DOI] [PubMed] [Google Scholar]
- Paznekas WA, et al. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–18. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleo G, et al. Cell proliferation and movement during early fin regeneration in zebrafish. Dev Dyn. 2001;221:380–90. doi: 10.1002/dvdy.1152. [DOI] [PubMed] [Google Scholar]
- Poss KD, et al. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–58. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- Quint E, et al. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A. 2002;99:8713–8. doi: 10.1073/pnas.122571799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, et al. Coupled mutagenesis screens and genetic mapping in zebrafish. Genetics. 2003;163:997–1009. doi: 10.1093/genetics/163.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe W, et al. Oculodentodigital dysplasia-causing connexin43 mutants are nonfunctional and exhibit dominant effects on wild-type connexin43. J Biol Chem. 2005;280:11458–66. doi: 10.1074/jbc.M409564200. [DOI] [PubMed] [Google Scholar]
- Santamaria JA, et al. Interactions of the lepidotrichial matrix components during tail fin regeneration in teleosts. Differentiation. 1992;49:143–50. doi: 10.1111/j.1432-0436.1992.tb00662.x. [DOI] [PubMed] [Google Scholar]
- Seki A, et al. Modifications in the biophysical properties of connexin43 channels by a peptide of the cytoplasmic loop region. Circ Res. 2004;95:e22–8. doi: 10.1161/01.RES.0000140737.62245.c5. [DOI] [PubMed] [Google Scholar]
- Shibayama J, et al. Functional characterization of connexin43 mutations found in patients with oculodentodigital dysplasia. Circ Res. 2005;96:e83–91. doi: 10.1161/01.RES.0000168369.79972.d2. [DOI] [PubMed] [Google Scholar]
- Smith A, et al. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblast differentiation and function. Dev Biol. 2006;299:438–54. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–89. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Thummel R, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–46. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- Wei CJ, et al. Connexins and cell signaling in development and disease. Annu Rev Cell Dev Biol. 2004;20:811–38. doi: 10.1146/annurev.cellbio.19.111301.144309. [DOI] [PubMed] [Google Scholar]
- Wei Y, et al. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish (Brachydanio rerio) University of Oregon Press; Eugene, OR: 1993. [Google Scholar]