Abstract
Mutations in the amyloid precursor protein (APP) gene cause early-onset familial Alzheimer disease (AD) by affecting the formation of the amyloid β (Aβ) peptide, the major constituent of AD plaques. We expressed human APP751 containing these mutations in the brains of transgenic mice. Two transgenic mouse lines develop pathological features reminiscent of AD. The degree of pathology depends on expression levels and specific mutations. A 2-fold overexpression of human APP with the Swedish double mutation at positions 670/671 combined with the V717I mutation causes Aβ deposition in neocortex and hippocampus of 18-month-old transgenic mice. The deposits are mostly of the diffuse type; however, some congophilic plaques can be detected. In mice with 7-fold overexpression of human APP harboring the Swedish mutation alone, typical plaques appear at 6 months, which increase with age and are Congo Red-positive at first detection. These congophilic plaques are accompanied by neuritic changes and dystrophic cholinergic fibers. Furthermore, inflammatory processes indicated by a massive glial reaction are apparent. Most notably, plaques are immunoreactive for hyperphosphorylated tau, reminiscent of early tau pathology. The immunoreactivity is exclusively found in congophilic senile plaques of both lines. In the higher expressing line, elevated tau phosphorylation can be demonstrated biochemically in 6-month-old animals and increases with age. These mice resemble major features of AD pathology and suggest a central role of Aβ in the pathogenesis of the disease.
The pathological hallmarks of Alzheimer disease (AD) are amyloid plaques and neurofibrillary tangles (1) composed of amyloid β (Aβ), a 39- to 43-amino acid peptide (2, 3) and hyperphosphorylated tau (4, 5). In addition, extensive neuritic degeneration has been described including a dramatic loss of cholinergic fibers (6, 7). The Aβ peptide is thought to play a central role in AD pathogenesis, as suggested by mutations in the gene encoding its precursor, amyloid precursor protein (APP) that are linked to forms of early-onset familial AD (FAD) (8, 9). These mutations either cause an elevated production of total Aβ (10, 11) or increase the more fibrillogenic Aβ-(1–42) (12, 13).
Numerous attempts focusing on transgenic expression of human APP (14–19) have been made to obtain a valid animal model of AD. However, only two transgenic mouse lines have been described that develop Aβ deposits characteristic of AD (20, 21). We have generated various transgenic mice that express human APP751. Two of these lines that differ in transgene-derived APP expression levels and FAD mutations develop plaque-like Aβ deposits in neocortex and hippocampus to different degrees. They also display additional aspects of AD pathology not commonly associated with the Aβ peptide.
MATERIALS AND METHODS
Expression Constructs and Transgenic Mice.
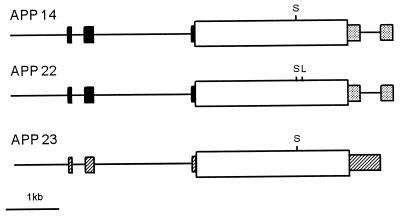
Transgenic constructs used in this study contain human (19) or murine (22) Thy-1 expression cassettes and human APP751 cDNAs starting with an optimized Kozak consensus sequence and extending to nucleotide 3,026 (HindIII site) as described (19). The human APP cDNAs carry either the Swedish double mutation at positions 670/671 (KM->NL) (8) alone or in conjunction with the London mutation (V717I) (9). Construct APP 14 utilizes the human Thy-1 cassette to drive human APP751 Swedish and was described in detail in a previous study (19). The APP 22 expression construct is identical to APP 14 except for the additional V717I mutation in the human APP cDNA. It was generated by replacing a 600-bp BglII–SpeI fragment of human APP751 cDNA with the corresponding fragment carrying the V717I mutation. To generate the construct for APP 23, the APP751 cDNA with the Swedish mutation was inserted into the blunt-ended XhoI site of the expression cassette containing the murine Thy-1.2 gene (22). Vector sequences were removed by NotI and NotI/PvuI digestion for APP 14/APP 22 and APP 23, respectively. Injection and manipulation of mice were identical to described procedures (19).
RNA Quantification.
Transgene mRNA was quantified by reverse transcription-coupled PCR (23). In brief, total RNA from brains of transgenic mouse lines APP 14, 22, and 23 was isolated and transcribed into cDNA. Fragments of APP harboring the Aβ region were amplified by PCR using primers 5′-ACCACCGTGGAGCTCCTTCCCGTGAA-3′ and 5′-GCAACTGCAGTGTGTACTGTTTCTTC-3′, specific for sequences common in human and murine APP. Fragments were directionally subcloned into M13 replicative form DNA, and dual filter lifts from plaque-containing plates were hybridized with oligonucleotides 5′-AATTCTGCATCCATCTTCACTTCCGA-3′ and 5′-AATTCTGCATCCAGATTCACTTCAGA-3′, discriminating mouse wild-type and mutated human APP.
In Situ Hybridization.
In situ hybridization was performed as described (24). A 33P-labeled oligonucleotide probe, 5′-AGCCTCTTCCTCTACCTCATCACCATCCTCATCGTCCTCG-3′, complementary to coding sequence of human APP between nucleotides 859 and 898 was used at a final concentration of 2 pmol/ml. The hybridized slides were exposed to β-max film (Amersham) for 3 days.
Western Blots.
Cerebral cortices were homogenized in 10 vol of 50 mM Tris⋅HCl, pH 8/1% Nonidet P-40/5 mM dithiothreitol/100 mM NaCl/5 mM EGTA/50 mM NaF/1 mM sodium orthovanadate/100 nM ocadaic acid/1× complete protease inhibitor mixture (Boehringer Mannheim) mixed with an equal volume of 2× SDS sample buffer and processed as described (25). Samples were electrophoresed on SDS/polyacrylamide minigels, the proteins were transferred to Immobilon P (Millipore) membranes, and the Western blots were probed with antibodies AT8 (Innogenetics, Gent, Belgium) (26, 27), R27 (28) and tau7, which was raised against tau purified from adult rat brain and recognizes tau from multiple species (29). Peroxidase-conjugated goat anti-mouse IgG (Dianova, Hamburg, F.R.G.) or peroxidase-coupled goat anti-rabbit IgG (Dianova) was used as a second antibody. The immunoblots were developed with the ECL system (Amersham).
Histology.
Heterozygous mice from line APP 22 were analyzed at 12 (n = 3) and 18 months (n = 3). Heterozygous APP 23 mice were sacrificed at 6 (n = 9), 9 (n = 12), 12 (n = 6), 15 (n = 4), 18 (n = 3), and 24 (n = 3) months. Equivalent numbers of nontransgenic littermates served as controls. Mice were perfused with 4% paraformaldehyde in phosphate-buffered saline. Paraffin sections (4 μm) of mouse brain were stained with hematoxilin/eosin, methenamine silver, Congo Red, Holmes luxol, and Gallyas silver iodide (30). Immunohistochemical staining was performed on deparaffinized brain sections. Anti-MAC-1, acetylcholinesterase (31), major histocompatibility complex (MHC) class II, complement C3, and anti-tau histochemistry was performed with 40-μm free-floating fixed frozen sections. To enhance antigenicity for NF200, glial fibrillary acidic protein (GFAP), and tau antibodies, dewaxed sections were rehydrated and treated for 30 min by immersion in citrate buffer at 90°C. Primary antibodies directed against the Aβ peptide (32) are summarized in Table 1. In addition, anti-heparan sulfate monoclonal antibody 10E4 (Seikagu, Tokyo); anti-neurofilament NF200 (NovoCastra, Newcastle, U.K.); anti-APP 474 raised against purified rat APPs; anti-apolipoprotein E antibody; anti-MAC-1 and anti-MHC class II (Serotec); anti-GFAP and anti-phosphotyrosine PT-66 (Sigma); anti-mouse complement C3 (Cappel, ICN); anti-PHF1 (33) recognizing tau phosphoserine 396 and 402; AT8 monoclonal antibody (Innogenetics) (26, 27) recognizing phosphoserine 202; R27 (28) raised against phosphorylated serine 396; R32 (28) raised against phosphorylated serine 262, N-tau 5 raised against a tau peptide as described (34), and monoclonal antibody Alz50 (35) were used. Sections were treated with normal serum to block nonspecific sites and incubated overnight at 4°C with the primary antibody. Bound antibody was visualized by using a Vectastain ABC Elite kit (Vector Laboratories).
Table 1.
Aβ antibodies recognizing mouse and human plaques
| Antibody | Antigen residues |
|---|---|
| Dako | 8–17 |
| NT11 | 1–40 |
| β1 | 1–40 |
| AS 1–5* | 1–5 |
| AS 12–28 | 12–28 |
| AS 40/18* | 17–40 |
| AS 42/14* | 36–42 |
| AS 43/15* | 36–43 |
These antibodies specifically recognize Aβ amino acids 1, 40, 42, and 43, respectively.
RESULTS
APP Overexpression in Brains of Transgenic Mouse Lines Controlled by the Thy-1 Promoter.
Expression cassettes containing the Thy-1 promoter were used to drive neuron-specific APP expression in transgenic mice (Fig. 1). The two mouse lines APP 14 (19) and APP 22 carry the human Thy-1 promoter to drive expression from different APP cDNAs. The APP 14 construct contains human APP751 with the Swedish double mutation at positions 670/671 (KM → NL), the APP 22 expression unit carries the London mutation (V717I) in addition. Both constructs result in comparable spatial expression patterns with highest transgene-derived mRNA levels in neocortex and hippocampus as shown for APP 22 mice (Fig. 2A). In both lines, mRNA levels exceed endogenous APP mRNA by 2-fold (data not shown) as determined by semiquantitative PCR. In a third line, APP 23, APP751 Swedish expression is driven by the murine Thy-1 promoter. The spatial expression pattern (Fig. 2B) is qualitatively similar to APP 14 and APP 22 mice; however, the transgene is 7-fold overexpressed. The degree of APP overexpression was confirmed by Western blotting for all three mouse lines (data not shown).
Figure 1.
Transcription units for APP transgenic mice. The diagrams are linear representations of the injected expression constructs and are labeled accordingly. Exons of the human Thy-1 gene are illustrated as solid boxes, and stippled boxes indicate sequences of simian virus 40 tumor antigen at the 3′ end of APP 14 and APP 22 constructs. Hatched boxes represent the murine Thy-1 exons used in the APP 23 construct. APP751 cDNAs are shown as open boxes with vertical lines marking the positions of the respective Swedish (S) and London (L) FAD mutations.
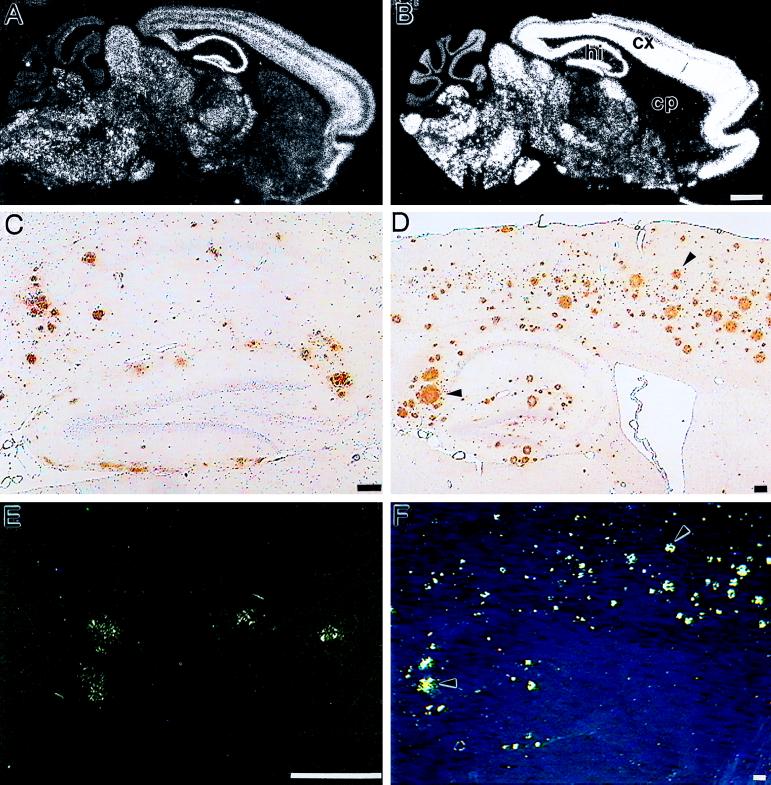
Figure 2.
Expression of APP mRNA and deposition of Aβ in brains of transgenic mice. (A, C, and E) Sections taken from 18-month-old APP 22 mice. (B, D, and F) Sections from 24-month-old APP 23 mice. In situ hybridization on sagittal sections of brains from mouse line APP 22 (A) and APP 23 (B) was performed with an oligonucleotide probe specific for human APP. cp, Caudate putamen; cx, cortex; hi, hippocampus. (Bars = 1 mm.) Immunohistochemistry (C and D) and Congo Red staining (E and F) of sagittal sections from brains of transgenic mice. Immunostaining was performed with an antibody raised against Aβ8–17 (Dako). (Bars = 100 μm.) Arrowheads point to identical deposits in D and F.
APP 22 and APP 23 Transgenic Mice Develop Aβ Deposits in Neocortex and Hippocampus.
Whereas APP 14 mice do not show any indication of AD pathology up to an age of 2 years (data not shown), Aβ deposits are detected in neocortex and hippocampus of 18-month-old but not 12-month-old APP 22 mice (Fig. 2C). The higher expressing APP 23 line shows first rare deposits at 6 months of age. They increase with age in size and number and eventually occupy a substantial area of the neocortex and hippocampus in 24-month-old mice (Fig. 2D). At this age deposits are also found in thalamus and olfactory nucleus and isolated in the caudate putamen. In both lines, all deposits show immunoreactivity with different antibodies specific for Aβ (Fig. 2 C and D and Table 1), which are comparable to the results obtained on brains from AD patients (data not shown). It is noteworthy that immunoreactivity with the end-specific Aβ42 antibody in APP 22 mice appears similar in intensity to that obtained in sections from the higher-expressing APP 23 mice, reflecting the impact of the mutation at position 717 on the production of the more amyloidogenic Aβ-(1–42) (12). Typical plaque-associated proteins in AD such as heparan sulfate proteoglycan and apolipoprotein E, which bind to Aβ, are also present in most deposits (data not shown). No differences are seen between male and female mice.
Differences in Plaque Anatomy in APP 22 and APP 23 Mice.
When compared directly, the intensity of the Aβ and heparan sulfate proteoglycan-specific immunostaining is much stronger in deposits of APP 23 mice. Consistent with this notion, the majority of deposits in APP 22 mice are of the “diffuse” type. Relatively few Congo Red-positive plaques are present in the neocortex and the subiculum (Fig. 2E). In contrast, almost all extracellular deposits in APP 23 mice are intensely stained with methenamine silver and at their first appearance already show Congo Red birefringence (Fig. 2F) indicative of dense core plaques.
Inflammatory Responses in Brains of APP Transgenic Mice.
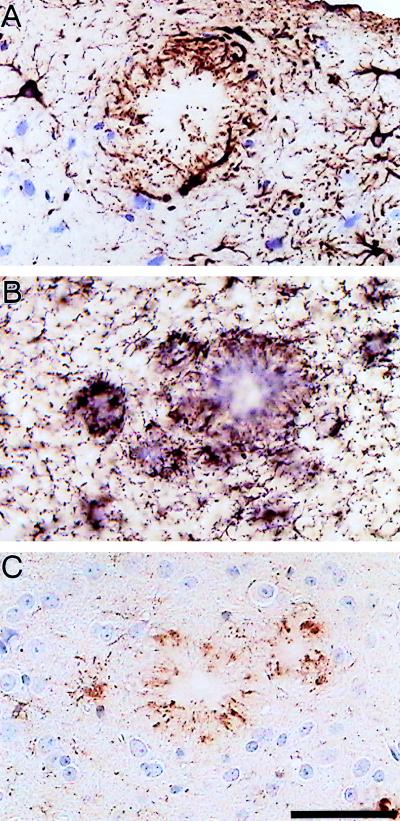
In addition to Aβ deposition, inflammatory processes reminiscent of AD occur in these mice. In both lines, a massive glial response can be demonstrated in brain areas with plaques, which is more pronounced in APP 23 mice and hence correlates with the higher burden of congophilic plaques. Immunocytochemistry for GFAP (Fig. 3A), phosphotyrosine (Fig. 3C), MAC-1 (Fig. 3B), MHC class II, and complement C3 suggest a contribution from both astrocytes and microglia.
Figure 3.
Characterization of inflammatory processes in 12-month-old APP 23 transgenic mice. Immunostaining with a GFAP-specific antibody (A) indicates hypertrophic astrocytes. Activated microglia are visualized by immunostaining with a MAC-1 antibody (B) and a phosphotyrosine antibody (C). Both glia cell types are intimately associated with Aβ deposits. (Bar = 50 μm.)
Neuritic Loss and Distortion of Cholinergic Fibers Are Observed in the Vicinity of Plaques.
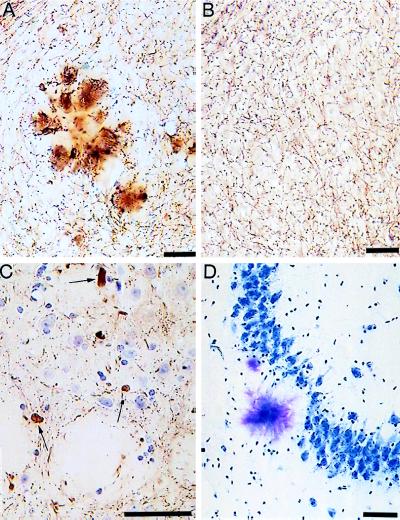
Plaques are surrounded by dystrophic neurites visualized by neurofilament (Fig. 4C), APP, and synaptophysin (data not shown) immunostaining that are only seen at the outer margin of dense compacted Aβ deposits. In areas of high cell density, such as hippocampal pyramidal cells, a reduction of cell bodies adjacent to the Aβ peptide deposition is apparent (Fig. 4D). Although this may simply be cell displacement, the lack of compression of the intracellular space suggests local loss of neurons in the plaque vicinity. When stained for acetylcholinesterase, a strong labeling of plaques and a local distortion of the cholinergic fiber network is observed (Fig. 4A). Acetylcholinesterase activity is found throughout the amyloid plaques and in enlarged neurites at the plaque periphery (Fig. 4A).
Figure 4.
Degenerative processes in APP 23 mice. (A and B) Staining for acetylcholinesterase in 12-month-old transgenic and control mice, respectively. A local distortion of cholinergic fibers in the plaque vicinity (A) compared with normal animals (B) can be noted. Neuritic spheroids (arrows) are stained with the neurofilament antibody NF200 (C). The loss of pyramidal neurons in the vicinity of Aβ deposits in area CA3 is shown in D by toluidine blue staining. (Bars = 50 μm.)
Congophilic Plaques Are Surrounded by Distorted Neurites Containing Hyperphosphorylated Tau.
When examined for aspects of neurofibrillary pathology, hyperphosphorylated microtubule-associated protein tau can be detected in transgenic mouse brains of both lines. Immunostaining with the AT8 antibody is exclusively associated with Congo Red-positive plaques in APP 22 and APP 23 mice. It highlights structures resembling distorted neurites surrounding the core of the deposits (Fig. 5 A and B). Similar reactions are obtained with the antibodies PHF1, R27, and R32 (data not shown), which recognize distinct phosphoepitopes of tau. Interestingly, the Alz50 antibody also shows a similar staining pattern (Fig. 5C). These antibodies do not stain sections from nontransgenic littermate controls. We failed to detect neurofibrillary tangles by the method of Gallyas.
Figure 5.
Phosphorylation of tau in the brains of 12-month-old APP 23 transgenic mice. Immunocytochemistry with antibody AT8 was performed on free floating sections (40 μm) (A and B). Immunostaining reveals fiber-like structures resembling distorted neurites. Qualitatively similar patterns are obtained with the Alz50 antibody (C). [Bars = 100 μm (A) and 10 μm (B and C).]
Hyperphosphorylated Tau Can Be Detected Biochemically in Brain Extracts of APP 23 Mice.
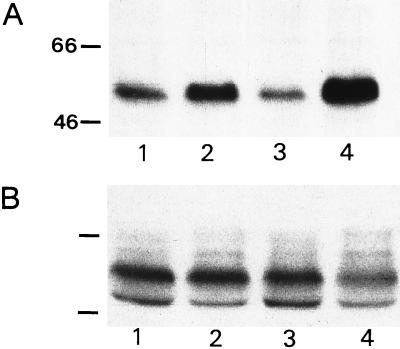
The increased phosphorylation of tau can also be demonstrated on immunoblots of brain extracts from APP 23 mice at 6 months and 15 months of age incubated with the AT8 (Fig. 6A) and the R27 antibody (data not shown). Age-matched littermate controls show much weaker immunoreactivity that does not increase with age. When incubated with phosphorylation-independent antibody tau7, no up-regulation of total tau protein is seen in transgenic mice (Fig. 6B). The tau phosphorylation therefore appears to occur parallel to Aβ peptide deposition.
Figure 6.
Biochemical detection of hyperphosphorylated tau protein. Western blots of brain extracts from APP 23 mice, 6 months (lane 2) and 15 months (lane 4) of age and littermate controls (lanes 1 and 3) are shown in A and B. Blots were stained with antibodies AT8 (A) and tau7 (B). Numbers indicate molecular weights of marker proteins in kDa.
DISCUSSION
Transgenic technology has led to recent advances in the generation of animal models developing aspects of AD pathology (20, 21). In this study we describe transgenic mouse lines overexpressing human APP with AD-linked mutations in a neuron-specific manner.
Increasing the total expression of human APP carrying the Swedish double mutation to 7-fold above endogenous APP leads to an early formation of amyloid plaques in neocortex and hippocampus as demonstrated in mice from line APP 23. A more than 10-fold overexpression was reported by Games et al. (20), whereas Hsiao and colleagues (21) describe transgene-derived APP levels exceeding endogenous APP by 6-fold. Interestingly, different promoter constructs, APP isoforms and FAD mutations were used in each study. Although it is difficult to compare the quantification of transgene expression in the individual studies, it is tempting to speculate that exceeding a certain threshold expression of human APP with a FAD-associated mutation is the trigger for plaque-like pathology.
Our results also demonstrate that the combination of two genetic lesions in the transgenic mice leads to the development of AD plaques at a significantly lower expression level. Mouse line APP 22 overexpressing APP 2-fold matches the quantitative and spatial expression of line APP 14 (19). APP 14 mice carry the Swedish double mutation only and do not develop any obvious pathology up to 2 years of age. APP 22 mice, however, which express human APP with the combined Swedish (8) and London (9) mutations, develop plaques by 18 months of age. These data also suggest a major contribution of the Aβ-(1–42) species to the development of AD pathology at a given expression level.
Interestingly, the majority of Aβ deposits in APP 22 mice is of a diffuse type with a few congophilic plaques in hippocampus and neocortex. In contrast, APP 23 mice develop almost exclusively congophilic plaques already at their first appearance. These results may be difficult to reconcile with the proposed hypothesis (1) that deposits of the diffuse type form a precursor for congophilic neuritic plaques. The two described mouse lines, therefore, offer the opportunity to study such proposed plaque maturation processes.
When examined for additional characteristics, the pronounced glial reaction is apparent in both lines. As in AD, proliferating astrocytes can be detected throughout the brain and accumulate around deposits (36). Phosphotyrosine, MAC-1, MHC class II, and complement C3 immunoreactivity is also found. Although considered indicative of activated microglia in the brains of AD patients (37), the precise cellular origin of these markers requires further investigation. These mice present a suitable model to investigate the pathways of inflammatory processes after Aβ formation in detail.
Besides the general detection of dystrophic neurites in the periphery of plaques by anti-neurofilament and anti-APP immunoreactivity, a local distortion of cholinergic fibers can be observed. The degeneration of cholinergic neurites and the association of cholinesterase activity with senile plaques is another well-described feature of AD (6, 7, 38) that is reproduced in the transgenic mice, suggesting a causal link between Aβ deposition and cholinergic degeneration.
Most importantly, hyperphosphorylated tau can be detected in distorted neurites associated only with congophilic plaques in both transgenic mouse lines with several antibodies recognizing distinct phosphoepitopes of tau. Fibrillary but not amorphous Aβ has been reported to induce hyperphosphorylation of tau and loss of microtubule binding in neuronal primary cultures (39). Similar changes in tau phosphorylation have also been described as early indicators of neurofibrillary changes in AD patients (40). The immunoreactivity with the Alz50 antibody is further evidence for tau-related pathology. This antibody was originally described to recognize an AD-specific epitope (35) and is now considered to react with a discontinuous epitope of tau that is formed after a conformational change of tau associated with polymerization into filaments (41). Therefore, one may expect neurofibrillary pathology in these mice despite the lack of tangles. This notion needs to be confirmed by ultrastructural analysis. A detailed ultrastructural study on APP transgenic mice with pathology (20), however, did not provide evidence for paired helical filaments (42).
Our two transgenic mouse lines develop distinct features of AD pathology and provide model systems to systematically investigate the role of the Aβ peptide in the pathogenesis of AD.
Acknowledgments
H. van der Putten supplied the murine Thy-1 expression cassette. M.J. was supported by grants from the Swiss National Science Foundation.
ABBREVIATIONS
- Aβ
amyloid β
- AD
Alzheimer disease
- APP
amyloid precursor protein
- FAD
familial AD
- MHC
major histocompatibility complex
- GFAP
glial fibrillary acidic protein
References
- 1.Probst A, Langui D, Ulrich J. Brain Pathol. 1991;1:229–239. doi: 10.1111/j.1750-3639.1991.tb00666.x. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D J. Annu Rev Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 3.Yankner B A. Neuron. 1996;16:921–932. doi: 10.1016/s0896-6273(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 4.Goedert M. Trends Neurosci. 1993;16:460–465. doi: 10.1016/0166-2236(93)90078-z. [DOI] [PubMed] [Google Scholar]
- 5.Trojanowski J Q, Schmidt M L, Shin R-W, Bramblett G T, Rao D, Lee V M-Y. Clin Neurosci. 1993;1:184–191. [Google Scholar]
- 6.Tago H, McGeer P L, McGeer E G. Brain Res. 1987;406:363–369. doi: 10.1016/0006-8993(87)90808-0. [DOI] [PubMed] [Google Scholar]
- 7.Geula C, Mesulam M. Neuroscience. 1989;33:469–481. doi: 10.1016/0306-4522(89)90399-0. [DOI] [PubMed] [Google Scholar]
- 8.Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, Lannfelt L. Nat Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 9.Goate A, Chartier-Harlin M-C, Mullan M, Brown J, Crawford F, et al. Nature (London) 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 10.Citron M, Oltersdorf T, Haass C, McConlogue L, Hung A Y, Seubert P, Vigo-Pelfrey C, Lieberburg I, Selkoe D J. Nature (London) 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- 11.Cai X-D, Golde T E, Younkin S G. Science. 1993;259:514–516. doi: 10.1126/science.8424174. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki N, Cheung T T, Cai X-D, Odaka A, Otvos L, Eckman C, Golde T E, Younkin S G. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 13.Tamaoka A, Odaka A, Ishibashi Y, Usami M, Sahara N, Suzuki N, Nukina N, Mizusawa H, Shoji S, Kanazawa I, Mori H. J Biol Chem. 1994;269:32721–32724. [PubMed] [Google Scholar]
- 14.Quon D, Wang Y, Catalano R, Scardina J M, Murakami K, Cordell B. Nature (London) 1991;352:239–241. doi: 10.1038/352239a0. [DOI] [PubMed] [Google Scholar]
- 15.Kammesheidt A, Boyce F M, Spanoyannis A F, Cummings B J, Ortegon M, Cotman C, Vaught J L, Neve R L. Proc Natl Acad Sci USA. 1992;89:10857–10861. doi: 10.1073/pnas.89.22.10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb B T, Sisodia S S, Lawler A M, Slunt H H, Kitt C A, Kearns W G, Pearson P L, Price D L, Gearhart J D. Nat Genet. 1993;5:22–30. doi: 10.1038/ng0993-22. [DOI] [PubMed] [Google Scholar]
- 17.Mucke L, Masliah E, Johnson W B, Ruppe M D, Alford M, Rockenstein E M, Forss-Petter S, Pietropaolo M, Mallory M, Abraham C R. Brain Res. 1994;666:151–167. doi: 10.1016/0006-8993(94)90767-6. [DOI] [PubMed] [Google Scholar]
- 18.Higgins L S, Rodems J M, Catalano R, Quon D, Cordell B. Proc Natl Acad Sci USA. 1995;92:4402–4406. doi: 10.1073/pnas.92.10.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrä K, Abramowski D, Duke M, Probst A, Wiederhold K-H, Bürki K, Goedert M, Sommer B, Staufenbiel M. Neurobiol Aging. 1996;17:183–190. doi: 10.1016/0197-4580(95)02066-7. [DOI] [PubMed] [Google Scholar]
- 20.Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, et al. Nature (London) 1995;373:523–528. doi: 10.1038/373523a0. [DOI] [PubMed] [Google Scholar]
- 21.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 22.Lüthi A, van der Putten H, Botteri F M, Mansuy I M, Meins M, Frey U, Sansig G, Portet C, Schmutz M, Schröder M, Nitsch C, Laurent J-P, Monard D. J Neurosci. 1997;17:4688–4699. doi: 10.1523/JNEUROSCI.17-12-04688.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sommer B, Köhler M, Sprengel R, Seeburg P H. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 24.Sola C, Mengod G, Probst A, Palacios J M. Neuroscience. 1993;53:267–295. doi: 10.1016/0306-4522(93)90304-x. [DOI] [PubMed] [Google Scholar]
- 25.Abramowski D, Staufenbiel M. J Neurochem. 1995;65:782–790. doi: 10.1046/j.1471-4159.1995.65020782.x. [DOI] [PubMed] [Google Scholar]
- 26.Biernat J, Mandelkow E M, Schroter C, Lichtenberg-Kraag B, Steiner B, Berling B, Meyer H, Mercken M, Vandermeeren A, Goedert M, Mandelkow E. EMBO J. 1992;11:1593–1597. doi: 10.1002/j.1460-2075.1992.tb05204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goedert M, Jakes R, Vanmechelen E. Neurosci Lett. 1995;189:167–169. doi: 10.1016/0304-3940(95)11484-e. [DOI] [PubMed] [Google Scholar]
- 28.Frey P, Sunderji S, Waridel C. Alzheimer’s Disease: Biology, Diagnosis and Therapeutics. New York: Wiley; 1997. pp. 449–455. [Google Scholar]
- 29.Probst A, Tolnay M, Langui D, Goedert M, Spillantini M G. Acta Neuropathol (Berlin) 1996;92:588–596. doi: 10.1007/s004010050565. [DOI] [PubMed] [Google Scholar]
- 30.Gallyas F. Acta Morphol Acad Sci Hung. 1971;19:1–8. [PubMed] [Google Scholar]
- 31.Hedreen J C, Bacon S J, Price D L. J Histochem Cytochem. 1985;33:134–140. doi: 10.1177/33.2.2578498. [DOI] [PubMed] [Google Scholar]
- 32.Paganetti P A, Lis M, Klafki H-W, Staufenbiel M. J Neurosci Res. 1996;46:283–293. doi: 10.1002/(SICI)1097-4547(19961101)46:3<283::AID-JNR1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg S G, Davies P, Schein J D, Binder L I. J Biol Chem. 1992;267:564–569. [PubMed] [Google Scholar]
- 34.Goedert M, Spillantini M G, Jakes R, Rutherford D, Crowther R A. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 35.Wolozin B L, Pruchnicki A, Dickson D W, Davies P. Science. 1986;232:648–650. doi: 10.1126/science.3083509. [DOI] [PubMed] [Google Scholar]
- 36.Mandybur T I, Chiurazzi B A. Neurology. 1990;40:635–639. doi: 10.1212/wnl.40.4.635. [DOI] [PubMed] [Google Scholar]
- 37.McGeer P L, Kawamata T, Walker D G, Akiyama H, Tooyama I, McGeer E G. Glia. 1993;7:84–92. doi: 10.1002/glia.440070114. [DOI] [PubMed] [Google Scholar]
- 38.Mesulam M M, Geula C H, Moran A. Ann Neurol. 1987;22:683–691. doi: 10.1002/ana.410220603. [DOI] [PubMed] [Google Scholar]
- 39.Busciglio J, Lorenzo A, Yeh J, Yankner B A. Neuron. 1995;14:879–888. doi: 10.1016/0896-6273(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 40.Braak E, Braak H, Mandelkow E M. Acta Neuropathol. 1994;87:554–567. doi: 10.1007/BF00293315. [DOI] [PubMed] [Google Scholar]
- 41.Carmel G, Mager E M, Binder L I, Kuret J. J Biol Chem. 1996;271:32789–32795. doi: 10.1074/jbc.271.51.32789. [DOI] [PubMed] [Google Scholar]
- 42.Masliah E, Sisk A, Mallory M, Mucke L, Schenk D, Games D. J Neurosci. 1996;16:5795–5811. doi: 10.1523/JNEUROSCI.16-18-05795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]