Abstract
Our laboratory recently molecularly characterized the type II secretion system (T2SS)- associated cytotoxic enterotoxin (Act) and the T3SS-secreted AexU effector from a diarrheal isolate SSU of Aeromonas hydrophila. The role of these toxin proteins in the pathogenesis of A. hydrophila infections was subsequently delineated in in vitro and in vivo models. In this study, we characterized the new type 6 secretion system (T6SS) from isolate SSU of A. hydrophila and demonstrated its role in bacterial virulence. Study of the role of T6SS in bacterial virulence is in its infancy, and there are, accordingly, only limited, recent reports directed toward a better understanding its role in bacterial pathogenesis. We have provided evidence that the virulence-associated secretion (vas) genes vasH (Sigma 54-dependent transcriptional regulator) and vasK (encoding protein of unknown function) are essential for expression of the genes encoding the T6SS and/or they constituted important components of the T6SS. Deletion of the vasH gene prevented expression of the potential translocon hemolysin coregulated protein (Hcp) encoding gene from bacteria, while the vasK gene deletion prevented secretion but not translocation of Hcp into host cells. The secretion of Hcp was independent of the T3SS and the flagellar system. We demonstrated that secreted Hcp could bind to the murine RAW 264.7 macrophages from outside, in addition to its ability to be translocated into host cells. Further, the vasH and vasK mutants were less toxic to murine macrophages and human epithelial HeLa cells, and these mutants were more efficiently phagocytosed by macrophages. We also provided evidence that the expression of the hcp gene in the HeLa cell resulted in apoptosis of the host cells. Finally, the vasH and vasK mutants of A. hydrophila were less virulent in a septicemic mouse model of infection, and animals immunized with recombinant Hcp were protected from subsequent challenge with the wild-type (WT) bacterium. In addition, mice infected with the WT A. hydrophila had circulating antibodies to Hcp, indicating an important role of T6SS in the pathogenesis of A. hydrophila infections. Taken together, we have characterized the T6SS from Aeromonas for the first time and provided new features of this secretion system not yet known for other pathogens.
Keywords: Aeromonas hydrophila, type VI secretion system, isogenic mutants, murine model of toxicity, translocation and secretion of effectors
1. Introduction
The genus Aeromonas, which is comprised of 17 species (spp.), was recently placed into its own family, namely the Aeromonadaceae [1]. These Gram- negative bacteria inhabit freshwater sources and produce a wide range of virulence factors, including surface molecules [2, 3]; extracellular enzymes [4–7]; adhesins, and various toxins [8].
Among the different species of Aeromonas, A. hydrophila is most commonly associated with a wide variety of human diseases, which include skin and wound infections and septicemia, that are often fatal [8–10]. Although Aeromonas spp. lead to gastroenteritis in young, elderly, or immunocompromised individuals [11], numerous cases of intestinal and extraintestinal infections in immunocompetent individuals have led to the suggestion that the virulence of this pathogen is not entirely dependent upon the immune status of the host [12]. As in the case of other forms of bacterial gastroenteritis, underlying factors such as liver and gastrointestinal diseases, as well as recent therapy with antimicrobials ineffective against aeromonads have been reported as relevant for the development of Aeromonas-associated diseases [13].
Evidence of the pathogenicity of Aeromonas spp. was recently noted in southern Thailand tsunami survivors, as 22% of all wounds in these patients were infected with this bacterium [14]. Furthermore, the floodwater samples collected after hurricane Katrina in New Orleans had elevated numbers of a variety of Aeromonas spp. [15]. In addition, the worldwide isolation rate of Aeromonas from diarrheic stool has been reported to be as high as 10.8%, compared to only 2.1% from the stools of healthy control subjects [16]. In a separate study, although it was noted that in the majority of the patients, only the small intestine seemed to be affected by this pathogen; up to one-third of the infected patient population also showed colitis by endoscopy [17, 18]. It has been documented that A. hydrophila is present in a wide variety of foods (introduced from water, animal feces containing organisms, or food handlers), and, thereby, it has the potential to be a significant food-borne pathogen and hence represents a serious public health concern [16]. With the high resistance of this organism to both water chlorination and multiple antibiotics [19], A. hydrophila has been categorized as an emerging human pathogen, and consequently, it has been placed on the Environmental Protection Agency’s (EPA) “Contaminant Candidate List” [20].
Our laboratory characterized two of the most potent virulence factors from a diarrheal isolate SSU of A. hydrophila, namely Act (Aeromonas cytotoxic enterotoxin) and a type III secretion system (T3SS) secreted effector protein, AexU [21, 22]. Act is secreted by the T2SS and possesses several biological activities, including its ability to lyse erythrocytes, inhibit phagocytosis by professional phagocytes, induce cytotoxicity in eukaryotic cells, and to evoke fluid secretory responses in the ligated ileal loops of animals [22, 23]. At sub-lethal doses, Act induces the production of pro-inflammatory cytokines, prostaglandins, and reactive oxygen species (ROS) from murine and human macrophages and human colonic epithelial cells by activating various kinase pathways [8, 9, 22, 24–28]. In addition, Act leads to mouse mortality when injected by the intravenous route with an LD50 dose of 27.5 ng [25].
AexU, on the other hand, leads to ADP-ribosylation of host cell proteins and actin reorganization resulting in HeLa cell rounding phenotype and eventual cell death via apoptosis [29]. AexU also inhibits phagocytosis, as AexU null mutant was phagocytosed more efficently by murine RAW 264.7 macrophages [21]. Likewise, both Δact and ΔaexU isogenic mutants caused less mortality (40–60%) in mice when injected via the intraperitoneal (i.p.) route, with Δact and ΔaopB (Aeromonas outer membrane protein B; an essential component of the T3SS) double knockout mutant causing only 10–20% mouse mortality in a septicemic model of A. hydrophila infection [21, 30]. Although our recent studies suggested that AexU is most likely the T3SS effector that leads to cell toxicity [21], we noted that the Δact/ΔaexU mutant of A. hydrophila SSU still caused the release of lactate dehydrogenase (LDH) enzyme from murine RAW 264.7 macrophages, albeit at a significantly lower level compared to that in the Δact background strain of the wild-type (WT) A. hydrophila for up to 5.5 hr of infection. Importantly after 6 hr of infection of macrophages with the Δact/ΔaexU mutant and the corresponding WT (parental) A. hydrophila SSU, no difference in LDH release was observed [21]. These data suggested the presence of some as yet unknown factor(s) that contributed to this cell toxicity associated with the Δact/ΔaexU mutant of A. hydrophila.
Protein secretion in Gram-negative bacteria presents a challenge because the secreted proteins must pass through at least two membranes before they can reach the extracellular milieu. To date, five secretion systems have been molecularly well-characterized in Gram-negative bacteria. These systems are highly conserved across different bacteria with unique characteristics which permit their differentiation from one another [31]. Recently, a new protein secretion mechanism called virulence-associated secretion (VAS), or type VI secretion system (T6SS), was described in Vibrio cholerae [32]. Previously, a gene cluster encompassing the T6SS was identified by bioinformatics analysis as being highly conserved among several gram-negative pathogens, and yet it exhibited differences in its organization in various bacteria [33]. It is believed that the primary function of the T6SS is to mediate the extracellular export of virulence factors [32]. This mechanism of secretion is different from T3- and T4- secretion systems because the T6SS represents an assembly of genes with a novel linkage that secretes proteins lacking the classical Sec-dependent signal sequences [34]. Several studies indicated participation of this cluster in the pathogenicity of different bacteria, such as Pseudomonas aeruginosa in which the role of T6SS-associated effector, hemolysin co-regulated protein (Hcp), has been demonstrated in cystic fibrosis [35]. Likewise, in Francisella tularensis and Salmonella enterica, these gene clusters are necessary for intracellular growth in eukaryotic cells [36, 37]. In Burkholderia mallei, it was found that the T6SS is required for virulence in the hamster model of glanders infection [38]. Most important, however, is that the role of this secretion system in the virulence of bacterial pathogens in general is still largely unknown.
In this paper, we report, for the first time, the presence of a functional T6SS gene cluster in a clinical isolate SSU of A. hydrophila. Our characterization of the T6SS showed that this cluster is able to secrete and translocate effector proteins into eukaryotic host cells and that mice immunized with a secreted component of this system (Hcp) were protected from a lethal challenge dose of the WT bacterium. Thus, the components of the T6SS in A. hydrophila SSU constitute exciting candidates for the potential development of preventative or therapeutic vaccines, as well as targets for antimicrobial drug development.
2. RESULTS
2.1. Type VI secretion system gene cluster is present in Aeromonas hydrophila SSU
DNA sequence analysis demonstrated the presence of a T6SS gene cluster in the genome of diarrheal isolate SSU of A. hydrophila (DQ667172) (Fig. 1A). We first noted the possible presence of the T6SS in this isolate by polymerase chain reaction (PCR)-amplification of the portion of vasH-, vasK-, and hcp-encoding genes based on the sequence of V. cholerae T6SS [32]. Our subsequent genome sequencing and annotation of the sequence from A. hydrophila ATCC 7966 [39] further provided evidence for the presence of T6SS in A. hydrophila SSU. We then designed primers based on the sequences of the corresponding T6SS gene cluster from A. hydrophila ATCC 7966 and V. cholerae to cover the entire T6SS gene cluster in isolate SSU of A. hydrophila. Most interesting are our findings that although the T6SS cluster is present in both A. hydrophila SSU and ATCC 7966 strains, the T3SS encoding genes are found only in the clinical isolate SSU, but not in the environmental isolate ATCC 7966 [39].
Figure 1.
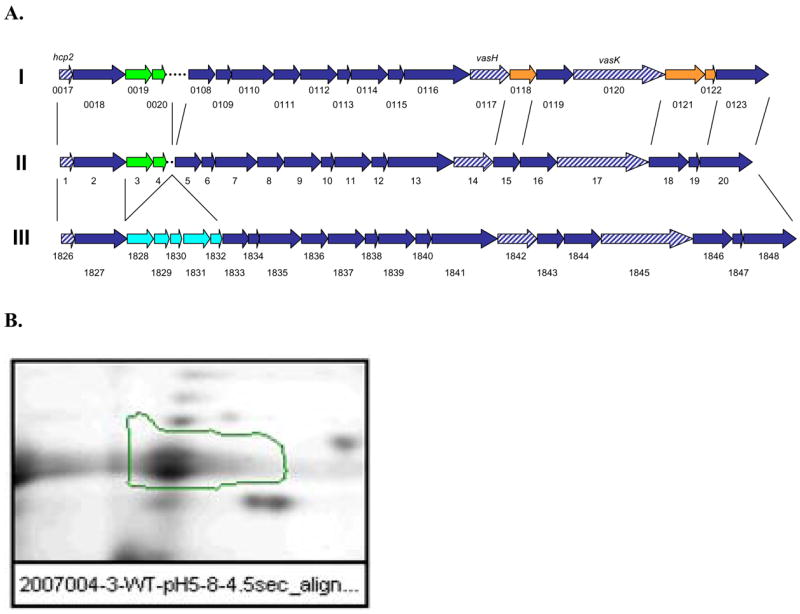
A. Diagram showing the genetic organization of the T6SS gene cluster of A. hydrophila SSU (II) in comparison with the similar cluster present in Vibrio cholerae N16691 (I) and A. hydrophila ATCC 7966 (III). Shown in green are the genes present only in V. cholerae and A. hydrophila SSU T6SS gene clusters. Dashed in blue are genes that were pursued during this study. Genes in cyan color are those that are present only in A. hydrophila ATCC 7966 strain (III). Genes in orange exhibit low identity (<25%) between V. cholerae and A. hydrophila SSU T6SS gene cluster. B. Two-dimensional gel electrophoresis of supernatants from A. hydrophila SSU after staining with Sypro Ruby. The spot for Hcp is highlighted, and its identity was confirmed by mass-spec analysis.
This T6SS gene cluster in A. hydrophila SSU contained 20 open reading frames (ORFs), out of which 18 genes had high identity (>80%) with genes present in a similar cluster that was identified in A. hydrophila ATCC 7966 [39]. We also noted that 10 ORFs had considerable identity (>50%) with genes present in the V. cholerae N16961 strain T6SS gene cluster (Fig. 1A), with the remaining 10 genes showing significant diversity in the sequence. Table 1 depicts the list of genes linked to the T6SS gene cluster present in both A. hydrophila strains (SSU and ATCC 7966) and in V. cholerae, the identity/homology between common genes/proteins, and the gene product description based on A. hydrophila SSU conserved domain analysis.
Table 1.
Comparison of T6SS gene cluster of A. hydrophila SSU with that of V. cholerae N16961 and A. hydrophila ATCC 7966 sequenced strain
| A. hydrophila SSU1 | V. cholerae2 | Identity/homology (%)1 vs. 2 | A. hydrophila ATCC 79663 | Identity/homology (%)1 vs. 3 | Gene product name | Conserved Domains |
|---|---|---|---|---|---|---|
| 1 | VCA0017 | 71/79 | AHA_1826 | 95/98 | Hemolysin coregulated protein Hcp | COG3157 |
| 2 | VCA0018 | 51/52 | AHA_1827 | 80/85 | VgrG protein | COG3501, DUF586 |
| 3 | VCA0019 | 52/38 | hypothetical protein | |||
| 4 | VCA0020 | 25/37 | hypothetical protein | |||
| AHA_1828 | hypothetical protein | |||||
| AHA_1829 | hypothetical protein | |||||
| AHA_1830 | hypothetical protein | |||||
| AHA_1831 | hypothetical protein | TRP repeat, Sel1 | ||||
| AHA_1832 | hypothetical protein | ImpG homolog | ||||
| 5 | VCA0108 | 74/82 | AHA_1833 | 92/97 | hypothetical protein | COG3517 |
| 6 | VCA0109 | 24/31 | AHA_1834 | 94/95 | hypothetical protein | COG3518, GPW/gp25 |
| 7 | VCA0110 | 58/52 | AHA_1835 | 96/98 | VasA | COG3519, DUF879 |
| 8 | VCA0111 | 51/43 | AHA_1836 | 93/97 | hypothetical protein | COG3520, Acyl-CoA dehydrogenase/oxidase C-term |
| 9 | VCA0112 | 18/34 | AHA_1837 | 92/94 | hypothetical protein | COG3456, SMAD/FHA |
| 10 | VCA0113 | 40/39 | AHA_1838 | 95/98 | Putative Lipoprotein | COG3521 |
| 11 | VCA0114 | 61/60 | AHA_1839 | 94/98 | hypothetical protein | COG3522 |
| 12 | VCA0115 | 41/45 | AHA_1840 | 9497 | VasF/IcmH | COG3455 |
| 13 | VCA0116 | 63/60 | AHA_1841 | 94/98 | CplA/ClpB family protein | COG0542, AAA, AAA_2, AAA+, chaperone |
| 14 | VCA0117 | 27/40 | AHA_1842 | 94/96 | Sigma-54 dependent transcriptional regulator/VasH | COG3829, AAA, Interaction RNA pol sigma factor 54, Fis-type Helix-turn-helix, core AAA+ ATPase, Homeodomain-like. |
| 15 | VCA0118 | 7/27 | AHA_1843 | 90/91 | hypothetical protein | |
| 16 | VCA0119 | 26/32 | AHA_1844 | 95/97 | ImpA related domain protein | COG3515, ImpA-related N-ter |
| 17 | VCA0120 | 53/41 | AHA_1845 | 96/99 | ImcF family protein/VasK | COG3523, DUF1215, IcmF-related, conserved hypothetical ATP binding protein |
| 18 | VCA0121 | 1/17 | AHA_1846 | 96/98 | ImpA related domain protein | COG3515, ImpA-related N-ter |
| 19 | AHA_1847 | 96/97 | paar motif protein | COG4104, PAAR | ||
| 20 | VCA0123 | 48/52 | AHA_1848 | 87/93 | Rhs element Vgr family protein | COG3501, DUF586, Phage_GPD |
COG: Cluster of Orthologous Groups of proteins; Most of these analysis data were obtained from the National Center of Biotechnology Information (http://www.ncbi.nlm.nih.gov/sites/entrez/) and the Integrated Microbial Genomes web page (http://img.jgi.doe.gov). The alignments were performed using the Clustalw software program (http://clustalw.genome.jp).
Previous studies in several Gram-negative bacteria have demonstrated the secretion of effector proteins via the T6SS [32, 35, 38, 40]. For example, V. cholerae secretes four proteins via the T6SS; one is a homolog of hemolysin-coregulated protein (Hcp) and the other three are members of the Vgr family of proteins [32]. A. hydrophila SSU T6SS gene cluster contains ORFs with a high identity to hcp (ORF 1, 71% identity,) as well as to two members of the vgr family (COG3501), VgrG2 (ORF 2; 51% identity with VCA0018) and VgrG3 (ORF 20; 48% identity with VCA0123) from V. cholerae (Table 1).
The clpB gene (VCA0116 [ORF 13]), present in the T6SS gene cluster of A. hydrophila SSU, is a member of the AAA+ (ATPase associated with diverse cellular activities) protein family and is a chaperon protein associated with thermotolerance and translocation of aggregated proteins in an energy-dependent manner [41]. Other genes in the T6SS gene cluster of A. hydrophila include homologs of the genes in V. cholerae, such as vasA, vasF, vasK and vasH (ORFs 7, 12, 17 and 14, respectively).
Recently, the function of 3 genes present in the P. aeruginosa T6SS gene cluster was described. These genes include ppkA, which has kinase activity; pppA, which has phosphatase activity; and fha1, a scaffold protein with a forkhead-associated (FHA) domain [42]. The T6SS gene cluster of A. hydrophila SSU has an ORF which contains an FHA domain (ORF 9) and could have a function similar to that of its homolog in P. aeruginosa. However, neither significant similarities nor kinase- or phosphatase-conserved domains were found after sequence alignments of ppkA and pppA genes from P. aeruginosa, with any of the 20 genes found in A. hydrophila SSU T6SS gene cluster (Table 1).
Other members of this T6SS gene cluster in A. hydrophila SSU include two ImpA N-terminal-related domain proteins (ORFs 16 and 18 [COG3515]) which could be associated with the export of proteins [43]; a putative lipoprotein (ORF 10 [COG3521]), and a PAAR motif protein (ORF 19 [COG4104]).
2.2. Hcp is secreted and translocated into human colonic epithelial cells via the T6SS in A. hydrophila SSU
Previous reports have shown that inactivation of the vasK gene in V. cholerae or its homologs in other bacteria blocks secretion of Hcp or other proteins associated with the T6SS gene cluster [32, 35, 40]. In a similar way, vasH, a sigma-54 dependent transcriptional regulator, in V. cholerae controls the expression of the hcp gene [32]. Mass spectrometry analysis, after 2- dimensional (2D)-gel electrophoresis, of culture supernatants from Luria-Bertani (LB)-grown WT A. hydrophila SSU revealed the presence of a protein which is highly homologous to Hcp from V. cholerae (Fig. 1B). These findings indicated the functionality of the T6SS gene cluster of A. hydrophila SSU under normal culture conditions, which consisted of overnight incubation in LB medium at 37°C with shaking (150 rpm).
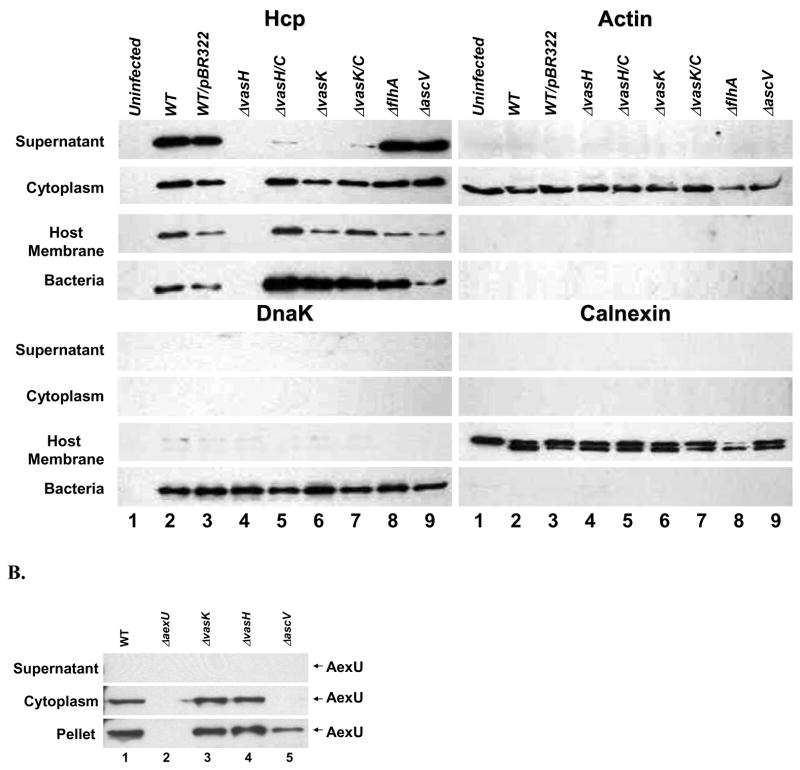
Based on above-mentioned studies, we decided to generate deletion mutants for vasH and vasK genes in A. hydrophila SSU to evaluate the effects of these mutations on the expression and secretion of Hcp. For these experiments, HT-29 human colonic epithelial cells were infected with the WT A. hydrophila SSU and its corresponding mutants, and Hcp was detected in various cellular fractions by Western blot analysis using Hcp-specific antibodies. As shown in Fig. 2A, deletion of the vasH gene abrogated the expression and secretion of Hcp (lane 4), whereas deletion of the vasK gene eliminated the secretion without affecting the expression and translocation of Hcp (lane 6). After complementation of both of these genes, vasH and vasK, in their respective mutants, the secretion and expression was restored, although the secretion was not fully complemented (lanes 3 versus 5 and 7). We demonstrated complementation of these mutants in vivo as well (section 2.7) indicating no potential polar effects of the mutation.
Figure 2.
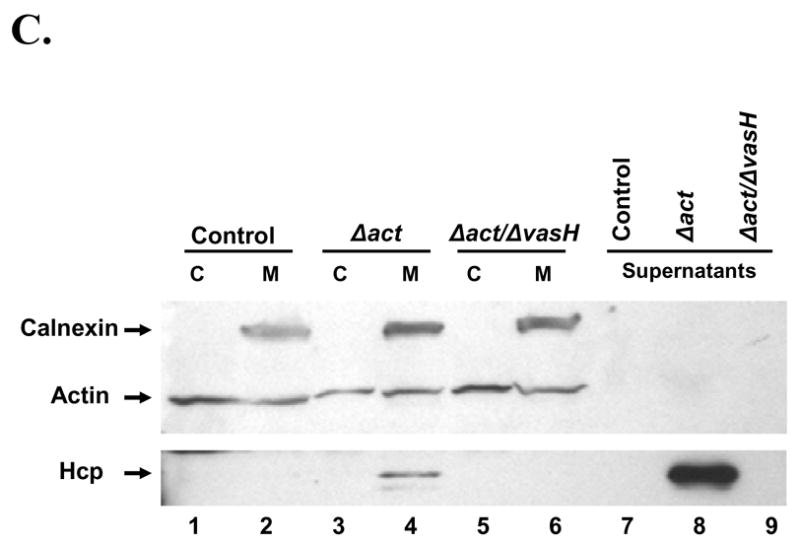
A. Production of Hcp from A. hydrophila SSU and the translocation of Hcp into host cells. HT-29 human colonic epithelial cells were infected with different A. hydrophila strains (MOI of 5) for 2 hr at 37°C in DMEM/0.5% FBS medium. The culture supernatants were TCA precipitated (supernatant fraction). The infected host cells were osmotically lysed and centrifuged to obtain soluble (cytoplasmic fraction representing translocated effectors) and insoluble fractions. The insoluble fraction was resuspended in cell lysis buffer containing 0.1% Triton X-100 and centrifuged to obtain soluble (host cell membrane fraction) and insoluble (intact bacterial pellet) fractions. The samples were run on 4–20% gradient SDS-PAGE and subjected to Western blot analyses using anti-Hcp, anti-actin, anti-DnaK and anti-calnexin antibodies. Lane 1: Untreated HT-29 cells; Lane 2: A. hydrophila WT; Lane 3: WT bacteria containing pBR322; Lane 4: ΔvasH mutant; Lane 5: ΔvasH complemented strain; Lane 6: ΔvasK mutant; Lane 7: ΔvasK complemented strain; Lane 8: ΔflhA mutant, and lane 9: ΔascV mutant B. Production and translocation of AexU are not affected by mutations in the T6SS components. HT-29 cells were infected with the WT A. hydrophila SSU (lane 1), ΔaexU mutant (lane 2), ΔvasK mutant (lane 3), ΔvasH mutant (lane 4), and ΔascV mutant (lane 5). The culture supernatants were TCA precipitated (supernatant fraction). The infected host cells were osmotically lysed and centrifuged to obtain soluble (cytoplasmic fraction) and insoluble fractions (pellet). The samples were run on 4–20% gradient SDS-PAGE and subjected to Western blot analyses using anti-AexU antibodies. C. Hcp binds to the cell membrane of RAW 264.7 murine macrophages. Supernatants from A. hydrophila Δact and Δact/ΔvasH mutants were added to RAW 264.7 cells and incubated for 2 hr at 37°C. The host cells were washed, osmotically lysed, and centrifuged to obtain soluble (cytoplasmic) and insoluble (membrane) fractions. Samples were run on a 4–20% gradient SDS-PAGE and subjected to Western blot analyses using the following antibodies: anti-calnexin, anti-actin and anti-Hcp. Control: Macrophages incubated with 1% FBS-DMEM (lanes 1 and 2); Δact: Macrophages incubated with supernatants from A. hydrophila act mutant (lanes 3 and 4); Δact/ΔvasH: Macrophages incubated with supernatants from A. hydrophila Δact/ΔvasH mutant (lanes 5 and 6). The supernatants from the Δact (lane 8) and Δact/ΔvasH (lane 9) mutants of A. hydrophila SSU were used as a control for the presence of Hcp. C=Cytoplasmic fraction from RAW 264.7 macrophages. M=Membrane fraction from RAW 264.7 macrophages.
Since it has been reported that cytotoxicity induced by V. cholerae in murine J774.1 macrophages requires direct cell-cell contact, and deletion of the hcp gene abrogates these morphological changes [32], we decided to evaluate the ability of A. hydrophila SSU WT and its various mutants to translocate Hcp into HT-29 cells. The above-mentioned bacteria were co-cultured with HT-29 cells, and, after 2 hr of infection, the supernatants were removed, and the cells were fractionated to obtain cytoplasmic proteins, eukaryotic host membrane proteins and the remaining pellet containing the bacterial cells [21]. To ensure that there was no cross-contamination of various cellular fractions, each one of the fractions was tested for the presence of actin, calnexin and DnaK as markers for host cytoplasmic proteins, host membrane proteins and bacterial intactness, respectively. We were able to detect the translocation of Hcp into the host cell cytoplasm by Western blot analysis of the HT-29 cells after infection with WT A. hydrophila SSU (Fig. 2A, lane 2). However, as expected, no translocated Hcp was seen in HT-29 cells infected with the ΔvasH strain (lane 4). In addition, although the ΔvasK mutant was unable to secrete Hcp into the extracellular media, translocation of Hcp was not affected (lane 6). We did not observe any cross-contamination of cellular fractions, and the bacterial integrity was intact, when antibodies to actin, calnexin, and DnaK were used for Western blot analysis (Fig. 2A)
2.3. T6SS in A. hydrophila SSU is independent of the Type III Secretion System (T3SS) and the flagellar system
The WT A. hydrophila SSU harbors different secretion systems, including the T3SS [30] and the flagellar system [44, 45], which are involved in the secretion as well as in the direct translocation of proteins into host eukaryotic cells. Therefore, we decided to test if shutting down these secretion systems would impact the secretion of Hcp. For these experiments, cellular fractions of HT-29 cells after infection with WT A. hydrophila SSU and its mutants for the T3SS (ΔascV) and for the flagellar system (ΔflhA) were used. The ΔascV mutant is able to express the aexU gene (encoding a T3SS effector protein) but unable to secrete the expressed protein [21], and the flhA gene codes for a protein that is believed to be part of the export apparatus for lateral flagellar assembly, as the flhA mutant of A. hydrophila showed reduced adherence and biofilm formation [46].
Our data indicated that translocation and secretion of Hcp was not affected in the ΔascV and ΔflhA mutants, indicating that, indeed, the T6SS is independent of the T3SS and the flagellar secretion system in mediating the secretion and translocation of Hcp (Fig. 2A, lanes 8 and 9). Furthermore, translocation of AexU (T3SS effector) was not affected in the ΔvasK and ΔvasH mutants, as shown in Fig. 2B (lanes 3 and 4). As positive controls, we infected HT-29 cells with ΔaexU and ΔascV mutants, and no translocation of AexU was noted (lanes 2 and 5). As expected, we noted the expression of the aexU gene in WT, vasH, vasK, and ascV mutants of A. hydrophila SSU (pellet fraction, lanes 1, 3, 4, and 5).
2.4. Binding of secreted Hcp to murine macrophages
Previous studies in P. aeruginosa indicated the ability of Hcp to form hexameric rings after secretion, and the findings suggested that these rings could be inserted into the eukaryotic membrane as a part of the “translocon” [35]. Since Hcp was detected in the culture supernatant of WT A. hydrophila SSU in abundant amounts (Fig. 1B), we decided to confirm if secreted Hcp was able to bind the cell membrane of RAW 264.7 murine macrophages to possibly initiate cell signaling from outside, in addition to the ability of Hcp to affect host cell signaling as a result of a translocated effector. For this experiment, a double mutant, Δact/ΔvasH, was generated to avoid the cytotoxic effects induced by Act present in the culture supernatants. Culture supernatants of A. hydrophila SSU Δact and Δact/ΔvasH mutants were collected after 2 hr of inoculation, filtered, and added to RAW 264.7 cells for 2 hr. After washing the host cells, cytoplasmic and membrane fractions were obtained. As shown in Fig. 2C, Western blot analysis demonstrated that Hcp present in the supernatant of the Δact A. hydrophila SSU mutant was able to bind to the cell membrane of RAW 264.7 cells (lane 4). In contrast, Hcp was not detected in the cytoplasmic fraction of these cells (lane 3), indicating no cross-contamination of the cytosolic and membrane fractions of the host cells. As expected, we did not observe Hcp binding in uninfected RAW 264.7 cells (lanes 1 and 2) and in host cells infected with the Δact/ΔvasH mutant (lanes 5 and 6). The latter mutant is unable to express the hcp gene. Lanes 7–9 show the presence or absence of Hcp in the culture medium of Δact and Δact/ΔvasH mutants of A. hydrophila SSU. Similar assays were performed using recombinant Hcp (rHcp) obtaining essentially the same results (data not shown).
2.5. A. hydrophila T6SS inhibits phagocytic activity and mediates cytotoxicity
Since infection by A. hydrophila SSU is not intracellular, and mutations in the vasH and vasK genes are able to alter expression and secretion of proteins (e.g., Hcp) associated with the T6SS, we decided to evaluate the effects on phagocytosis caused by mutation in those two genes, namely vasH and vasK of A. hydrophila SSU.
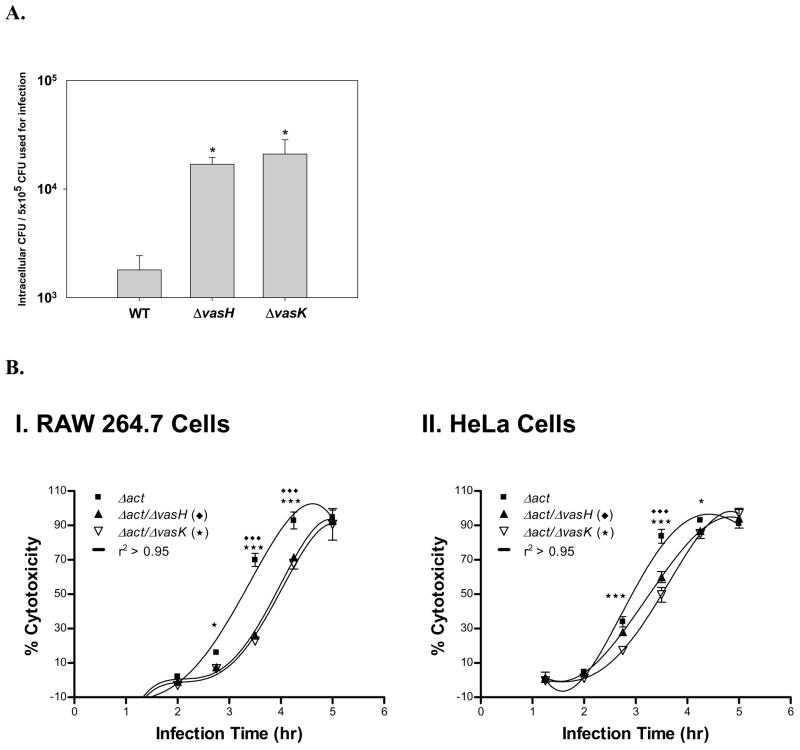
To test for phagocytic activity, RAW 264.7 macrophages were infected with WT A. hydrophila SSU and ΔvasH and ΔvasK mutants, and the intracellular colony forming units (cfu) were estimated as an indicator of phagocytosis. As noted from Fig. 3A, phagocytosis was significantly increased in RAW 264.7 macrophages infected with ΔvasH and ΔvasK mutants when compared to host cells infected with the WT A. hydrophila.
Figure 3.
A. Phagocytosis is enhanced in A. hydrophila ΔvasH and ΔvasK mutants. RAW 264.7 murine macrophages were infected at an MOI of 5 with A. hydrophila strains, namely, WT, ΔvasH and ΔvasK mutants. Thirty minutes after infection, the cells were washed and treated with 100 μg/ml of gentamicin for 1 hr. Then, RAW 264.7 cells were washed and lysed with water. The bacteria were plated at different dilutions and the colony forming units were determined.* denotes statistically significant values (p<0.01) compared to the parent strain (WT). B. Cytotoxicity associated with the T6SS. RAW 264.7 cells (Panel I) and HeLa cells (Panel II) were infected with Δact/ΔvasH mutant (solid triangle) and Δact/ΔvasK mutant (open triangle) double-knockout mutants and their parental strain Δact mutant (solid square) at an MOI 0.5. At different time points, cytotoxicity was measured by the lactate dehydrogenase (LDH) enzyme release assay. *** or ♦♦♦ denotes statistically significant values (p<0.001) compared to the parent strain Δact. * denotes statistically significant values (p<0.05) compared to the parent strain Δact. Three independent experiments in duplicate wells were performed.
The cytotoxic effect of A. hydrophila SSU ΔvasH and ΔvasK mutants was then tested in RAW 264.7 macrophages and HeLa cells by measuring the release of lactate dehydrogenase (LDH) enzyme. RAW 264.7 and HeLa cells were incubated with either the A. hydrophila mutant deleted for the act gene (as a control) or the ΔvasH and ΔvasK mutants at a multiplicity of infection (MOI) of 0.5 for different time points, and supernatants were collected for measuring the LDH release. Significant differences in cytotoxicity between the Δact and Δact/ΔvasK or Δact/ΔvasH strains were detected in RAW 264.7 and HeLa cells after 3–4.5 hr of infection (Fig. 3B). Although the Δact background strain was used in this experiment as a control to remove the strong cytotoxic effects associated with this protein, five hours after infection, the percentage of cytotoxicity induced by the different bacterial mutant strains was similar to that of the control strain (Δact of A. hydrophila SSU) as a consequence of other virulence factors produced (i.e., AexU). These data indicated that mutations in the vasH and vasK genes were able to alter the biological effects associated with the T6SS of A. hydrophila. Although differences in cytotoxicity induced by the parental versus mutant strains may appear small, we believe they are biologically meaningful, as there are other A. hydrophila virulence factors that also lead to cell toxicity.
2.6. Expression of the hcp gene in HeLa Tet-Off cells and induction of apoptosis
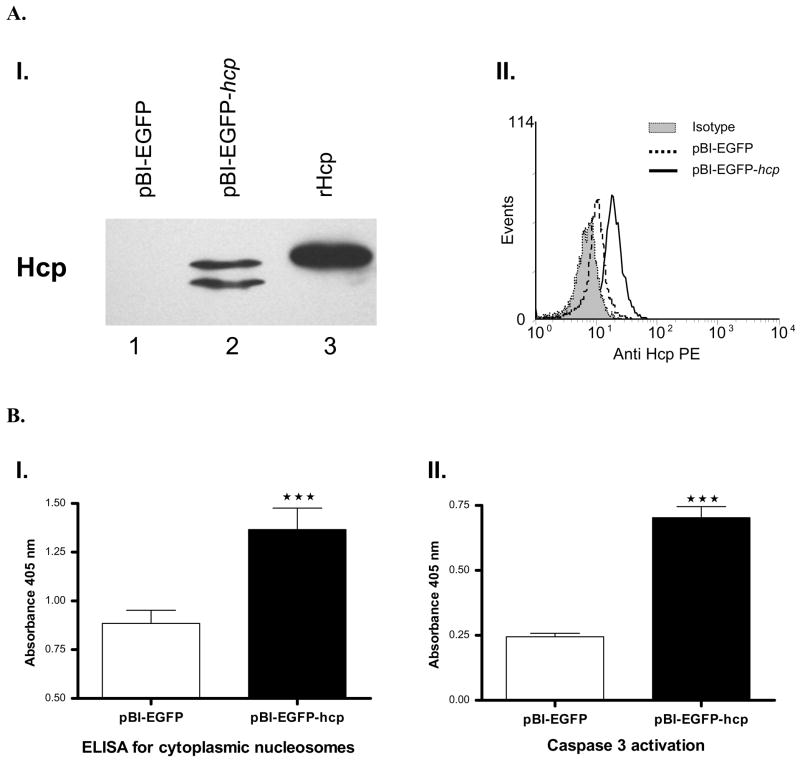
Since we showed that Hcp is translocated into eukaryotic cells, we decided to express the hcp gene into HeLa cells from pBI-EGFP vector using the HeLa cell Tet-Off system. HeLa Tet-Off cells were transfected by electroporation with the pBI-EGFP vector containing the hcp gene (519 bp), and the transfection efficiency was measured by flow cytometry as the percentage of EGFP (enhanced green fluorescent protein)-positive cells. Expression and production of Hcp were evaluated by Western blot and flow cytometry analyses. A band of ~20 kDa was observed by Western blot analysis in the HeLa Tet-Off cells transfected with pBI-EGFP-hcp plasmid (Fig. 4A, panel I). The lower band probably represented a degradation product of Hcp. These results were confirmed by flow cytometry by performing intracellular staining of cells transfected with pBI-EGFP-hcp plasmid (Fig. 4A, panel II). As negative controls, we used host cells transfected with pBI-EGFP vector alone and isotype antibody.
Figure 4.
A. Expression and production of Hcp in transfected HeLa Tet-Off cells. Panel I. Western blot analysis showing production of Hcp in whole cell lysates of HeLa Tet-Off cells after 24 hr of transfection with pBI-EGFP-hcp (lane 2) or pBI-EGFP (empty vector) (lane 1) plasmid. Recombinant Hcp was used as a positive control (lane 3). Panel II. Expression of the hcp gene in HeLa Tet-Off cells after permeabilization and intracellular staining using anti-Hcp antibodies. Mouse pre-immune serum was used as an isotype control. The cells were acquired using a FACScan flow cytometer and analyzed using WinMDI software, gated on EGFP-positive cells. B. Induction of apoptosis in HeLa Tet-Off cells transfected with the hcp gene. Panel I. Detection by ELISA of cytoplasmic nucleosomes in HeLa Tet-Off cells transfected with hcp after 24 hr. *** denotes statistically significant values (p<0.001) compared to those in cells transfected with the pBI-EGFP (empty vector) plasmid. Standard deviations were calculated from duplicate samples from one representative experiment. A minimum of three experiments were performed with similar results. Panel II. Colorimetric caspase 3 detection in total lysates of HeLa Tet-Off cells transfected with the hcp gene after 24 hr. Figures are representative of three independent experiments. *** denotes statistically significant values (p<0.001) compared to those of cells transfected with the pBI-EGFP (empty vector) plasmid. Standard deviations were calculated from duplicate assays from one experiment. A minimum of three experiments were performed with similar results.
We then evaluated cytotoxicity and mitochondrial activity by colorimetric MTT assays in the HeLa cells transfected with the pBI-EGFP-hcp plasmid, and there was no significant difference between the cells transfected with the vector alone (pBI-EGFP) and the cells transfected with the vector containing the hcp gene after 24 hr of transfection (data not shown). Likewise, incorporation of 7-amino actinomycin D (7-ADD), which permeates the membranes of dead and dying cells and stains their DNA, was not significantly different between vector alone and the hcp transfected HeLa cells after 24 hr of infection (data not shown). These studies led us to examine any apoptosis of the host cells that might be associated with Hcp.
To evaluate the apoptotic rate in the HeLa Tet-Off cells transfected with the hcp gene for 24 hr, we measured the cytoplasmic histone-associated DNA fragments (nucleosomes) by ELISA. As shown in Fig. 4B (panel I), cells producing Hcp had a significantly higher rate of apoptosis compared to HeLa cells transfected with the vector alone (p<0.001). To confirm these results, we assessed caspase 3 activity in these cells by a colorimetric activity assay. The activation of caspase 3 was significantly increased in the HeLa cells producing Hcp (p<0.001) when compared to cells transfected with the vector alone (Fig. 4B, panel II). We could not perform similar experiments in which host cells were treated with rHcp as most of the protein produced from E. coli was membrane bound, thus requiring harsh conditions for its solubilization, which resulted in a loss in biological activity.
2.7. T6SS is important for the virulence of A. hydrophila SSU in the mouse model
To evaluate the importance of the hcp gene during in vivo infection, we challenged mice with WT A. hydrophila at a sub-lethal dose, and collected the sera from the surviving mice. Subsequently, the sera were probed by Western blot analysis against the purified rHcp. As shown in Fig. 5A, the surviving mice developed specific antibodies against Hcp. As a control, sera obtained before infection was also tested. The presence of anti-Hcp specific antibodies indicated that Hcp was produced during in vivo infection and that it highlighted the immunogenic potential of this protein. Therefore, we decided to test the ability of Hcp to protect mice from challenge with ~3 LD50 (5 × 107 cfu) of WT A. hydrophila SSU. Mice were immunized with 10 μg of rHcp, boosted after 3 weeks, and challenged with WT bacteria. Then their survival rates were monitored for 16 days. All of the mice immunized with rHcp survived after challenge, whereas all of the control (non-immunized) animals died within 48 hr (Table 2). We used only 3 LD50 dose initially, as in addition to the T6SS, other toxins produced A. hydrophila can lead to mice killing. Therefore, it is important that other virulence-associated genes should also be deleted to develop a safer vaccine.
Figure 5.
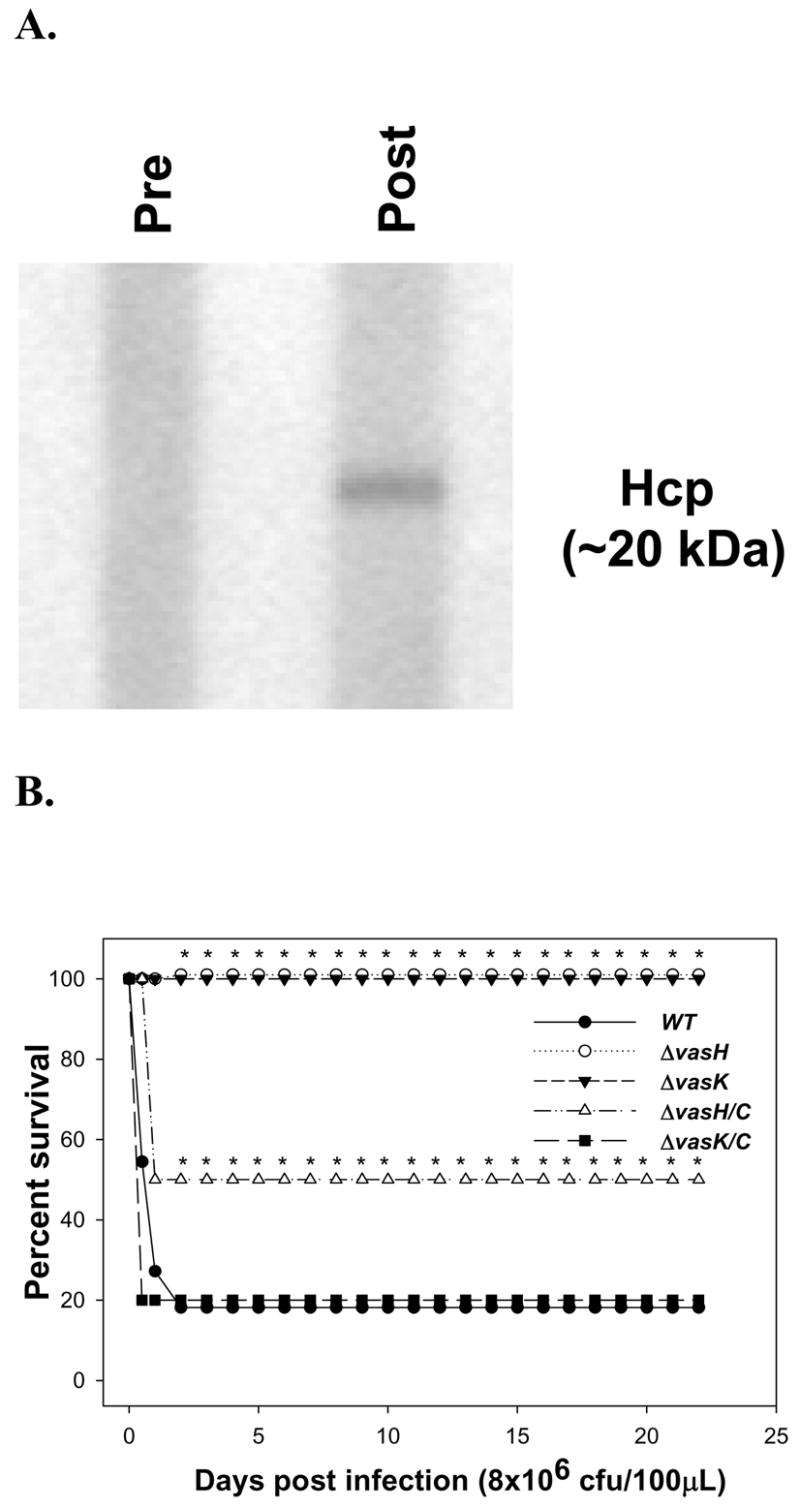
Role of Hcp during A. hydrophila infection in a mouse model. A. Swiss Webster mice were infected i.p., with WT A. hydrophila at a dose of 1 LD50 (1 × 105 cfu/100μl). After 2 weeks of infection, sera from the surviving mice were collected and pooled. The sera was diluted 1:100 and used as a source of primary antibodies in Western blot analysis against rHcp. Pre-immune serum was used as a negative control. B. Groups of 10 Swiss Webster mice were infected i.p. (8 × 106 cfu) with A. hydrophila: WT (solid circles), ΔvasH mutant (open circle) and ΔvasK mutant (solid triangle). Same doses of the complemented strains were also used, ΔvasH/C (open triangle) and ΔvasK/C (solid square). Deaths were recorded for 16 days post-infection. The bacterial doses represented approximately 2 LD50 of WT A. hydrophila. * denotes statistically significant values (p<0.001) of the mutants (vasH and ΔvasK) compared to the WT bacterium and of ΔvasH mutant compared to the ΔvasH/C strain (p<0.05) using the Fisher exact test. The death curve for WT A. hydrophila with pBR322 vector alone was similar to that of the WT bacterium (without the vector) (not shown).
Table 2.
Protection of mice immunized with rHcp against lethal challenge of WT A. hydrophila SSU
| Survivals | |
|---|---|
| Immunized | 5/5 |
| Non Immunized | 0/5 |
Animals were challenged with 3LD50 (5×107 cfu/100μl)
Finally, to evaluate the role of T6SS during infection, we infected mice via the i.p route with WT, as well as with the ΔvasH, ΔvasK, and their respective complemented mutant strains (ΔvasH/C and ΔvasK/C) of A. hydrophila SSU (8 × 106 cfu), and the survival rate of the animals was recorded. After 48 hr, 80% of the animals infected with the WT and ΔvasK/C, and 50% of the animals infected with ΔvasH/C strain of A. hydrophila died (Fig. 5B), while, in contrast, 100% of the animals infected with ΔvasH and ΔvasK mutant strains survived 16 days postinfection (Fig. 5B). Importantly, at higher doses of the WT bacterium (5 × 107 cfu), no difference in the survival rate of mice was noted when compared to the ΔvasH and ΔvasK mutant strains. Although the ΔvasK mutant is still able to translocate Hcp, its virulence was attenuated in the mouse model similar to that of the ΔvasH mutant, which does not produce Hcp (Fig. 2A, lanes 4 and 6).
3. DISCUSSION
Although the presence of a T6SS was recently described in V. cholerae [32], bioinformatic analysis demonstrated that many gram-negative bacteria harbor the T6SS gene cluster [33]. Some of these bacteria that carry the T6SS gene cluster, which has been variously named in different pathogens, include Yersinia pestis, Legionella pneumophila (the T6SS gene cluster named as IAHP [IcmF-associated homologous protein]), Pseudomonas aeruginosa (the gene cluster known as HSI [Hcp secretion Island]), Enteroaggregative Escherichia coli (EAEC) O42 (the gene cluster known as aai [AggR-Activaded Island]), Salmonella enterica (the gene cluster known as Sci), Agrobacterium tumefaciens, Edwardsiella ictaluri, Edwardsiella tarda, Rhizobium leguminosarum, Burkholderia mallei [33, 37, 38, 40, 47–49], and recently in A. hydrophila SSU (GenBank accession number DQ667172) and A. hydrophila ATCC 7966 [39]. The latter is an environmental isolate (type strain ATCC 7966 of A. hydrophila subspecies hydrophila), and we recently annotated its genome sequence in collaboration with The Institute for Genome Research (TIGR). However, the role of this secretion system in the virulence of bacterial pathogens, in general, is largely unknown.
Hcp and VgrG family of proteins are known effectors of T6SS [32, 35, 38, 40]. It has been reported that a member (VgrG1) of the Vgr family from V. cholerae shares with RtxA toxin a subdomain that mediates actin crosslinking and cytotoxicity when it is expressed in the cytoplasm of eukaryotic cells. However, this activity has been associated with a second domain of this protein which is not present in VgrG2 and VgrG3 found in A. hydrophila (Table 1) [50]. Further, studies have shown that mutations affecting the ATPase domain of clpV1 (a member of the clpB family) in P. aeruginosa abrogates Hcp secretion [35]. These results suggest that the clpB gene in A. hydrophila T6SS gene cluster (Table 1) could play a similar role in unfolding proteins to be secreted, as well as in providing energy for this process.
The T6SS gene cluster in A. hydrophila SSU contains homologs of vasA, vasF, vasK, and vasH genes found in V. cholerae (ORF 7, 12, 17, and 14 respectively (Table 1). For the first three genes, their functions remain poorly characterized (COG3519, COG3455, and COG3523, respectively). The vasH gene encodes a sigma-54 factor-dependent transcriptional regulator carrying a factor for inversion stimulation (Fis)-type helix-turn-helix and homeodomain-like motifs, which are involved in DNA-binding and ATPase activity (AAA+ core). Approximately one-half of the proteins that interact with RNA polymerase sigma factor-54 domain can be phosphorylated by a sensor kinase. The ATPase activity in most of these proteins has been associated with conformational changes that promote interaction with Sigma-54 factor, and the Fis-type domain and the homeodomain are directly associated with DNA-binding [51–53]. Thus, vasH in A. hydrophila SSU could be a key component necessary for the expression of components of T6SS which mediates the association between RNA polymerase and DNA.
Hcp and VgrG proteins are proposed effectors associated with the T6SS gene cluster as they are associated with cytotoxicity in some in vitro models, such as the rounded phenotype in the J774.1 murine macrophages, and killing of Dictyostelium amoeba in V. cholerae [32]. However, it has also been reported that expression of the hcp gene is required for VgrG secretion [32]. This, together with the ability of Hcp to form hexameric rings after secretion, suggest that Hcp could be part of the translocon and that VgrG then passes through the Hcp channel [32, 35, 41].
In Fig. 2A, we showed importance of vasH and vasK genes in the T6SS gene cluster of A. hydrophila SSU. Likewise, studies conducted by Mougous et al. (2006) demonstrated by fluorescence microscopy that IcmF (homolog to VasK) in P. aeruginosa could be required (but not totally essential) for the efficient assembly of the T6- secretion apparatus. Therefore, we consider that VasK may constitute a structural protein of the T6SS in A. hydrophila SSU that provides the stability necessary to secrete Hcp into the extracellular milieu in absence of cell- to- cell contact. However, when the T6SS apparatus makes a cell- to-cell contact with eukaryotic membranes, its (VasK) function could be redundant due to the stability that the translocon (e.g., Hcp) could render, thus still allowing Hcp to be translocated into the host cell (Fig. 2A). Finally, the ability of A. hydrophila SSU Hcp to bind to murine macrophages (Fig. 2C) supported the contention proposed for Hcp from P. aeruginosa that Hcp alone could be able to attach to the cellular membrane [35].
Previous reports indicated that a homolog of vasK, called icmF and sciS in L. pneumophila and S. enterica, respectively, is involved in the intracellular survival and replication of these bacteria [37, 54]. Similarly, our findings indicated that mutations in components of the T6SS gene cluster (e.g., vasH and vasK) could alter the virulence of A. hydrophila by altering the ability of bacteria to be phagocytosed. Our future studies will delineate how deletion of vasH and vasK genes leads to increased phagocytosis of the bacteria.
Finally, we provided evidence that expression of the hcp gene in HeLa cells led to host cell apoptosis (Fig. 4). It has been proposed that Hcp from P. aeruginosa could be able to form a channel allowing the release of ions as well as the transport of macromolecules [35]. Bacterial pore-forming proteins like alpha toxin from Staphylococcus aureus, listeriolysin O from Listeria monocytogenes and alpha-hemolysin from E. coli are able to induce apoptosis by the selective release of ions leading to DNA fragmentation [55]. However, alpha toxin can induce apoptosis or necrosis depending upon its concentration [55]. The induction of apoptosis by Hcp in A. hydrophila might be related to the formation of pores in the cell membrane, and, since the amount of Hcp produced by the HeLa cells is not high because the pBI-EGFP vector possesses a minimal cytomegalovirus promoter which lacks the enhancer, the effects that we are seeing are consistent with the induction of apoptosis rather than necrosis. Our future studies will be targeted at studying Hcp-induced apoptosis in details.
In Fig. 5A, we demonstrated circulating antibodies to Hcp in mice after infection with the WT A. hydrophila SSU. Following infections with P. aeruginosa, similar results were reported in patients with chronic respiratory infections in that they showed a high titer of antibodies against Hcp [35]. In addition, animals of different species showed, following infection with glanders disease, the presence of anti-Hcp antibodies [38]. Since anti-Hcp antibodies were present in the sera of mice infected with the sub-lethal dose of A. hydrophila and these antibodies were protective against subsequent challenge of animals with the WT bacterium (Table 2), we speculate that Hcp could be a potential target for vaccine development.
Importantly, our data reported in Figs. 2A and 5B indicated that although ΔvasK mutant could translocate Hcp, it was attenuated in a mouse model of infection. These results raise the question whether the secreted Hcp plays a more important role in bacterial virulence than the translocated one and will be studied in our future studies. The susceptibility of the ΔvasH and ΔvasK mutants to be phagocytosed, compared to that of the WT A. hydrophila, could explain the increased survival of the mice infected with these mutants. Thus, ΔvasH and ΔvasK mutants could be cleared from the intraperitoneal space before infection becomes systemic. Further, since these mutants killed mice at higher doses (5 × 107 cfu compared to 8 × 106 cfu), these data implied that the presence of T3SS effectors, as well as Act could be contributing to animal lethality, which confirms our previous studies [21, 30].
It is also interesting to note that both ΔvasH and ΔvasK mutants showed increased phagocytosis (Fig. 3A) in spite of the fact that the ΔvasK mutant could translocate Hcp efficiently (Fig. 2A). These data could indicate: i) secreted Hcp might play an important role in phagocytosis, and ii) the translocated Hcp might have been altered in its function in the ΔvasK mutant. Our future studies will be focused on these aspects as it relates to Hcp.
In summary, we reported the presence of a functional T6SS gene cluster in A. hydrophila SSU and showed, for the first time, the ability of this secretion system to translocate Hcp into the eukaryotic cells and to induce apoptosis that was mediated by caspase 3 activation. Additionally, we showed that immunization of mice with Hcp protected animals from subsequent challenge with the lethal dose of the WT bacterium.
4. Materials and Methods
4.1. Cell Lines and transfections
HT-29, a human colonic intestinal epithelial cell line, and RAW 264.7, a murine macrophage cell line, were obtained from American Type Culture Collection (Manassas, VA). These cells were grown as previously described [29]. HeLa Tet-Off™, a human cervical epithelial cell line, was obtained from Clontech (Mountain View, CA). Cells were cultured in Dulbecco’s modified eagle medium (DMEM) with high glucose (Invitrogen-Gibco, Carlsbad, CA), supplemented with 10% tetracycline free fetal bovine serum (FBS) (Clontech) and 100 μg/ml of G-418 (Cellgro, Herndon, VA) as described previously [29].
HeLa Tet-off™ cells were transfected by electroporation with a pBI-EGFP plasmid (Clontech) that separately controls expression of the gene encoding EGFP and the gene of interest. The DNA fragment encoding Hcp (519 bp) was cloned at the NheI and MluI restriction enzyme sites of the vector pBI-EGFP. The efficiency of transfection was determined as the percentage of cells expressing EGFP by fluorescent microscopy (Axiovert 200M, Carl Zeiss, Thornwood, NY) and flow cytometry (FACScan, Becton Dickinson, San Diego, CA). HeLa Tet-Off cells transfected with the pBI-EGFP vector alone (without any insert) were used as a negative control in different assays. Western blot analysis and flow cytometry, after intracellular staining using specific antibodies, were also performed to examine the production of Hcp in HeLa Tet-Off cells.
4.2. Bacterial cultures and vectors
Escherichia coli HMS174-DE3 cells, obtained from Novagen (Madison, WI), were grown after transformation with appropriate recombinant plasmid DNA in Luria Bertani (LB) medium supplemented with appropriate antibiotics [29]. The plasmid DNA was isolated from E. coli DH5α clones transformed with pBI-EGFP recombinant plasmids, which were similarly cultivated [29]. The gene encoding Hcp was cloned into a pET-30a vector for hyper-expression and purification purposes. Briefly, the appropriate DNA fragment was cloned at the XhoI and BglII restriction enzyme sites of the vector, resulting in the fusion of Hcp with the 6x-histidine (His)-tag. The other cultures and plasmids used are listed in Table 3.
Table 3.
Various bacterial strains and plasmids used in this study.
| Strain or Plasmid | Relevant Characteristic(s) | Component of | Source or Reference |
|---|---|---|---|
| A. hydrophila SSU | Diarrheal isolate of A. hydrophila | CDC* | |
| SSU-R | Rifampin resistant (Rifr) strain of A. hydrophila SSU | Laboratory Stock | |
| ΔascV | ascV mutant of A. hydrophila SSU-R; Rifr, Spr/Smr | T3SS | [61] |
| Δact | act mutant of A. hydrophila SSU-R; Rifr, Kmr | T2SS toxin | [60] |
| ΔaexU | aexU toxin mutant of A. hydrophila SSU-R; Rifr, Kmr | T3SS effector | [21] |
| ΔflhA | flhA mutant of A. hydrophila SSU-R; Rifr, Spr/Smr | Flagellar system | This study |
| ΔvasH | vasH mutant of A. hydrophila SSU-R; Rifr, Spr/Smr | T6SS | This study |
| ΔvasK | vasK mutant of A. hydrophila SSU-R; Rifr, Spr/Smr | T6SS | This study |
| Δact/ΔvasH | act and vasH double knockout mutant of A. hydrophila SSU-R; Rifr, Kmr, Spr/Smr | T2SS/T6SS | This study |
| Δact/ΔvasK | act and vasK double knockout mutant of A. hydrophila SSU-R; Rifr, Kmr,Spr/Smr | T2SS/T6SS | This study |
| ΔvasH/C | vasH mutant of A. hydrophila SSU-R complemented with pBR322-vasH; Rifr, Spr/Smr, Tcr | T6SS | This study |
| ΔvasK/C | vasK mutant of A. hydrophila SSU-R complemented with pBR322-vasK; Rifr,Spr/Smr, Tcr | T6SS | This study |
| WT/pBR322 | A. hydrophila SSU-R transformed with pBR322 vector alone; Rifr, Tcr, Apr | Laboratory Stock | |
|
| |||
| E.coli | |||
| DH5α | recA, gyrA | Life Technologies | |
| TOP10 | mcrA, hsdR, lacZ M15,endA1, recA1 | Invitrogen | |
| SM10 | λ pir | [60] | |
| HMS174 (DE3) | recA mutated K-12 background, that carries a chromosomal copy of the T7RNA polymerase under control of lacUV5 promoter. | Novagen | |
|
| |||
| Plasmids | |||
| pBR322 | Low copy plasmid used for complementation; Tcr, Apr | Pierce | |
| pET30a | pBR322-derived expression vector with T7 lac promotor, up- and down- stream His tags; Apr. | Novagen | |
| pDMS197 | Suicide vector; R6K ori, sacB, Tcr. | [62] | |
| pBI-EGFP | Eukaryotic expression vector regulated by a tetracycline element response (TER) and a minimal CMV promoter; Apr | Clontech | |
| pBluescript | Vector used for cloning proposes; Apr. | Stratagen | |
| pUC4K | Contains 1.2 Kb kanamycin resistance gene cassette; Kmr | Amersham | |
| pHPΩ45 | Contains a 2.0 Kb streptomycin/spectinomycin resistance gene cassette; Smr/Spr. | [63] | |
| pCR2.1 | Used for PCR cloning proposes; Apr, Kmr | Invitrogen | |
Centers for Diseases Control and Prevention
4.3. Generation of ΔvasK and Δact/ΔvasK knockout mutants
Two pairs of primers (VasKup5/VasKup3 and VasKdn5/VasKdn3) (Table 4) were used to PCR amplify the upstream and downstream flanking DNA sequences to the vasK gene from the genomic DNA (gDNA) of A. hydrophila SSU. After the PCR reactions, the resulting up- and down- stream flanking DNA fragments (1472- and 1198-bp, respectively) were ligated together through the introduced common BglII enzyme site and cloned into the pBluescript vector at XbaII/KpnI restriction enzyme sites, generating a recombinant plasmid pBluevasKUD. Subsequently, a streptomycin/spectinomycin (Smr/Spr) gene cassette flanked by the BamHI site was removed from plasmid pHP45Ω and inserted at the BglII site (compatible with the BamHI site) of pBluevasKUD which generated the recombinant plasmid pBluevasKUDSmr/Spr. After digestion with XbaII/KpnI restriction enzymes, the DNA fragment containing the up- and down- vasK flanking DNA sequences, as well as the Smr/Spr gene cassette, was removed from the above plasmid and ligated into the pDMS197 suicide vector (harbors tetracycline resistance [Tcr] and the sacB gene [encodes levansucrase, which is lethal to bacteria when the sacB gene is induced with sucrose]) at the compatible restriction enzyme sites, and the resulting plasmid (pDMS197vasKUDSm/Sp) was transformed into E. coli SM10 (λpir). The recombinant E. coli [pDMS197vasKUDSm/Sp] clone was conjugated with either the WT A. hydrophila SSU-R (rifampin [Rif] resistant) or its Δact mutant to generate ΔvasK single- and Δact/ΔvasK double- knockout mutants of A. hydrophila SSU, respectively. The transconjugants were selected based on their resistance to appropriate antibiotics and sucrose and subjected to further analyses. Briefly, transconjugants were plated onto LB agar plates with Rif (200 μg/ml), Smr/Spr (50 μg/ml) and 15% sucrose. Single colonies that replicated on plates with Sm and Sp antibiotics, but were sensitive to Tc, were verified by Southern blot analysis [56] using the vasK gene probe.
Table 4.
Sequences of the primers used in this study
| Primer sequence | Restriction site | Description |
|---|---|---|
| 5′-GCTCTAGACCGGTGAACCCATCAAGCGCGTCCACT-3′
5′-TCCCCCCGGGCTGGTGGCCAGCAGCAGAGGCAATA-3′ |
XbaI
XmaI |
for cloning vasH gene into pDMS197 |
| 5′-CGGGGTACCGAATTCCCGGGGATCCGGTGATTGAT-3′
5′-CGGGGTACCGAATTCCCGGGGATCCGGTGATTGAT-3′ |
KpnI
KpnI |
Amplification of Sm/Sp cassette |
| 5′-CTGTCTAGACGCCTGCTCACCAGCCTGC-3′
5′-CCTAGATCTCATAAAATGTGATCCAACTCCGACTC-3′ |
XbaI
BglII |
vasK upstream flanking sequence |
| 5′-GTCAGATCTGCCTGAATCCCTCTATTGATCGCG-3′
5′-ATCGGTACCAAGCTGCGTCTGCCACTGC-3′. |
BglII
XbaI |
vasK downstream flanking sequence |
| 5′-CGCGGATCCCCGGTGAACCCATCAAGCGCGTCCAC-3′
5′-CCGGAATTCCTGGTGGCCAGCAGCAGAGGCAATAG-3′ |
BamHI
EcoRI |
for cloning vasH gene into pBR322 |
| 5′-TCATAGATCTAATGCCAACTCCATGTTATTCAG-3′
5′-GGTCCTCGAGTTAGGCCTCGATCGGCGCGCG-3′ |
BglII
XhoI |
for cloning hcp in pET30a |
| 5′-ACGTACGCGTATGCCAACTCCATGTTATATCAGC-3′
5′-GTGAGCTAGCTTAGGCCTCGATCGGCGCGCGCCAGT-3′ |
MluI
NheI |
for cloning hcp in pBI-EGFP |
4.4. Generation of ΔvasH and Δact/ΔvasH knockout mutants
To generate the ΔvasH knockout mutant of A. hydrophila SSU in the WT background strain, first the recombinant plasmid pDMS197vasH was constructed. To clone the vasH gene in the pDMS197 vector, this gene was PCR amplified using gDNA of A. hydrophila SSU-R as a template and a pair of specific primers (vasH-N-XbaI, vasH-C-XmaI) (Table 4). The PCR product and vector were digested with XbaI/XmaI enzymes, ligated and electroporated into the E. coli SM10 (λpir) strain. To clone the Sm/Spr gene cassette into the vasH gene of pDMS197vasH plasmid, the cassette was PCR amplified using the pHP45Ω plasmid and N-KpnI and C-KpnI primers (Table 4). Then E. coli SM10 strain containing the final recombinant plasmid pDMS197vasHSm/Sp was conjugated with A. hydrophila WT and its Δact mutant strain (SSU-R). Transconjugants were plated as described above, and the correct identity of the clone verified by Southern blot analysis using the vasH gene probe.
4.5. Complementation of A. hydrophila SSU ΔvasH and ΔvasK knockout mutants
To complement the vasH gene, the latter was PCR amplified using gDNA of A. hydrophila as a template and proper primers (vasH-N/BamHI and vasH-C/EcoRI) (Table 4). This DNA fragment (1.5 kb) was cloned in the pBR322 vector (Tcr and Apr) at BamHI-EcoRI sites and transformed into the E. coli TOP10 strain. The pBR322-vasH (Tcs and Apr) recombinant plasmid was isolated from the E. coli strain and electroporated in the A. hydrophila ΔvasH mutant. To complement the vasK gene in the A. hydrophila mutant, pCR2.1-vasK plasmid was digested with EcoRI restriction enzyme and the gene (3.5-kb fragment) was excised from gel, ligated with pBR322 vector and transformed into E. coli TOP10 cells. The pBR322-vasK recombinant plasmid was isolated from the E. coli strain and electroporated into the A. hydrophila mutant strain. We then generated as a control the A. hydrophila WT strain containing the pBR322 vector alone. To test the presence of the pBR322-vasH and pBR322-vasK recombinant plasmids and of pBR322 vector in A. hydrophila, the plasmid DNA was isolated from all of the above-mentioned strains and digested with appropriate restriction endonucleases.
4.6. Electroporation
Electroporation of E. coli as well as of the HeLa Tet-Off cells with various plasmids was performed as we recently described [29]. Well-isolated bacterial colonies were selected and grown in LB medium for the plasmid DNA isolation using a QIAprep® Miniprep Kit (Qiagen, Inc., Valencia, CA). The DNA was subjected to restriction enzyme analysis and DNA sequencing at the Biomolecular Resource Facility at UTMB to confirm target gene sequences. HeLa cells after electroporation were recovered in complete medium (DMEM/10% FBS/G-418), plated, and grown under standard tissue culture conditions [29].
4.7. Two-dimensional gel electrophoresis
Supernatants from overnight cultures (10 ml grown in LB medium at 37°C/8 hr with shaking [150 rpm]) of A. hydrophila WT were prepared after centrifugation and filtering of the culture filtrates through 0.22-μm filters. These supernatants were concentrated by TCA precipitation (final concentration 10%). Two-dimensional gel electrophoresis was performed using an Ettan IPGphor isoelectric focusing system (GE Healthcare, Piscataway, NJ), per the manufacturer’s instructions and as previously described in detail [57, 58]. After electrophoresis, the gels were washed (10% methanol and 7% acetic acid) for 30 min and stained with SYPRO Ruby protein stain (BioRad, Hercules, CA). The images were taken with a Fluochem 8800 (Alpha Innotech, San Leandro, CA), and the data were analyzed using a Progenesis Work station (Nonlinear Dynamics, Durham, NC) at the Protein Chemistry Core Laboratory, UTMB. Based on the molecular mass and isoelectric point (pI) of the proteins, several spots in the vinicity of 15–25 kDa and a pI of 4.9–5.4, which corresponded to the size (19 kDa) and pI (5.24) of Hcp, were robotically excised from the stained gels, subjected to trypsin digestion, and the tryptic fragments were then analyzed by matrix-assisted laser desorption ionization/time of flight (MALDI-TOF) mass spectrometry using 4800 MALDI-TOF/TOF analyzer (Applied Biosystems, Foster City, CA). Importantly, Hcp was produced in relatively higher amounts than the other spots that were excised from the gel.
4.8. Recombinant Hcp (rHcp) purification
Purification of the Hcp was achieved using the Probond™ purification system (Invitrogen-Gibco) as previously described [29]. The proteins from the nickel column were eluted with 250 mM imidazole and subjected to sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE) to verify the identity of hyper-produced Hcp protein by performing Coomassie blue staining of the gels. Fractions containing the protein were mixed and dialyzed against phosphate-buffered saline (PBS) overnight at 4ºC. The protein concentration was measured by a Bradford Assay (BioRad) and the samples stored at −20ºC.
4.9. Antibody production
Swiss Webster mice (Taconic Farms, Germantown, NY) were immunized i.p. with 10 μg of rHcp protein mixed with a synthetic adjuvant as previously described [21]. Sera were obtained from mice and the antibody titers determined by an enzyme-linked immunosorbent assay (ELISA) using rHcp protein as the source of antigen [59]. Dilutions of the sera that were in the middle of the logarithmic portion (~1:1000) of the ELISA titration curves were tested by Western blot analysis for their reactivity to the antigen.
4.10. Western blot analysis
For assessment of antibody reactivity in sera, rHcp protein (2 μg/lane) was subjected to SDS-PAGE and transferred to Hybond™-ECL™ nitrocellulose membranes (GE Healthcare) following the standard Western blot procedure [59]. Membranes were cut into strips corresponding to the lanes of the gel, blocked with 1% bovine serum albumin [BSA]/5% skim milk, and were subsequently incubated with anti-Hcp sera, taken from individual mice (1:1000) and diluted in Tris-buffered saline (TBS), pH 7.6, and 0.5% skim milk for 1 hr with constant shaking at room temperature. The strips were then incubated for 1 hr with secondary antibody (Goat α-mouse IgG [diluted 1:10000] conjugated with horse-radish peroxidase ([HRP] [Southern Biotechnology Associates, Inc., Birmingham, AL]). Five washes of strips were performed between various steps using TBS/0.05% Tween 20 for 10 min each. The blots were developed with Super Signal® West Pico Chemiluminescent substrate (Pierce, Rockford, IL) followed by X-ray film exposure.
Likewise, HeLa cells were lysed in SDS-Tris-Glycine buffer [59] after 24 hr of transfection and subjected to electrophoresis and Western blot analysis as described above.
4.11. Intracellular staining
HeLa Tet-Off cells after 24 hr of transfection were permeabilized using CytoFix™/CytoPerm™ (Becton Dickinson, San Diego, CA). The α-Hcp antibody containing the hyper-immune serum (diluted 1:100), as well as the pre-immune serum (diluted 1:100), was used as the source of primary antibodies. Hcp in HeLa cells was then visualized using phycoerythrin (PE)-conjugated α-mouse IgG antibody (Santa Cruz, Santa Cruz, CA) as previously described [29]. The samples were acquired in a FACScan™ (Becton Dickinson) and analyzed using CellQuest™ (Becton Dickinson) software and WinMDI©.
4.12. Translocation assays
For these assays, we followed the methodology reported by Sha et al (2007) with minor modifications [21]. Briefly, bacterial strains grown to log phase were washed and re-suspended in PBS, and their turbidity measured at OD600 nm. Human colonic epithelial cells, HT-29, were grown in 6 well plates to ~80% confluence in DMEM medium supplemented with 10% FBS before infection with bacteria, and infection was performed at an MOI of 5 in DMEM/0.5% FBS medium. The bacteria and host cells were co-cultured for 2 hr at 37ºC in 5% of CO2. Subsequently, four fractions were collected: i) supernatant fraction, ii) cytoplasmic fraction, iii) eukaryotic host membrane fraction, and iv) whole bacterial lysates. The supernatant fraction was collected by removing the medium and centrifuging it at 1000 x g for 10 min. The supernatants were separated from the pellet and filtered through a 0.22-μm membrane filter. Proteins present in the supernatant fraction were precipitated with trichloroacetic acid (TCA) (10% final concentration) and pelleted by high- speed centrifugation at 14000 x g for 15 min at 4ºC. The pellet was re-suspended in 1X Lammeli loading buffer [59].
The cytoplasmic fraction was collected following lysis of the host cells with 500 μl of sterile water. The cells were disrupted by gentle pipetting and then centrifuged at 6000 x g for 10 min at 4ºC. The eukaryotic membrane fraction was obtained by extraction of the proteins from the above pellet with 500 μl of the cell lysis buffer (200 mM NaCl, 5 mM EDTA, 10% glycerol, 1 mM PMSF, 0.1% Triton X-100, 10 mM tris-HCl pH 7.0). Only eukaryotic cell membranes were affected by this method, while the bacterial membrane integrity was maintained. After centrifugation, the remaining pellet was re-suspended directly in 1X loading buffer. This fraction was considered as the whole bacterial lysate fraction. Samples as obtained above were separated by 4–20% gradient SDS-PAGE, and then proteins were electro-transferred to nitrocellulose membranes for performing Western blot analysis as described earlier. As controls of contamination between fractions, antibodies to actin (eukaryotic cytoplasmic protein), calnexin (eukaryotic membrane protein), and DnaK (bacterial cytosolic protein) were run in parallel for Western blot analysis. Although less likely, the cytoplasmic fraction could contain bacterial secreted proteins that are present in endocytic vesicles.
4.13. Hcp binding to RAW 264.7 macrophages
To perform this experiment, supernatants of A. hydrophila SSU Δact and double knockout mutant ΔactΔvasH were used to avoid the cytotoxic effects mediated by Act. Briefly, bacteria were grown overnight in the LB medium containing appropriate antibiotics. The next day, the cells were washed 3X with PBS, quantified by optical density measurements at 600nm and grown in DMEM supplemented with 1% FBS at a concentration of 5 × 106 cfu/ml. After 2 hr, the medium was centrifuged and filtered through a 0.22-μm filter. Subsequently, 2 × 106 RAW 264.7 cells/well were grown in 6-well plates. After the cells were attached, the medium was exchanged with 2 ml of conditioned medium used to grow bacteria (see above) and incubated for 2 hr at 37ºC. Then, the host cells were washed 3X with PBS and lysed with 500 μl of water. The supernatant was collected as a cytoplasmic fraction, and the pellet as a membrane fraction. These samples were subjected to SDS-4–20%-PAGE, transferred to nitrocellulose membranes and tested by Western blot analysis for the presence of Hcp using specific antibodies. Similar assays using rHcp were preformed. For these assays, DMEM supplemented with 1% FBS containing rHcp (5 μg/ml) was used instead of bacteria-grown conditioned DMEM.
4.14. Phagocytosis
RAW 264.7 murine macrophages were plated and grown at a density of 1 × 105 cell/well in a 96-well plate in DMEM with 10% FBS under normal tissue culture conditions for 2 hr. Subsequently, 5 × 105 bacteria that were previously washed and re-suspended in PBS were added. The plate was centrifuged at 300 x g for 5 min to facilitate contact between macrophages and bacteria and then incubated for 30 min at 37ºC. Next, gentamicin was added at a final concentration of 100 μg/ml for 1 hr to kill extracellular bacteria. Subsequently, the cells were washed twice with PBS, and lysated in 200 μl of water. Different dilutions were plated on LB agar plates and incubated overnight at 37ºC. The colony formation units (cfu) were calculated based on the number of bacteria used for infection by determining the number of colonies inside the macrophages multiplied by the dilution factor.
4.15. Host cell apoptosis
Apoptosis of host cells expressing the hcp gene was assessed after 24 and 48 hr post-transfection by detection of cytoplasmic nucleosomes and measurement of caspase 3 activities as previously described [29]. The color reaction was measured in a microplate reader at 405 nm.
4.16. Host cell viability
To determine host cell viability, we performed the incorporation of 7-amino actinomycin D (7-AAD) and the colorimetric MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrasodium bromide] assay for cell survival as we previously described [29]. For 7-AAD assays, the percentage of positive cells was determined by flow cytometry using CellQuest™ software. For the MTT assay, the color reaction was measured with a microplate reader at 570 nm [29].
4.17. LDH release assay
RAW 264.7 murine macrophages and HeLa cells were infected at an MOI of 0.5 with WT A. hydrophila, and Δact, Δact/ΔvasH and Δact/ΔvasK mutants. During infection, cell morphology was monitored, and at various time points after infection, host cell cytotoxicity was measured by the release of lactate dehydrogenase (LDH) enzyme using CytoTox 96® kit (Promega, Madison, WI) in the tissue culture supernatant [30]. The percentage of cytotoxicity was calculated as recommended by the manufacturer using the following formula: [(OD490 Sample − OD490 Spontaneous)/(OD490 Maximum Release − OD490 Spontaneous)] × 100. OD490 spontaneous indicated LDH release from uninfected cells into the culture supernatant and maximum release denoted LDH release obtained by lysis of the uninfected cells. Three independent experiments were performed in duplicate wells.
4.18. Detection of specific Hcp antibodies from sera of mice infected with WT A. hydrophila SSU
A group of 10 Swiss Webster mice were infected i.p. with WT A. hydrophila at a dose of approximately 1 LD50. After 2 weeks of infection, sera from the surviving mice were pooled and used as the source of primary antibodies in the Western blot analysis with purified rHcp as an antigen [21]. As a negative control, pre-immune sera were used.
4.19. Animal experiments
Groups of 10 Swiss Webster mice were infected i.p. with WT A. hydrophila, ΔvasH, and ΔvasK mutants as well as their complemented strains in accordance with an approved IACUC protocol. We also used WT A. hydrophila with pBR322 vector alone as a control in these experiments. Deaths were recorded for 16 days post-infection. The bacterial doses used represented approximately 2 LD50s of WT A. hydrophila [60]. In another experiment, animals were immunized with purified rHcp (as described for the antibody production experiment) and then challenged with the WT A. hydrophila at a dose of 3 LD50s by the i.p. route after 1 month of immunization. Control animals included those that were given the adjuvant alone (without the antigen) and then infected with the WT bacterium. Deaths were recorded for 16 days post-infection. Fisher’s exact test was used for statistical analysis, and two independent experiments were performed.
4.20. Statistical analysis
Two-way ANOVA and Bonferroni posttests were used for statistical analysis of the data using GraphPad Prism® version 4.02 for windows™ (Software MacKiev, San Diego, CA). The animal mortality data were analyzed by the Fisher exact test.
Acknowledgments
The research was supported by grants from the NIH/NIAID (AI041611) and the Environmental Protection Agency. The NSF grant (NSF EF-0334247) was the conduit for getting the ATCC 7966 A. hydrophila genome sequenced. We thank the Protein Chemistry Core Laboratory at UTMB for providing their expertise in planning studies related to two-dimensional gel electrophoresis. We thank Ms. Mardelle Susman for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martin-Carnahan AM, Joseph SW. The Proteobacteria, Part B the Gammaproteobacteria. Vol. 2. New York, NY: Springer-Verlag; 2005. Aeromonadaceae. [Google Scholar]
- 2.Kay WW, Trust TJ. Form and functions of the regular surface array (S-layer) of Aeromonas salmonicida. Experientia. 1991;47:412–4. [PubMed] [Google Scholar]
- 3.Merino S, Rubires X, Aguilar A, Alberti S, Hernandez-Alles S, Benedi VJ, et al. Mesophilic Aeromonas sp. serogroup O:11 resistance to complement-mediated killing. Infect Immun. 1996;64:5302–9. doi: 10.1128/iai.64.12.5302-5309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anguita J, Rodriguez Aparicio LB, Naharro G. Purification, gene cloning, amino acid sequence analysis, and expression of an extracellular lipase from an Aeromonas hydrophila human isolate. Appl Environ Microbiol. 1993;59:2411–7. doi: 10.1128/aem.59.8.2411-2417.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gobius KS, Pemberton JM. Molecular cloning, characterization, and nucleotide sequence of an extracellular amylase gene from Aeromonas hydrophila. J Bacteriol. 1988;170:1325–32. doi: 10.1128/jb.170.3.1325-1332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merino S, Aguilar A, Nogueras MM, Regue M, Swift S, Tomas JM. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect Immun. 1999;67:4008–13. doi: 10.1128/iai.67.8.4008-4013.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivero O, Anguita J, Paniagua C, Naharro G. Molecular cloning and characterization of an extracellular protease gene from Aeromonas hydrophila. J Bacteriol. 1990;172:3905–8. doi: 10.1128/jb.172.7.3905-3908.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galindo CL, Sha J, Fadl AA, Pillai L, Chopra AK. Host immune responses to Aeromonas virulence factors. Current Immunology Reviews. 2006;2:13–26. [Google Scholar]
- 9.Chopra AK, Houston CW. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1999;1:1129–37. doi: 10.1016/s1286-4579(99)00202-6. [DOI] [PubMed] [Google Scholar]
- 10.Merino S, Rubires X, Knochel S, Tomas JM. Emerging pathogens: Aeromonas spp. Int J Food Microbiol. 1995;28:157–68. doi: 10.1016/0168-1605(95)00054-2. [DOI] [PubMed] [Google Scholar]
- 11.Trower CJ, Abo S, Majeed KN, von Itzstein M. Production of an enterotoxin by a gastro-enteritis-associated Aeromonas strain. J Med Microbiol. 2000;49:121–6. doi: 10.1099/0022-1317-49-2-121. [DOI] [PubMed] [Google Scholar]
- 12.Clark NM, Chenoweth CE. Aeromonas infection of the hepatobiliary system: report of 15 cases and review of the literature. Clin Infect Dis. 2003;37:506–13. doi: 10.1086/376629. [DOI] [PubMed] [Google Scholar]
- 13.von Graevenitz A. The role of Aeromonas in diarrhea: a review. Infection. 2007;35:59–64. doi: 10.1007/s15010-007-6243-4. [DOI] [PubMed] [Google Scholar]
- 14.Hiransuthikul N, Tantisiriwat W, Lertutsahakul K, Vibhagool A, Boonma P. Skin and soft-tissue infections among tsunami survivors in southern Thailand. Clin Infect Dis. 2005;41:e93–6. doi: 10.1086/497372. [DOI] [PubMed] [Google Scholar]
- 15.Presley SM, Rainwater TR, Austin GP, Platt SG, Zak JC, Cobb GP, et al. Assessment of pathogens and toxicants in New Orleans, LA following Hurricane Katrina. Environ Sci Technol. 2006;40:468–74. doi: 10.1021/es052219p. [DOI] [PubMed] [Google Scholar]
- 16.Edberg SC, Browne FA, Allen MJ. Issues for microbial regulation: Aeromonas as a model. Crit Rev Microbiol. 2007;33:89–100. doi: 10.1080/10408410601172180. [DOI] [PubMed] [Google Scholar]
- 17.Farraye FA, Peppercorn MA, Ciano PS, Kavesh WN. Segmental colitis associated with Aeromonas hydrophila. Am J Gastroenterol. 1989;84:436–8. [PubMed] [Google Scholar]
- 18.George WL, Nakata MM, Thompson J, White ML. Aeromonas-related diarrhea in adults. Arch Intern Med. 1985;145:2207–11. [PubMed] [Google Scholar]
- 19.Palu AP, Gomes LM, Miguel MA, Balassiano IT, Queiroz ML, Freitas-Almeida AC, et al. Antimicrobial resistance in food and clinical Aeromonas isolates. Food Microbiol. 2006;23:504–9. doi: 10.1016/j.fm.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Chauret C, Volk C, Creason R, Jarosh J, Robinson J, Warnes C. Detection of Aeromonas hydrophila in a drinking-water distribution system: a field and pilot study. Can J Microbiol. 2001;47:782–6. [PubMed] [Google Scholar]
- 21.Sha J, Wang SF, Suarez G, Sierra JC, Fadl AA, Erova TE, et al. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila-Part I. Microb Pathog. 2007;43:127–46. doi: 10.1016/j.micpath.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Chopra AK, Xu X, Ribardo D, Gonzalez M, Kuhl K, Peterson JW, et al. The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect Immun. 2000;68:2808–18. doi: 10.1128/iai.68.5.2808-2818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galindo CL, Sha J, Ribardo DA, Fadl AA, Pillai L, Chopra AK. Identification of Aeromonas hydrophila cytotoxic enterotoxin-induced genes in macrophages using microarrays. J Biol Chem. 2003;278:40198–212. doi: 10.1074/jbc.M305788200. [DOI] [PubMed] [Google Scholar]
- 24.Wong CY, Heuzenroeder MW, Flower RL. Inactivation of two haemolytic toxin genes in Aeromonas hydrophila attenuates virulence in a suckling mouse model. Microbiology. 1998;144(Pt 2):291–8. doi: 10.1099/00221287-144-2-291. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson MR, Xu XJ, Houston CW, Peterson JW, Coppenhaver DH, Popov VL, et al. Hyperproduction, purification, and mechanism of action of the cytotoxic enterotoxin produced by Aeromonas hydrophila. Infect Immun. 1997;65:4299–308. doi: 10.1128/iai.65.10.4299-4308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribardo DA, Kuhl KR, Boldogh I, Peterson JW, Houston CW, Chopra AK. Early cell signaling by the cytotoxic enterotoxin of Aeromonas hydrophila in macrophages. Microb Pathog. 2002;32:149–63. doi: 10.1006/mpat.2001.0490. [DOI] [PubMed] [Google Scholar]
- 27.Chopra AK, Pham R, Houston CW. Cloning and expression of putative cytotonic enterotoxin-encoding genes from Aeromonas hydrophila. Gene. 1994;139:87–91. doi: 10.1016/0378-1119(94)90528-2. [DOI] [PubMed] [Google Scholar]
- 28.Galindo CL, Fadl AA, Sha J, Gutierrez C, Jr, Popov VL, Boldogh I, et al. Aeromonas hydrophila cytotoxic enterotoxin activates mitogen-activated protein kinases and induces apoptosis in murine macrophages and human intestinal epithelial cells. J Biol Chem. 2004;279:37597–612. doi: 10.1074/jbc.M404641200. [DOI] [PubMed] [Google Scholar]
- 29.Sierra JC, Suarez G, Sha J, Foltz SM, Popov VL, Galindo CL, et al. Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila-Part II. Microb Pathog. 2007;43:147–60. doi: 10.1016/j.micpath.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Sha J, Pillai L, Fadl AA, Galindo CL, Erova TE, Chopra AK. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect Immun. 2005;73:6446–57. doi: 10.1128/IAI.73.10.6446-6457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desvaux M, Parham NJ, Henderson IR. The autotransporter secretion system. Res Microbiol. 2004;155:53–60. doi: 10.1016/j.resmic.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–33. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das S, Chaudhuri K. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 2003;3:287–300. [PubMed] [Google Scholar]
- 34.Williams SG, Varcoe LT, Attridge SR, Manning PA. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect Immun. 1996;64:283–9. doi: 10.1128/iai.64.1.283-289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–30. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Bruin OM, Ludu JS, Nano FE. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 2007;7:1. doi: 10.1186/1471-2180-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons DA, Heffron F. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect Immun. 2005;73:4338–45. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–85. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 39.Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, et al. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol. 2006;188:8272–82. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–82. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 41.Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–65. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 42.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007 doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 43.Mintz KP, Fives-Taylor PM. impA, a gene coding for an inner membrane protein, influences colonial morphology of Actinobacillus actinomycetemcomitans. Infect Immun. 2000;68:6580–6. doi: 10.1128/iai.68.12.6580-6586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young BM, Young GM. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J Bacteriol. 2002;184:1324–34. doi: 10.1128/JB.184.5.1324-1334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young GM, Schmiel DH, Miller VL. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci U S A. 1999;96:6456–61. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canals R, Altarriba M, Vilches S, Horsburgh G, Shaw JG, Tomas JM, et al. Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J Bacteriol. 2006;188:852–62. doi: 10.1128/JB.188.3.852-862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bladergroen MR, Badelt K, Spaink HP. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- 48.Moore MM, Fernandez DL, Thune RL. Cloning and characterization of Edwardsiella ictaluri proteins expressed and recognized by the channel catfish Ictalurus punctatus immune response during infection. Dis Aquat Organ. 2002;52:93–107. doi: 10.3354/dao052093. [DOI] [PubMed] [Google Scholar]
- 49.Roest HP, Mulders IH, Spaink HP, Wijffelman CA, Lugtenberg BJ. A Rhizobium leguminosarum biovar trifolii locus not localized on the sym plasmid hinders effective nodulation on plants of the pea cross-inoculation group. Mol Plant Microbe Interact. 1997;10:938–41. doi: 10.1094/MPMI.1997.10.7.938. [DOI] [PubMed] [Google Scholar]
- 50.Sheahan KL, Cordero CL, Satchell KJ. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc Natl Acad Sci U S A. 2004;101:9798–803. doi: 10.1073/pnas.0401104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morett E, Bork P. Evolution of new protein function: recombinational enhancer Fis originated by horizontal gene transfer from the transcriptional regulator NtrC. FEBS Lett. 1998;433:108–12. doi: 10.1016/s0014-5793(98)00888-6. [DOI] [PubMed] [Google Scholar]
- 52.Austin S, Dixon R. The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. Embo J. 1992;11:2219–28. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morett E, Segovia L. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–74. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci U S A. 1998;95:1669–74. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinrauch Y, Zychlinsky A. The induction of apoptosis by bacterial pathogens. Annu Rev Microbiol. 1999;53:155–87. doi: 10.1146/annurev.micro.53.1.155. [DOI] [PubMed] [Google Scholar]
- 56.Sha J, Kozlova EV, Chopra AK. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect Immun. 2002;70:1924–35. doi: 10.1128/IAI.70.4.1924-1935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brasier AR, Spratt H, Wu Z, Boldogh I, Zhang Y, Garofalo RP, et al. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected a549 cells identified by high-resolution two-dimensional gel electrophoresis. J Virol. 2004;78:11461–76. doi: 10.1128/JVI.78.21.11461-11476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forbus J, Spratt H, Wiktorowicz J, Wu Z, Boldogh I, Denner L, et al. Functional analysis of the nuclear proteome of human A549 alveolar epithelial cells by HPLC-high resolution 2-D gel electrophoresis. Proteomics. 2006;6:2656–72. doi: 10.1002/pmic.200500652. [DOI] [PubMed] [Google Scholar]
- 59.Coligan JE, Dunn BE, Speicher DW, Wingfield PT. Current protocols in protein science. New York: John Wiley & Sons, Inc.; 2002. [Google Scholar]
- 60.Xu XJ, Ferguson MR, Popov VL, Houston CW, Peterson JW, Chopra AK. Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect Immun. 1998;66:3501–9. doi: 10.1128/iai.66.8.3501-3509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fadl AA, Galindo CL, Sha J, Erova TE, Houston CW, Olano JP, Chopra AK. Deletion of the genes encoding the type III secretion system and cytotoxic enterotoxin alters host responses to Aeromonas hydrophila infection. Microb Patho. 2006;40:198–210. doi: 10.1016/j.micpath.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analize 987P fimbria gene expression. Gene. 1998;207:149–57. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 63.Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–13. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]