Abstract
Plasmodium falciparum resistance to the former first-line antimalarials chloroquine and sulfadoxine/pyrimethamine has reached critically high levels in many malaria-endemic regions. This has spurred the introduction of several new artemisinin-based combination therapies (ACTs) that display excellent potency in treating drug-resistant malaria. Monitoring for the emergence of drug resistant P. falciparum is important for maximizing the clinically effective lifespan of ACTs. Here, we provide a commentary on the article by Kaddouri et al., published in this issue of the International Journal of Parasitology, which documents the levels of susceptibility to ACT drugs and chloroquine in P. falciparum isolates from Mali. These authors report that some isolates approached a proposed in vitro threshold of resistance to monodesethyl-amodiaquine (the principal effective metabolite of amodiaquine, an important ACT partner drug), and establish baseline levels of susceptibility to the ACT drugs dihydroartemisinin and lumefantrine. In contrast, the majority of clinical isolates manifested in vitro resistance to chloroquine. The authors also show good concordance between field-based assays employing a non-radioactive lactate dehydrogenase-based method of determining in vitro drug IC50 values and the optimal [3H]-hypoxanthine-based radioactive method. This work illustrates a good example of drug resistance surveillance, whose global coordination is being championed by the World Antimalarial Resistance Network. Our commentary also more generally discusses the complexities inherent to conducting in vitro investigations with P. falciparum patient isolates and correlating these findings with treatment outcome data.
Keywords: Plasmodium falciparum malaria, In vitro assays, Drug resistance, Chloroquine, Artemisinin-based combination therapies
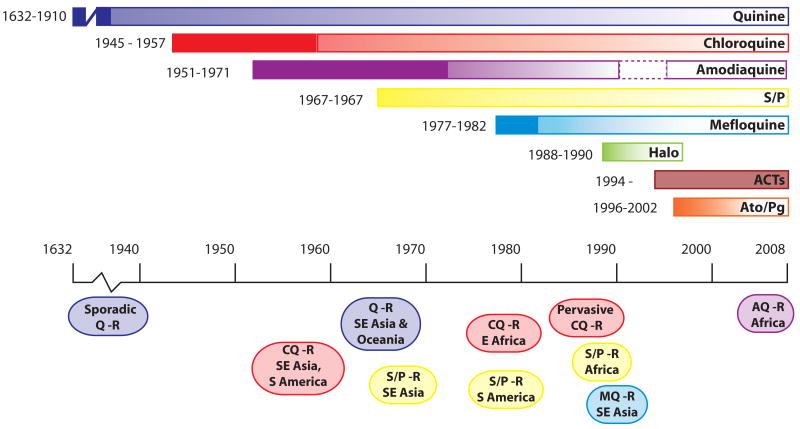
Malaria inflicts a heavy toll on the health and quality of life of the residents of intertropical countries, affecting one third of the world's human population (Sachs and Malaney, 2002; Guerra et al., 2008). Communities in Africa bear the brunt of this parasitic, blood-borne disease, with an estimated 90% of all malarial deaths occurring in African children below the age of 5 years (Hay et al., 2005). Efforts to control malaria have repeatedly faltered, in part as a result of the selection and spread of drug resistant Plasmodium falciparum parasites. These parasites have developed at least partial resistance to nearly every antimalarial regimen introduced to date, including the former first-line drugs chloroquine and sulfadoxine-pyrimethamine (Fig. 1) (Wongsrichanalai et al., 2002). In 2005, the World Health Organization (WHO) began recommending the use of artemisinin-based combination therapies (ACTs) as the preferred first-line antimalarials. These pair an extremely potent but short-lived artemisinin derivative (typically artesunate or artemether) with a longer-acting antimalarial such as lumefantrine, amodiaquine or mefloquine. Substantial financial support is being put into place to facilitate their distribution throughout endemic areas (Ashley and White, 2005; Bathurst and Hentschel, 2006; Grabowsky, 2008). While ACTs currently demonstrate excellent clinical efficacy, the history of antimalarial chemotherapy predicts that it is only a matter of time before parasite resistance emerges (Chretien et al., 2007).
Fig. 1.
Emergence of resistance to the principal antimalarials. Each colored bar represents an antimalarial monotherapy or combination. Years to the left of each bar represent the date the drug was introduced and the first reported instance of resistance. Ovals below the time line denote the approximate periods when resistance spread though different geographical regions. Quinine, chlororoquine and sulfadoxine/pyrimethamine remained effective for considerable periods after the first reported instances of resistance. Halofantrine has had limited usage since about 1998 due to cardiotoxicity concerns. Amodiaquine was removed from the list of approved antimalarials in 1990 due to concerns over serious side effects, but was reinstated in 1996 because the perceived benefits outweighed the risks (dashed border). Chemically pure artemisinin (Qinghaosu) and its derivatives were first used in field trials in China in the early 1970s. Artemisinin has a low radical cure rate when used alone in a short course, presumably due to its very short half-life in vivo. Since 1994, artemisinin and its derivatives have been used in combination therapies (ACTs). No clinical resistance to ACTs has yet been demonstrated. Timelines were derived from (Wongsrichanalai et al., 2002; Hyde, 2005; Tinto et al., 2008) and references therein. ACTs: Artemisinin-based Combination Therapies, AQ, amodiaquine; Ato/Pg: Atovaquone/Proguanil, CQ: Chloroquine, Halo: Halofantrine, MQ, mefloquine; Q: Quinine, R: Resistance, S/P: Sulfadoxine/Pyrimethamine.
The paper by Kaddouri et al. (2008) in this issue of the International Journal for Parasitology uses an in vitro approach to assay clinical isolates of P. falciparum for their degrees of susceptibility to chloroquine and several ACT component drugs (lumefantrine, dihydroartemisinin and the active monodesethyl metabolite of amodiaquine). This study provides baseline efficacy data that are necessary for subsequent detection of the emergence of resistance to ACT drugs. Reflecting back on chloroquine, it is striking that four decades elapsed between the early reports of emerging clinical resistance and the identification of the primary genetic determinant, pfcrt (Fidock et al., 2000; Wellems and Plowe, 2001). Only then could a simple PCR-based molecular assay be designed to determine the prevalence of chloroquine-resistant strains of P. falciparum, revealing their origins and widespread dissemination throughout nearly all malaria-endemic regions (Djimdé et al., 2001; Wootton et al., 2002). Advances in recent years have now equipped us with powerful genetic and genomic tools (that include allelic exchange and transgene expression, quantitative trait loci analysis, haplotype mapping, microarrays and whole genome sequencing (Ekland and Fidock, 2007)), a better understanding of the molecular mechanisms of antimalarial drug resistance (Woodrow and Krishna, 2006), and the capacity to exchange and analyze data as never before. Translating this into more effective management and control of drug-resistant malaria presents an incredible scientific challenge. Meeting this challenge will be further complicated by issues such as the distribution of ineffective or counterfeit drugs, regional differences in implementation programs, and the limited funds available for basic malaria research. In this article we will discuss some of the issues confronting researchers investigating antimalarial drug resistance in vitro with patient isolates and how the paper by Kaddouri et al. (2008) fits into this context.
A high rate of clinical failure defines the ultimate measure of resistance, however, there are many challenges associated with clinical studies of drug resistance, including costs and logistical difficulties. The ability to detect drug resistant parasites can be obscured by multiple factors including poor patient compliance, patient variability in drug absorption or metabolism, the general health and nutritional state of the patients, the rate at which re-infections are acquired, the genetic complexity of the resistance phenotype, and the degree of pre-existing immunity to plasmodial parasites. As an example of the apparent impact of immunity, a study conducted in Mali in 2001 estimated the prevalence of the chloroquine resistant PfCRT K76T mutation at 41% of the sampled pre-treatment parasite population, yet clinical treatment failure was observed in only 14% of the patients (Djimdé et al., 2001; all of these treatment failures harbored P. falciparum infections carrying the PfCRT K76T mutation). Age stratification of treatment outcomes revealed striking differences. Comparing children < 10 with those ≥ 10 years of age, in vivo failure rates with parasites bearing the mutant 76T allele were 68% and 35%, respectively (Djimdé et al., 2001). The authors attributed these differences to the acquisition of immunity to malaria in the older patients. These types of confounding factors can make it challenging for clinical efficacy studies to detect the emergence of drug resistant parasites early on, when these are present at low levels in the population.
Studies measuring the presence of genetic markers of drug resistance provide an attractive method for monitoring the prevalence and spread of resistant parasites, as they are relatively fast, quantitative and inexpensive compared with clinical studies involving more patient care and follow-up. In a recent application, researchers documented that the PfCRT K76T marker for chloroquine resistance disappeared in the years after Malawi enforced a change of first line antimalarial treatment from chloroquine to sulfadoxine/pyrimethamine (Kublin et al., 2003; Mita et al., 2003). A subsequent clinical study concurred that chloroquine has since regained its efficacy in Malawi (Laufer et al., 2006). Unfortunately, these molecular methods can only be used to track established resistance markers. Several groups are striving to identify genes that could potentially mediate artemisinin resistance using heterologous systems or rodent malaria models (Uhlemann et al., 2005; Afonso et al., 2006; Krishna et al., 2006; Hunt et al., 2007). However, in the absence of clear clinical failures it is still too early to identify which genetic markers will accurately predict the failure of ACT therapies.
Culturing clinical isolates and measuring their susceptibility to antimalarial compounds in vitro removes variables such as patient compliance, nutritional status, immune status, re-infection and pharmacokinetics, thereby providing a powerful technique for detecting the emergence of drug resistant parasites. In vitro studies have the additional benefit that cultured isolates can potentially be frozen down for use in future genotyping or drug susceptibility assays. Notably, small shifts in 50% inhibitory concentration (IC50) values can be sufficient to affect clinical outcomes for many of the current antimalarials. As an example, Price and Uhlemann (2004) observed that Thai patients harboring P. falciparum infections with an increased copy number of pfmdr1 had a five-fold increased risk of mefloquine treatment failure by day 28, with a three-fold increase in mefloquine IC50 values observed in vitro, compared with infections carrying single copy pfmdr1 alleles. Subtler shifts in IC50 values might indicate genetic changes that confer a degree of parasite tolerance to drug and might on occasion foretell the appearance of bone fide clinical resistance.
It is also important to note some of the caveats of in vitro assays. Not all of the primary parasites will adapt to in vitro culture conditions and hence these conditions may themselves impose certain biases in terms of the parasite genotypes that will propagate and yield IC50 data. Other factors that can affect in vitro assays and IC50 measurements include the presence of mixed genotypes in patient isolates (especially in high transmission settings in Africa), sample handling, the assay chosen for quantifying growth and even the particular growth conditions used, such as the gas mixture conditions, culture duration and culture medium (Bacon et al., 2007). As an example of the latter, variable concentrations of folate or para-aminobenzoic acid in the medium can substantially influence antifolate IC50 values recorded in vitro (Wang et al., 1997). Finally, while an in vitro assay may detect a shift in IC50 values, there are no clear criteria for establishing IC50 thresholds of resistance in the absence of clinical failure (Ringwald and Basco, 1999). Even for drugs that encounter clinical failure, identifying in vitro IC50 values that can predict in vivo resistance has proven exceptionally difficult. One relevant example is chloroquine, for which the IC50 value of 100 nM has been commonly cited as a threshold of in vitro resistance (Ringwald and Basco, 1999; Reed et al., 2000; Legrand et al., 2008). This value was selected following comparisons of chloroquine IC50 values of 11 geographically distinct culture-adapted isolates, without in vivo correlates (Cremer et al., 1995). In a separate study, Brasseur and colleagues set the in vitro threshold at 80 nM, based on in vitro assays of isolates from patients with known clinical outcomes (Brasseur et al., 1992).
Establishing a clinically relevant IC50 value, which represents a threshold above which the risk of clinical treatment failure rises substantially, would ideally require detailed pharmacokinetic/pharmacodynamic studies in humans and comparison of these data with ex vivo values obtained with patient isolates, as well as a sufficient number of treatment failures to compare against cases of successful treatment outcomes. The scarcity of such studies reflects the difficulty in approaching this question. Some of the confounding parameters include differences in patient immune status and drug metabolism, initial biomass of the infection (i.e. parasitemia), sampling limitations, mixed infections and difficulties in defining clinical cure rates (Price et al., 2007). Another complication pertains to sampling bias. Generating in vitro drug susceptibility data is dependent on certain prerequisites including venipuncture, which biases selection towards adults, and high parasitemias. These restrictions potentially bias the representation of parasites in the in vitro analysis compared with the complete in vivo data set.
One example of a study that has surmounted these difficulties was recently published by Price et al. (2006), who identified a lumefantrine plasma concentration of 175 ng/ml, measured 7 days after initiation of treatment, as a threshold for increased risk of parasite recrudescence. This pharmacokinetic parameter showed good sensitivity and specificity (75% and 84%, respectively) when predicting treatment failure. Interestingly, an increase in pfmdr1 copy number, which was also associated with an increased risk of treatment failure, only increased the mean IC50 value from an average of ∼17 ng/ml in parasites harboring one copy of pfmdr1 to ∼34 ng/ml in parasites with two or more pfmdr1 copies (values that are noticeably lower than the lumefantrine plasma levels achieved at the day seven time point following initiation of treatment). One conclusion is that in vitro thresholds should not be seen as absolute values or predictors of a particular treatment outcome, but rather as indicators that a parasite laboratory line or field isolate has decreased its degree of in vitro susceptibility, which in turn is likely to increase the likelihood of clinical resistance or recrudescence.
In an effort to coordinate global monitoring efforts, a consortium of researchers has established the World Antimalarial Resistance Network (WARN). The goals of WARN include establishing a centralized databank, defining global baseline sensitivities of parasites to antimalarials, and standardizing methods in order to make data conducted by different groups more comparable (Sibley et al., 2008). The study by Kaddouri et al. (2008) is commendable in its treatment of these issues. In measuring the in vitro susceptibility of primary P. falciparum isolates to ACT partner drugs, the authors established an important baseline reference point for future studies looking for the emergence of resistance in Mali. Their analysis of 96 Malian isolates (51 tested in Mali and 45 collected from French travelers returning from Mali to Paris) revealed no evidence of in vitro resistance to dihydroartemisinin or lumefantrine, using proposed thresholds of 10 nM and 150 nM, respectively. The authors also report that some isolates had monodesethyl-amodiaquine IC50 values that nearly attained their proposed in vitro resistance cut-off of 80 nM. It is important to note however that these proposed thresholds have not been validated in terms of their ability to predict in vitro resistance or in vivo failure. Using an in vitro threshold of 100 nM for chloroquine, the authors observed resistance in 60–69% of isolates, and found good concordance with the presence of the PfCRT K76T mutation. These data also showed a continuum of IC50 values for each drug. Multiple factors could contribute to this – physiological differences between isolates that alter their basal level of susceptibility, single versus mixed infections, differences in rates of growth of non-adapted isolates under in vitro conditions, experimental variability between wells, and the degree of genetic complexity of the resistance phenotype. This variability contributes to the difficulty of establishing in vitro thresholds and calls for caution when interpreting values obtained from single assays performed with non-adapted isolates. Importantly, the authors used two culture adapted strains, 3D7 and W2, obtained from MR4-ATCC (www.mr4.malaria.org) as reference strains. Comparisons to these reference strains should assist other researchers when comparing their own data to these results.
Kaddouri et al. (2008) also compared results from an ELISA-based drug susceptibility assay, based on detection of Plasmodium lactate dehydrogenase (pLDH) and performed by researchers in Mali, with results from the optimal [3H]-hypoxanthine assay performed in Paris. Both sites used the same batches of plates containing predosed, lyophilized concentrations of drug. This minimized the contribution of plates and drugs to experimental variability between the two-site comparisons, while highlighting differences between drugs in their degree of stability post-lyophilization. While the absolute IC50 values differed slightly between sites, the global values showed parallel trends. These results confirmed the utility of the nonradioactive pLDH assay for use in field settings.
Recent years have seen a dramatic change in antimalarial policy worldwide as ACTs become the prevailing first-line therapies. In addition to the most widely used current ACTs artemether-lumefantrine, artesunate-amodiaquine and artesunate-mefloquine, additional combinations are undergoing clinical evaluation including artesunate-pyronaridine and dihydroartemisinin-piperaquine. Their pharmacokinetic mismatching of a short-lived artemisinin with a longer-lived partner drug exposes the latter to a substantial risk of resistance. Thus, it will be critical to identify early signs of resistance before these strains become prevalent and compromise the clinical utility of the ACTs. That such a situation can arise is evidenced by recent reports from the Thai-Cambodian border where increased IC50 values with artemisinin derivatives and delayed times to parasite clearance are being noted (H. Noedl, personal communication). Maximizing the efficacy and longevity of ACTs as a tool to treat and control malaria will critically depend on pursuing intensive research into identifying in vitro markers of resistance as well as implementing the types of in vitro surveillance programs described in the paper by Kaddouri et al (2008).
Acknowledgments
The research of David A. Fidock, Ph.D., is supported in part by the Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund and by the NIH (R01 AI50234). Eric H. Ekland, Ph.D., is a Hoffman-LaRoche Fellow of the Life Sciences Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonso A, Hunt P, Cheesman S, Alves AC, Cunha CV, do Rosario V, Cravo P. Malaria parasites can develop stable resistance to artemisinin but lack mutations in candidate genes atp6 (encoding the sarcoplasmic and endoplasmic reticulum Ca2+ ATPase), tctp, mdr1, and cg10. Antimicrob Agents Chemother. 2006;50:480–489. doi: 10.1128/AAC.50.2.480-489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, White NJ. Artemisinin-based combinations. Curr Opin Infect Dis. 2005;18:531–536. doi: 10.1097/01.qco.0000186848.46417.6c. [DOI] [PubMed] [Google Scholar]
- Bacon DJ, Jambou R, Fandeur T, Le Bras J, Wongsrichanalai C, Fukuda MM, Ringwald P, Sibley CH, Kyle DE. World Antimalarial Resistance Network (WARN) II: in vitro antimalarial drug susceptibility. Malaria J. 2007;6:120. doi: 10.1186/1475-2875-6-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathurst I, Hentschel C. Medicines for Malaria Venture: sustaining antimalarial drug development. Trends Parasitol. 2006;22:301–307. doi: 10.1016/j.pt.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Brasseur P, Kouamouo J, Moyou-Somo R, Druilhe P. Multi-drug resistant falciparum malaria in Cameroon in 1987-1988. I. Stable figures of prevalence of chloroquine- and quinine-resistant isolates in the original foci. Am J Trop Med Hyg. 1992;46:1–7. doi: 10.4269/ajtmh.1992.46.1. [DOI] [PubMed] [Google Scholar]
- Chretien JP, Fukuda M, Noedl H. Improving surveillance for antimalarial drug resistance. J Am Med Assoc. 2007;297:2278–2281. doi: 10.1001/jama.297.20.2278. [DOI] [PubMed] [Google Scholar]
- Cremer G, Basco LK, Le Bras J, Camus D, Slomianny C. Plasmodium falciparum: detection of P-glycoprotein in chloroquine- susceptible and chloroquine-resistant clones and isolates. Exp Parasitol. 1995;81:1–8. doi: 10.1006/expr.1995.1086. [DOI] [PubMed] [Google Scholar]
- Djimdé A, Doumbo MD, Cortese JF, Kayentao K, Doumbo S, Diourté Y, Coulibaly D, Dicko A, Su X-z, Nomura T, Fidock DA, Wellems TE, Plowe CV. A molecular marker for chloroquine resistant falciparum malaria. New Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- Ekland EH, Fidock DA. Advances in understanding the genetic basis of antimalarial drug resistance. Curr Opin Microbiol. 2007;10:363–370. doi: 10.1016/j.mib.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu ABS, Naude B, Deitsch K, Su X-z, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowsky M. The billion-dollar malaria moment. Nature. 2008;451:1051–1052. doi: 10.1038/4511051a. [DOI] [PubMed] [Google Scholar]
- Guerra CA, Gikandi PW, Tatem AJ, Noor AM, Smith DL, Hay SI, Snow RW. The limits and intensity of Plasmodium falciparum transmission: implications for malaria control and elimination worldwide. PLoS Med. 2008;5:e38. doi: 10.1371/journal.pmed.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay SI, Guerra CA, Tatem AJ, Atkinson PM, Snow RW. Urbanization, malaria transmission and disease burden in Africa. Nat Rev Microbiol. 2005;3:81–90. doi: 10.1038/nrmicro1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P, Afonso A, Creasey A, Culleton R, Sidhu AB, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol. 2007;65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JE. Drug-resistant malaria. Trends Parasitol. 2005;21:494–498. doi: 10.1016/j.pt.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddouri H, Djimdé A, Dama S, Kodio A, Tekete M, Hubert V, Koné A, Maiga H, Yattara O, Fofana B, Sidibe B, Sangaré CPO, Doumbo O, Le Bras J. Baseline in vitro efficacy of ACT component drugs on Plasmodium falciparum clinical isolates from Mali. Int J Parasitol. 2008;38 doi: 10.1016/j.ijpara.2007.12.002. in this issue. [DOI] [PubMed] [Google Scholar]
- Krishna S, Woodrow CJ, Staines HM, Haynes RK, Mercereau-Puijalon O. Re-evaluation of how artemisinins work in light of emerging evidence of in vitro resistance. Trends Mol Med. 2006;12:200–205. doi: 10.1016/j.molmed.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimde AA, Kouriba B, Taylor TE, Plowe CV. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- Lakshmanan V, Bray PG, Verdier-Pinard D, Johnson DJ, Horrocks P, Muhle RA, Alakpa GE, Hughes RH, Ward SA, Krogstad DJ, Sidhu AB, Fidock DA. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV. Return of chloroquine antimalarial efficacy in Malawi. New Engl J Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- Legrand E, Volney B, Meynard JB, Mercereau-Puijalon O, Esterre P. In vitro monitoring of Plasmodium falciparum drug resistance in French Guiana: a synopsis of continuous assessment from 1994 to 2005. Antimicrob Agents Chemother. 2008;52:288–298. doi: 10.1128/AAC.00263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T, Kaneko A, Lum JK, Bwijo B, Takechi M, Zungu IL, Tsukahara T, Tanabe K, Kobayakawa T, Bjorkman A. Recovery of chloroquine sensitivity and low prevalence of the Plasmodium falciparum chloroquine resistance transporter gene mutation K76T following the discontinuance of chloroquine use in Malawi. Am J Trop Med Hyg. 2003;68:413–415. [PubMed] [Google Scholar]
- Price RN, Uhlemann A-C, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, van Vugt M, Brockman A, Hutagalung R, Nair S, Nash D, Singhasivanon P, Anderson TJ, Krishna S, White NJ, Nosten F. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis. 2006;42:1570–1577. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Dorsey G, Ashley EA, Barnes KI, Baird JK, d'Alessandro U, Guerin PJ, Laufer MK, Naidoo I, Nosten F, Olliaro P, Plowe CV, Ringwald P, Sibley CH, Stepniewska K, White NJ. World Antimalarial Resistance Network I: clinical efficacy of antimalarial drugs. Malaria J. 2007;6:119. doi: 10.1186/1475-2875-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Ringwald P, Basco LK. Comparison of in vivo and in vitro tests of resistance in patients treated with chloroquine in Yaounde, Cameroon. Bull WHO. 1999;77:34–43. [PMC free article] [PubMed] [Google Scholar]
- Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002;415:680–685. doi: 10.1038/415680a. [DOI] [PubMed] [Google Scholar]
- Sibley CH, Barnes KI, Watkins WM, Plowe CV. A network to monitor antimalarial drug resistance: a plan for moving forward. Trends Parasitol. 2008;24:43–48. doi: 10.1016/j.pt.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Tinto H, Guekoun L, Zongo I, Guiguemde RT, D'Alessandro U, Ouedraogo JB. Chloroquine-resistance molecular markers (Pfcrt T76 and Pfmdr-1 Y86) and amodiaquine resistance in Burkina Faso. Trop Med Int Health. 2008;13:238–240. doi: 10.1111/j.1365-3156.2007.01995.x. [DOI] [PubMed] [Google Scholar]
- Uhlemann AC, Cameron A, Eckstein-Ludwig U, Ho W-Y, Chan WC, Fischbarg J, Iserovich P, Zuniga FA, East M, Lee A, Brady L, Haynes RK, Krishna S. A single amino acid residue can determine sensitivity of SERCAs to artemisinins. Nature Struct Mol Biol. 2005;12:628–629. doi: 10.1038/nsmb947. [DOI] [PubMed] [Google Scholar]
- Wang P, Read M, Sims PF, Hyde JE. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- Woodrow CJ, Krishna S. Antimalarial drugs: recent advances in molecular determinants of resistance and their clinical significance. Cell Mol Life Sci. 2006;63:1586–1596. doi: 10.1007/s00018-006-6071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]