Abstract
Of the six fatty acid elongase (Elovl) subtypes expressed in mammals, adult rat liver expresses four subtypes: Elovl-5 > Elovl-1 = Elovl-2 = Elovl-6. Overnight starvation and fish oil-enriched diets repressed hepatic elongase activity in livers of adult male rats. Diet-induced changes in elongase activity correlate with Elovl-5 and Elovl-6 mRNA abundance. Adult rats fed the peroxisome proliferator-activated receptor α (PPARα) agonist WY14,643 have increased hepatic elongase activity, Elovl-1, Elovl-5, Elovl-6, Δ5, Δ6, and Δ9 desaturase mRNA abundance, and mead acid (20:3,n-9) content. PPARα agonists affect both fatty acid elongation and desaturation pathways leading to changes in hepatic lipid composition. Elovl activity is low in fetal liver but increases significantly after birth. Developmental changes in hepatic elongase activity paralleled the postnatal induction of Elovl-5 mRNA and mRNAs encoding the PPARα-regulated transcripts, Δ5 and Δ6 desaturase, and cytochrome P450 4A. In contrast, Elovl-6, Δ9 desaturase, and FAS mRNA abundance paralleled changes in hepatic sterol regulatory element binding protein 1c (SREBP-1c) nuclear content. SREBP-1c is present in fetal liver nuclei, absent from nuclei immediately after birth, and reappears in nuclei at weaning, 21 days postpartum. In conclusion, changes in Elovl-5 expression may account for much of the nutritional and developmental control of fatty acid elongation activity in the rat liver.
Supplementary key words: fatty acid desaturase, postnatal development, polyunsaturated fatty acids, peroxisome proliferator-activated receptor α, sterol regulatory element binding protein-1c
Most cells have the capacity to synthesize fatty acids from glucose de novo. This pathway uses products from glycolysis and along with the two enzymes, acetyl-CoA carboxylase and fatty acid synthase, generates palmitate (16:0). Insulin, 3,5,3′-triiodothyronine, glucocorticoids, and glucose induce de novo lipogenesis (DNL), whereas C20 PUFAs, glucagon, and epinephrine suppress DNL (1–3). Many cells also have the capacity to modify fatty acid structure through metabolic pathways that include desaturation, elongation, mono-oxidation, and peroxisomal β-oxidation (chain shortening). Such modifications occur to fatty acids generated de novo as well as fatty acids derived from the diet. These metabolic pathways play an important role in the maintenance of membrane lipid composition and the generation of precursors for certain signaling molecules, such as eicosanoids. These pathways may also contribute to the control of cellular fatty acids that affect specific nuclear receptors [e.g., peroxisome proliferator-activated receptor α (PPARα)] (2, 4, 5).
Of the various pathways known to affect fatty acid structure, physiological control of fatty acid elongation remains poorly defined. The predominant pathway for fatty acid elongation occurs in the endoplasmic reticulum and uses malonyl-CoA and fatty acyl-CoA as substrates for C2 additions to fatty acids. Elongases are condensing enzymes that interact with 3-keto acyl-CoA reductase, a dehydratase, and trans-2,3-enoyl-CoA reductase to elongate fatty acids (6–8). The rate of fatty acid elongation is determined by the activity of the elongase (condensing enzyme) and not the reductases or the dehydratase. Six distinct fatty acid elongase (Elovl) subtypes (Elovl-1 through Elovl-6) are present in the mouse, rat, and human genomes. Elovl-1 (Ssc1) and Elovl-6 (LCE, FACE, rElo2) elongate saturated and monounsaturated fatty acids. Disorders of sphingolipid metabolism, such as the Quaking and Jimpy phenotypes (9), have been associated with impaired Elovl-1 activity. Elovl-6 is induced in transgenic mice overexpressing sterol regulatory element binding protein 1 (SREBP-1) (10–12). Elovl-2 (Ssc2) substrates include C20–22 PUFAs, whereas Elovl-5 (FAE1, Relo1, Helo1) may use a broad substrate array, C16–22 (10, 13). Elovl-2 and Elovl-5 likely play a role in endogenous PUFA synthesis, that is, the conversion of the essential fatty acid precursors linoleic acid (18:2,n-6) and α-linolenic acid (18:3,n-3) to arachidonic acid (20:4,n-6) and docosahexaenoic acid (22:6,n-3) (11). Elovl-3 (Cig30, Elo3) and Elovl-4 (Elo4) are expressed in the skin (14) and retina (15), respectively. Both Elovl-3 and Elovl-4 elongate a broad array of fatty acids (≤C26). Elovl-3 is induced in brown adipose tissue after exposure of animals to the cold (16). Stargardt-like macular dystrophy and autosomal dominant macular dystrophy are associated with defective Elovl-4 expression (17).
Many studies have examined the regulation of mammalian elongases at the level of enzyme activity (6, 8–10, 12, 13, 16, 18–31). Overlapping substrate/product profiles for these enzymes make it difficult to ascertain which enzyme subtype is regulated under specific physiological conditions. More recent studies have examined nutritional and tissue-specific regulation of elongase expression (9, 10, 12, 14, 16, 18). We are unaware of any study that has correlated nutritional and developmental changes in fatty acid elongation activity with the expression of specific elongase subtypes. We have cloned five of the six Elovls known to be expressed in mammals and examined their expression in rat liver. Elovl-4 was not examined because of its retina-specific expression. Our studies included an analysis of the tissue-specific expression of the five elongases as well as dietary and developmental regulation of hepatic elongase expression and fatty acid elongation activity. Elongases and desaturases work in concert to synthesize monounsaturated and polyunsaturated fatty acids (32). Accordingly, we examined the expression of Δ5, Δ6, and Δ9 desaturase to determine the physiological conditions leading to coordinated control of elongase and desaturase expression. Our studies show that of the four Elovl subtypes expressed in rat liver, Elovl-5 is the predominant elongase. Rat hepatic Elovl-5 and Δ5 and Δ6 desaturase are coordinately regulated by diet and during post-natal development. Changes in Elovl-5 expression may account for much of the nutritional and developmental control of fatty acid elongation activity in the rat liver.
MATERIALS AND METHODS
Cloning of mouse and rat Elovls and desaturases
cDNAs for Elovls and desaturases were cloned by RT-PCR. Primers corresponding to the open reading frame of each elongase and desaturase (Table 1) were designed based on sequences obtained from GenBank and the location of the open reading frame as determined using DNA-Star. Mouse liver was used as a template to clone all desaturases and elongases, except Elovl-5; Elovl-5 was cloned from rat liver. The RT-PCR products were inserted into TOPO cloning plasmids (Invitrogen, Carlsbad, CA) and screened by blue-white selection. Positive clones were selected and sequenced at the Genomic Technology Support Facility at Michigan State University. Sequences were verified by alignment to sequences in GenBank (Local Alignment Search Tool; http://www.ncbi.nlm.nih.gov/bl2seq/bl2.html). Rat and mouse elongases are ≥85% homologous at the protein sequence level.
TABLE 1.
Primers used to clone Elovls and desaturases from mouse or rat liver
| Enzyme | Accession No. | Primer Sequence |
|---|---|---|
| Elongase | ||
| Elovl-1 | BC006735 | Sense: ATGGAGGCTGTTGTCAACTTG
Antisense: TCAGTTGGCCTTGACCTTGGT |
| Elovl-2 | NM_019423 | Sense: ATGGGCGGCCGCATGGAGCAGCTGAAGGCCTTT
Antisense: TTATTGAGCCTTCTTGTCCGT |
| Elovl-3 | AF054504 | Sense: ATGGACACATCCATGAATTTC
Antisense: TCATTGGCTCTTGGATGCAAC |
| Elovl-5 | NM_134382 | Sense: ATGGAACATTTCGATGCGTCA
Antisense: TCAATCCTTCCGCTGCTTCCT |
| Elovl-6 | AY053453 | Sense: ATGAACATGTCAGTGTTGACT
Antisense: TCTAGACTACTCAGCCTTCGTGGCTTTCTT |
| Desaturase | ||
| Δ5 desaturase | NM_146094 | Sense: ATGGCTCCCGACCCGGTGCCG
Antisense: CTATTGGTGAAGGTAAGCGTC |
| Δ6 desaturase | BC057189 | Sense: AGTCGACATGGGGAAGGGAGGTAACCAG
Antisense: TCATTTATGGAGGTAAGCATC |
| Δ9 desaturase | NM_009127 | Sense: ATGCCGGCCCACATGCTCCAA
Antisense: TCAGCTACTCTTGTGACTCCC |
Elovl, fatty acid elongase.
Animals
All procedures for the use and care of animals for laboratory research were approved by the All University Committee for Animal Use and Care at Michigan State University. Rats were maintained on Harlan-Teklad laboratory chow (#8640) and water ad libitum. Tissues for the analysis of elongase expression were derived from 60 day old Sprague-Dawley male rats maintained on a chow diet ad libitum. Nutritional regulation of elongases used a meal-feeding protocol (33). Briefly, rats were acclimated to meal feeding of a high-carbohydrate (HiCHO; glucose) diet (ICN Biochemicals, Aurora, OH) supplemented with olive oil (Pompeian, Baltimore, MD) at 10% (w/w) for 7 days. The meal began at 8 AM and ended at noon. After the acclimation period, rats were either maintained on the olive oil diet or switched to a HiCHO diet supplemented with fish oil (Dyets, Inc., Bethlehem, PA) at 10% (w/w) or a HiCHO-olive oil diet supplemented with WY14,643 at 0.1% (w/w) (34). Animals were maintained on the olive oil, fish oil, or olive oil-WY14,643 diet for 7 days. Fasted rats were meal fed the HiCHO-olive oil diet for 7 days but were fasted for 24 h before euthanasia. Two hours after completion of the final meal, animals were euthanized for tissue and blood collection.
The developmental studies used timed pregnant female Sprague-Dawley rats from Charles River Laboratories (Kalamazoo, MI). Fetal livers were obtained at 18–19 days after coitus and pooled for analysis. Male suckling pups (1–20 days of age) and weaned male animals (30 days of age) were used as a source of liver. Rats were weaned onto Harlan-Teklad laboratory chow at 21 days postpartum.
Primary hepatocytes
Rat primary hepatocytes from Harlan-Teklad chow-fed (ad libitum) male Sprague-Dawley rats were prepared and treated with fatty acids or WY14,643 as previously described (5, 33).
In vitro fatty acid elongation assay
Rat liver microsomes were isolated by differential centrifugation (35). Elongation reactions were carried out with modifications to the procedure described by Moon et al. (10). Briefly, reaction mixtures contained 50 μg of microsomal proteins in a total reaction volume of 100 μl. The reaction constituents were 50 mM potassium phosphate buffer, pH 6.5, 5 μM rotenone (Sigma, St. Louis, MO), 40 μM fatty acyl-CoA (Avanti Polar Lipids, Alabaster, AL, and Sigma), 60 μM malonyl-CoA (Sigma), 6.5 dpm/pmol [2-14C]malonyl-CoA (Perkin-Elmer), 1 mM NADPH (Sigma), and 20 μM BSA (fatty acid free). Reactions (at 37°C) were initiated with the addition of NADPH. Reactions were terminated after 20 min with the addition of 100 μl of 5 N KOH + 10% methanol; lipids were saponified for 1 h at 65°C. The saponification reaction was acidified with 100 μl of 5 N HCl; 100 μl of ethanol was added to aid hexane extraction of fatty acids. Elongated fatty acids were collected by two independent extractions with hexane (800 μl). Hexane extracts were pooled, and 14C radioactivity was quantified by β-scintillation counting. Results are expressed as elongase activity units (nanomoles of [14C]malonyl-CoA incorporated per milligram of protein per 20 minutes). The formation of reaction products was dependent on the presence of NADPH and the fatty acid-CoA. Fatty acid elongation products were verified by reversed-phase HPLC using a flow-through β-scintillation counter (5).
RNA extraction, Northern blot analysis, and RT-PCR
RNA was extracted and separated on denaturing (formaldehyde) agarose gels, transferred to nitrocellulose, and hybridized with [32P]cDNAs. The cDNAs for the various transcripts have been described previously (5, 33, 35). Hybridization was visualized by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, CA). To ensure an accurate assessment of all elongases, RT-PCR was used. Transcript-specific primers are described in Table 1.
Immunoblotting
Hepatic nuclear and microsomal proteins were separated by SDS-PAGE and transferred to nitrocellulose. SREBP-1, SREBP-2, and cytochrome P450 4A (CYP4A) were detected as previously described (35). PPARα was measured using antibodies obtained from Santa Cruz Biotechnology. Anti-goat antibodies were also obtained from Santa Cruz. The detection system used the Super Signal West Pico chemiluminescence kit (Pierce).
Quantitation of hepatic and plasma mead acid levels
Total lipid was extracted from liver or plasma in chloroform-methanol (2:1) plus 1 mM butylated hydroxytoluene (5). 7-Non-adecenoic acid (19:1) was added as a recovery standard at the time of extraction. Protein (Bio-Rad) was measured in extracts after the initial homogenization step. Total lipids were saponified, fractionated, and quantified by reversed-phase HPLC using a YMC J-Sphere (ODS-H80) column and a sigmoidal gradient starting at 86.5% acetonitrile + acetic acid (0.1%) and ending at 100% acetonitrile + acetic acid (0.1%) over 50 min with a flow rate of 1.0 ml/min using a Waters 600 controller. Fatty acids were detected using both ultraviolet absorbance at 192 nm (Waters model 2487) and evaporative light scatter (Waters model 2420). Fatty acid composition and structures were confirmed at the Michigan State University Mass Spectrometry Facility by GC-MS (www.bch.msu.edu/facilities/massspec/index.html). Fatty acid standards for reversed-phase HPLC were obtained from Nu-Chek Prep (Elysian, MN). Mead acid (20:3,n-9) was obtained from Sigma-Aldrich (St. Louis, MO).
Statistical analysis
Statistical analysis involved Student’s t-test. P values were calculated using Microsoft Excel t-test for two samples with unequal variance.
RESULTS
Tissue-specific expression of rat Elovls
Northern blot and RT-PCR approaches were used to determine the profile of Elovl expression in several rat tissues (Fig. 1). Elovl-1 was well expressed in lung, brain, kidney, and heart but was barely detectable in liver, brown adipose tissue, and skin. Because Northern blot analysis of hepatic Elovl-2 indicated that this transcript was expressed at low levels (Fig. 2), RT-PCR was used to examine its expression in various rat tissues. Elovl-2 was expressed in liver, lung, brain, and kidney; its expression was not detected in other tissues. Elovl-3 was only detected in skin. Elovl-5 expression was readily detected in all tissues examined. Elovl-6 mRNA was also expressed at low levels in rat liver (Fig. 2). RT-PCR was used to show that Elovl-6 was expressed in all tissues; its highest expression was in brain. The tissue distribution of Elovl-1 (9), Elovl-2 (9), Elovl-3 (9, 14), Elovl-5 (13), and Elovl-6 (10, 13) was similar to that in previous reports. Rat liver expressed four distinct Elovls: Elovl-1, Elovl-2, Elovl-5, and Elovl-6. The relative mRNA abundance of elongase subtypes expressed in livers of adult male rats fed Harlan-Teklad chow ad libitum was Elovl-5 > Elovl-1 = Elovl-2 = Elovl-6 (Figs. 1, 2).
Fig. 1.
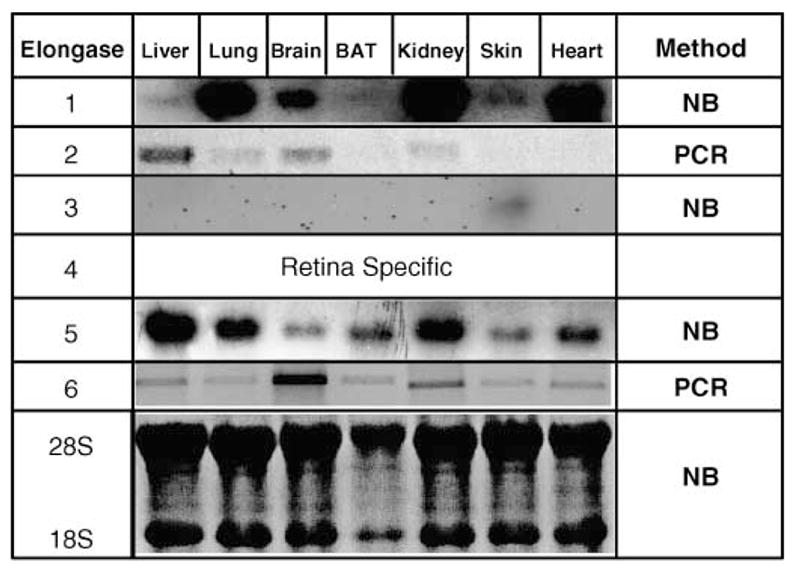
Tissue-specific expression of rat fatty acid elongases (Elovls). RNA was extracted from rat liver, lung, brain, brown adipose tissue (BAT), kidney, skin, and heart. These tissues were obtained from a 60 day old male rat maintained on a Harlan-Teklad chow diet ad libitum. Elovl-1, Elovl-3, and Elovl-5 mRNAs were detected by Northern blot analysis (NB) using cDNAs cloned as described in Materials and Methods. Elovl-2 and Elovl-6 were detected by RT-PCR (35 cycles) (PCR).
Fig. 2.
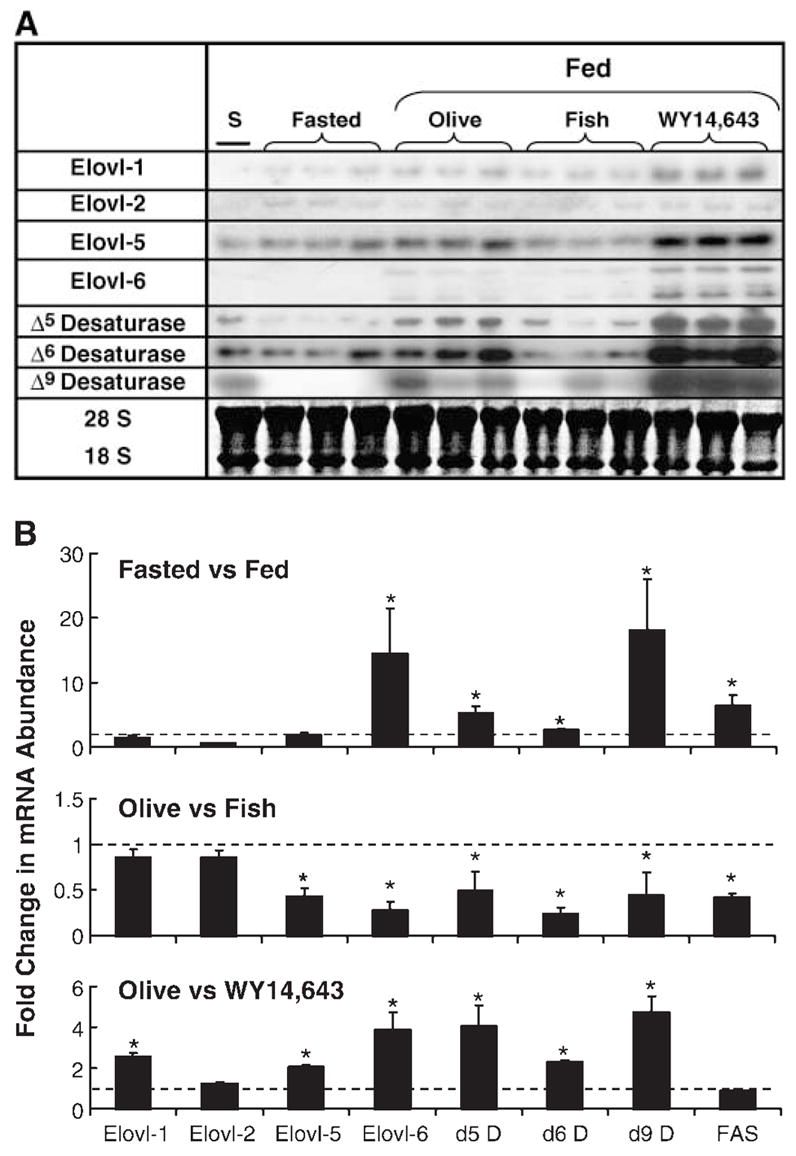
Effect of diet on rat hepatic elongase and desaturase expression. Male Sprague-Dawley rats (50 days of age) were meal-fed a high carbohydrate (HiCHO) diet supplemented with olive oil (Olive; 10%, w/w), fish oil (Fish; 10%, w/w), or olive oil plus WY14,643 (WY14,643; 0.1%, w/w) for 7 days. Fasted animals were meal-fed the HiCHO-olive oil diet as described above but were fasted for 24 h before euthanasia. A: Northern blots of elongase and desaturase mRNAs. A rat liver standard (S) derived from a 90 day old rat fed Harlan-Teklad chow was included in all Northern blots for Elovl-1, Elovl-2, Elovl-5, and Elovl-6 and Δ5, Δ6, and Δ9 desaturase mRNA measurements. Single transcripts were detected for each mRNA, except Elovl-6. Two transcripts have been reported for Elovl-6 (10). In all cases, the size of the mRNA corresponded with the reported size of the transcript. A representative ethidium bromide stain of 18S and 28S mRNA documents equal loading of the RNA samples. Elovl-3 was not detected in any treatment (not shown). B: Fold changes in Elovl, fatty acid desaturase (D), and FAS mRNA abundance after dietary challenge. Top panel: Effect of fasting and refeeding on mRNA expression. Results are expressed as means ± SD; n = 3/animals/group. * P ≤ 0.05, fasted versus fed. Middle panel: Effect of fish oil feeding on mRNA expression. Results are expressed as means ± SD; n = 6/group. * P ≤ 0.05, olive oil versus fish oil. Bottom panel: Effect of WY14,643 feeding on mRNA expression. Results are expressed as means ± SD; n = 6/group. * P ≤ 0.05, olive oil versus olive oil + WY14,643. The dashed lined in each panel represents a fold change of 1.0.
Dietary effects on hepatic elongase and desaturase gene expression
Because elongases act in concert with desaturases for MUFA and PUFA synthesis (32), we examined the effect of diet on the hepatic expression of four elongases (Elovl-1, -2, -5, and -6), three desaturases (Δ5, Δ6, and Δ9), and FAS. Our goal was to determine if diet coordinately regulates fatty acid elongation, fatty acid desaturation, and DNL pathways. Adult male rats were meal-fed HiCHO diets supplemented with olive oil (10%, w/w), fish oil (10%, w/w), or the PPARα agonist WY14,643 (0.1%, w/w) for 7 days. Figure 2A, B illustrates the effect of these treatments on mRNA abundance of the various transcripts.
Fasting and refeeding
Fasting suppresses the expression of enzymes involved in DNL, fatty acid desaturation (1, 3, 36), and at least one elongase, Elovl-6 (12, 13). To determine if other hepatic Elovls are regulated by fasting, rats meal-fed a HiCHO diet supplemented with olive oil (10%, w/w) were fasted for 24 h and refed the same diet for 4 h (Fig. 2). mRNAs encoding Elovl-1 and Elovl-5 were induced marginally (1.5-and 1.9-fold) by refeeding fasted rats. Only Elovl-6 mRNA displayed a robust (14-fold) induction in response to re-feeding fasted rats. In contrast, the desaturase (Δ5, Δ6, and Δ9) mRNAs and FAS mRNA were induced 5.2-, 2.6-, 18-, and 6.5-fold, respectively. Unlike the desaturases, the elongases did not display a uniform response to fasting and refeeding.
Fish oil feeding
Feeding rats diets enriched in n3 PUFAs (i.e., fish oil) inhibits DNL and the expression of all fatty acid desaturases (36–38). Although fish oil suppressed FAS and each desaturase mRNA by ≥50%, fish oil feeding had no significant effect on Elovl-1 and Elovl-2 mRNA abundance. Only Elovl-5 and Elovl-6 mRNAs were significantly (≥50%) suppressed by fish oil (Fig. 2). Therefore, fish oil feeding does not uniformly suppress the expression of all hepatic Elovls.
WY14,643 feeding
All hepatic fatty acid desaturases are induced by PPARα agonists, such as WY14,643 (36, 39, 40). To determine if the elongases are regulated by WY14,643, rats were fed WY14,643 (0.1%, w/w) in the HiCHO-olive oil diet for 7 days (Fig. 2). mRNAs encoding all of the desaturases and three of the elongases were induced by WY14,643. Elovl-1 and Elovl-5 were induced 2- and 2.5-fold; Elovl-6 was induced nearly 4-fold. Elovl-2 mRNA was not affected by WY14,643 feeding. Δ5, Δ6, and Δ9 desaturase mRNAs were induced 4.1-, 2.3-, and 4.8-fold, respectively, whereas FAS mRNA was not affected by WY14,643. The effect of WY14,643 on both elongase and desaturase expression suggests that PPARα agonists may induce long-chain poly-unsaturated synthesis.
Dietary effects on hepatic elongase activity
We next determined whether changes in hepatic elongase mRNA abundance correlated with hepatic fatty acid elongation activity. Elongase assays used seven substrates (i.e., 16:0-CoA, 18:0-CoA, 18:1,n-9-CoA, 20:0-CoA, 20:4,n-6-CoA, 22:0-CoA, and 24:0-CoA) in separate assays (Fig. 3). Based on reports by others, these substrates can be used by one or more hepatic elongase. Specifically, 16:0-CoA is a substrate for Elovl-1 and Elovl-6 but not Elovl-2 (9, 10, 12). 18:1-CoA is not a substrate for Elovl-6 (12) but is a substrate for other elongases. 20:4,n-6 is a substrate for Elovl-2 and Elovl-5 but not Elovl-6 (10, 13). 20:0-CoA, 22: 0-CoA, and 24:0-CoA are substrates for Elovl-1 (9).
Fig. 3.
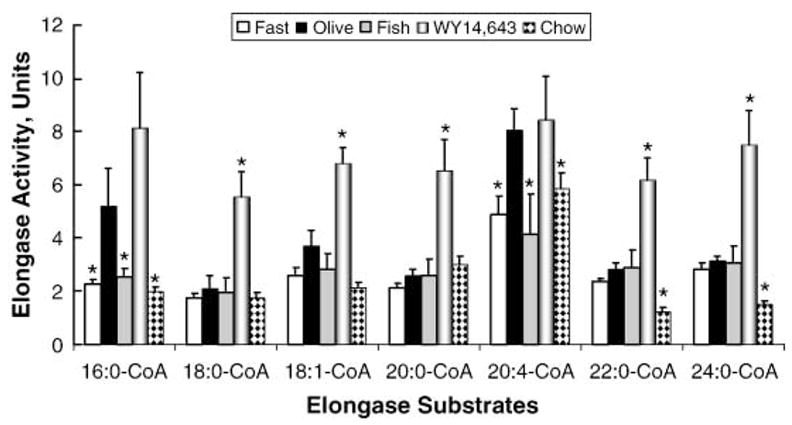
Effect of diet on hepatic Elovl activity. In vitro assays for elongase activity used microsomes isolated from livers of rats fasted or fed a HiCHO diet containing olive oil, fish oil, or WY14,643 as described in Materials and Methods. Separate reactions were run using 16:0-CoA, 18:0-CoA, 18:1,n-9-CoA, 20:0-CoA, 20:4,n-6-CoA, 22:0-CoA, and 24:0-CoA as substrates. Results are expressed as elongase activity units (nmol [14C]malo-nyl-CoA incorporated/mg protein), means ± SD; n = 3/group. * P ≤ 0.05, olive oil-fed rats versus fasted, fish oil fed-, WY14,643 fed-, or Harlan-Teklad chow-fed rats.
Compared with the olive oil-fed rats, hepatic microsomes isolated from fasted rats had significantly lower (~50%) elongase activity when using 16:0-CoA and 20:4-CoA as substrates. Elongation of other fatty acyl-CoAs was not significantly affected by fasting and refeeding. This effect correlates with the suppression of Elovl-6, Elovl-1, and Elovl-5 mRNAs in starved animals (Fig. 2). A comparison of the elongase activity in microsomes isolated from olive oil- and fish oil-fed animals indicates that fish oil feeding suppressed (~60%) the elongation of both 16:0-CoA and 20:4-CoA. This change in elongase activity correlated with the fish oil suppression of Elovl-6 and Elovl-5 mRNA abundance (Fig. 2). Feeding rats WY14,643 increased the elongation of all saturated and monounsaturated fatty acyl-CoA substrates except 16:0-CoA by 2- to 3-fold. Increased elongase activity after WY14,643 feeding correlated with increased hepatic abundance of mRNAs encoding Elovl-1, Elovl-5, and Elovl-6 (Fig. 2).
Effect of fish oil and WY14,643 on hepatic and plasma fatty acid composition
We next determined whether changes in elongase gene expression and elongase activity altered hepatic or plasma fatty acid composition (Fig. 4). We focused on mead acid (20:3,n-9) because it is not a dietary fat; its presence in cells is the result of the elongation and desaturation of 18: 1,n-7 or 18:1,n-9 (41).
Fig. 4.
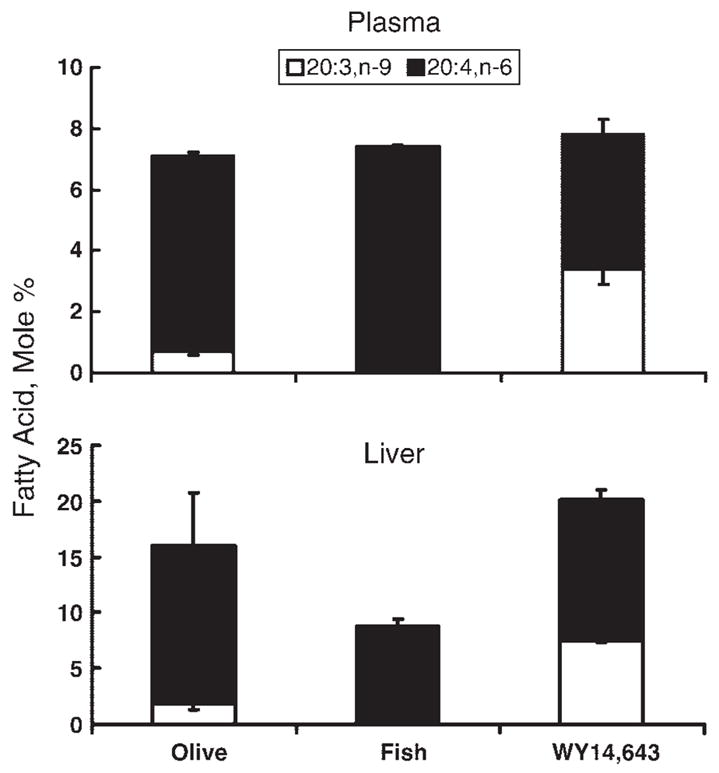
Effect of diet on plasma and liver mead acid levels. Total lipids extracted from rat liver and plasma of animals fed olive oil, fish oil, or WY14,643 were saponified, separated by reversed-phase HPLC, and quantified (see Materials and Methods). The identities of all fatty acids were confirmed by gas chromatography-mass spectrometry. Levels of mead acid (20:3,n-9) and arachidonic acid (20: 4,n-6) were quantified and the results are presented as mol% of the total saponified lipid fraction (means ± SD; n = 3/animals/group). The mol% of 20:3,n-9 in olive oil-fed versus fish oil-fed animals and in olive oil-fed versus olive oil + WY14,643-fed animals was significantly different (P < 0.01).
Fish oil and WY14,643 feeding significantly affected tissue and plasma levels of 20:3,n-9 (Fig. 4). In olive oil-fed rats, the mol% of 20:3,n-9 and 20:4,n-6 in the plasma was 0.7 and 6.4; in liver, it was 1.8 and 14.2, respectively. The ratio of 20:3,n-9 to 20:4,n-6 in plasma and liver was 0.1 and 0.13, respectively. These values are in agreement with essential fatty acid sufficiency (42). Rats fed the fish oil diet had a >95% reduction in plasma and hepatic 20:3,n-9. In contrast, olive oil- and WY14,643-fed rats had a 4- and 5-fold increase in 20:3,n-9 levels in plasma and liver, respectively. The 20:3,n-9/20:4,n-6 ratio in the WY14,643-fed rats was 0.8 and 0.6 in plasma and liver; these values are found in essential fatty acid deficiency (EFAD). The increase in 20:3,n-9 levels can be correlated to increased elongation of 18:1-CoA (Fig. 3) and increased Elovl-1 and Elovl-5 and Δ5 and Δ6 desaturase mRNA abundance (Fig. 2). The increased dietary intake of 18:1,n-9 in the olive oil diet likely contributes to increased 20:3,n-9 synthesis.
Developmental regulation of hepatic Elovl and desaturase gene expression
SREBP-1c, a key transcription factor controlling hepatic lipid synthesis (43), has been implicated in the regulation of at least one Elovl (Elovl-6) (10–12). In an effort to determine whether changes in hepatic SREBP-1c nuclear content correlate with hepatic elongase activity and expression, we used the model of early postnatal development. In this model, SREBP-1c is present in fetal hepatic nuclei (Fig. 5A). Soon after birth, nuclear SREBP-1c levels decrease to undetectable levels and increase again when the animals are weaned at 21 days postpartum (35).
Fig. 5.
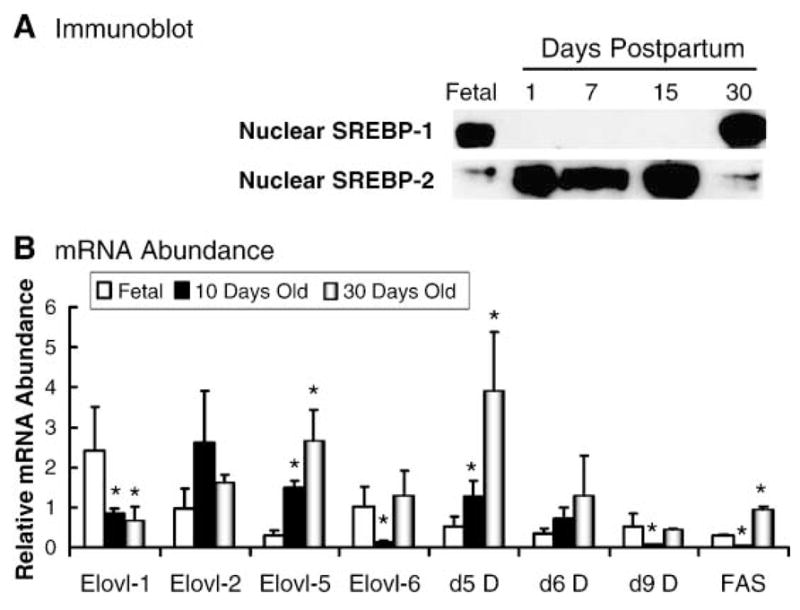
Developmental regulation of rat hepatic sterol regulatory element binding protein 1 (SREBP-1) and SREBP-2 nuclear content and Elovl, fatty acid desaturase, and fatty acid synthase mRNA abundance. A: Immunoblot. Nuclear proteins isolated from livers at 18 days after coitus (Fetal) and 1, 7, 15, and 30 days postpartum were prepared, separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies for SREBP-1 and SREBP-2 (35). The inclusion of SREBP-2 shows that hepatic SREBP-1 and SREBP-2 nuclear contents are regulated differently during development. B: mRNA abundance. Rat liver mRNAs from 18 days after coitus (Fetal) and 10 and 30 days postpartum were used to quantify levels of Elovls (Elovl-1, Elovl-2, Elovl-5, and Elovl-6), fatty acid desatu-rases (Δ5 D, Δ6 D, and Δ9 D), and FAS. The results are represented as relative RNA abundance and are normalized to the level of expression in a 90 day old male rat (means ± SD; n = 3/group). * P ≤ 0.05, fetal versus 10 or 30 days.
Figure 5B illustrates changes in the abundance of elongase and desaturase mRNAs in fetal liver (18 days after coitus) and livers derived from male rats at 10 days post-partum (suckling) and 30 days postpartum (weaned). All four elongases (Elovl-1, -2, -5, and -6), all three desaturases (Δ5, Δ6, and Δ9), and FAS were detected in fetal liver. Elovl-1 decreased to adult values after birth. Elovl-2 remained unchanged from 18 days after coitus to 30 days postpartum. Fetal Elovl-5 expression was <10% of the 30 day value and increased 5-fold by 10 days and 10-fold by 30 days postpartum. Elovl-6 mRNA was expressed in fetal and adult liver but was undetectable by 10 days postpartum. Elovl-6 mRNA levels increased after weaning.
The developmental regulation of Elovl-6, Δ9 desaturase, and FAS parallels changes in SREBP-1c nuclear content during early development. However, at no time in this analysis was Elovl-6 mRNA found to be highly expressed; it remained a low-abundance transcript. In contrast, mRNAs encoding Elovl-5 and Δ5 and Δ6 desaturase were very low in fetal liver and increased after parturition. Developmental changes in Elovl-5 and Δ5 and Δ6 desaturase do not parallel changes in SREBP-1c nuclear content.
At parturition, animals ingest a high-fat milk diet in which 65% of the calories are fat (44). The ingestion of the high-fat milk diet is associated with the activation of PPARα-regulated transcripts (45, 46). Nuclear PPARα content changed little in livers of fetal, neonatal, or weaned animals (Fig. 6). The PPARα-regulated transcript, CYP4A, was induced (~10-fold) within 1 day of birth; both CYP4A mRNA and microsomal CYP4A protein increased by >10-fold. The induction of CYP4A mRNA paralleled the induction of Elovl-5 mRNA after birth. Both Δ5 and Δ6 desaturase mRNAs also rapidly increased after birth (not shown). This observation, coupled with the fact that Elovl-5 mRNA was induced by WY14,643 in adult rats (Fig. 2), suggests that PPARα and the high-fat milk diet contribute to the postnatal increase in Elovl-5 mRNA.
Fig. 6.
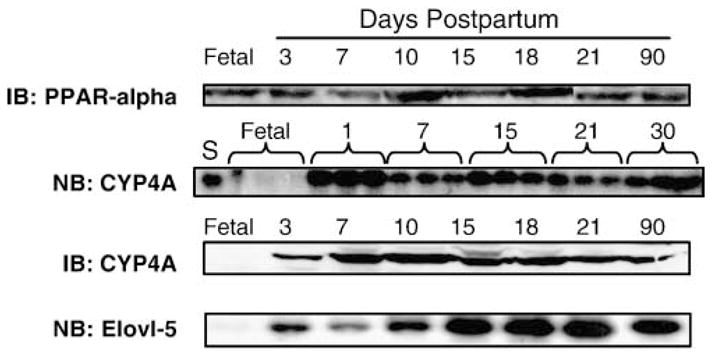
Rapid induction of hepatic Elovl-5 and cytochrome P450 4A (CYP4A) mRNA after parturition. Immunoblot (IB) CYP4A and PPARα (peroxisome proliferator-activated receptor α): Nuclear (PPARα) or microsomal (CYP4A) proteins were isolated from livers at 18 days after coitus (Fetal) and 3, 7, 10, 15, 18, 21, and 30 days postpartum. Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies for PPARα or CYP4A (35). Northern blot (NB) CYP4A: Total RNA was extracted from rat livers at 18 days after coitus (Fetal) and 1, 7, 15, 21, and 30 days postpartum (n = 3/group). S (standard), RNA from 90 day old rat liver. RNA electrophoretically separated and transferred to nitrocellulose was probed for CYP4A (50). Northern blot (NB) Elovl-5: Total RNA was extracted from livers of fetal rats and from male rats 3, 7, 10, 15, 18, 21, and 90 days old. RNA electrophoretically separated and transferred to nitrocellulose was probed for Elovl-5.
Developmental regulation of hepatic Elovl activity
Fatty acid elongation activity was examined in microsomes isolated from fetal liver and livers of 10 and 30 day old male rats (Fig. 7). Fatty acid elongation activity for all substrates was low in fetal liver and increased 5- to 20-fold by 10 days postpartum. Elongation of C22–24 saturated fatty acids was ~2- to 3-fold higher than that of C16–18 saturated fatty acids in fetal liver, a pattern consistent with increased Elovl-1 expression in fetal liver. Elongation activity for each substrate was induced significantly by 10 days postpartum. The level of elongase activity at 10 days postpartum was at or near adult (90 day) values. Of all the fatty acyl-CoA substrates used, two to three times more 20:4-CoA was converted to its elongation product (22:4,n-6) than any other fatty acyl-CoA tested.
Fig. 7.
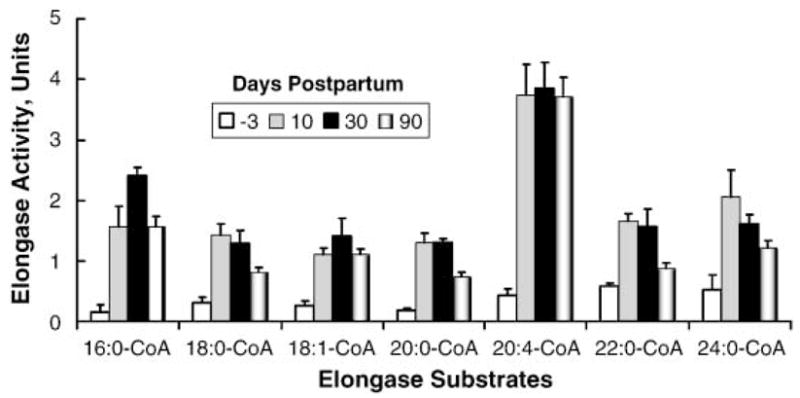
Developmental regulation of hepatic Elovl activity. In vitro assays for elongase activity used microsomes isolated from livers of fetal rats (18 days after coitus) and 10, 30, and 90 days postpartum. Separate reactions were run using 16:0-CoA, 18:0-CoA, 18:1,n-9-CoA, 20:0-CoA, 20:4,n-6-CoA, 22:0-CoA, and 24: 0-CoA as substrates. Results are expressed as elongase activity units (nmol [14C]malonyl-CoA incorporated/mg protein), means ± SD; n = 3/group. Elongase activity in fetal liver was significantly lower than in postpartum liver for all substrates examined (P ≤ 0.05).
The low elongase activity in fetal livers correlates with the low level of expression of all hepatic elongases. Note that Elovl-1, Elovl-2, and Elovl-6 are expressed at low levels in both fetal and adult livers (Figs. 1, 2, 5). The postnatal increase in hepatic elongase activity correlates with only one elongase transcript, Elovl-5. Elovl-5 mRNA abundance increased 5-fold by 10 days postpartum (Fig. 5, 6). The finding that no other elongase was significantly induced by 10 days postpartum suggests that Elovl-5 is likely the elongase responsible for a major fraction hepatic fatty acid elongation. If so, then Elovl-5 is capable of elongating a broad range of fatty acid substrates, including saturated (16:0, 18:0, 20:0, 22:0, and 24:0), monounsaturated (18: 1, n-9), and polyunsaturated (20:4, n-6) fatty acids (Fig. 7).
DISCUSSION
The goal of this study was to examine the dietary and developmental regulation of Elovl expression in rat liver. This study was prompted by earlier studies in which C20 PUFAs were rapidly elongated to C22 PUFAs in primary hepatocytes (5), Hek293 cells (4), and FTO-2b hepatoma cells. C20–22 PUFAs regulate the activity of fatty acid-regulated nuclear receptors, such as PPARα and liver X receptor α (LXRα) (4, 5). As such, changes in C20 PUFA elongation might affect PPARα and LXRα action. The studies reported here revealed new information on the regulation of elongase expression and which elongase likely contributes to C20 PUFA metabolism in rat liver.
Of the four Elovl subtypes (Elovl-1, Elovl-2, Elovl-5, and Elovl-6) expressed in liver (Figs. 1, 2), only Elovl-1, Elovl-2, and Elovl-5 use C20 fatty acid substrates (9, 10, 13). Compared with Elovl-5, mRNAs encoding Elovl-1 and Elovl-2 were low-abundance transcripts in rat liver (Figs. 1, 2). Moreover, Elovl-1 and Elovl-2 mRNAs did not increase during postnatal development, a time in which the liver substantially increased its capacity for C20 PUFA elongation (Fig. 7). Elovl-5 was the only Elovl readily detected in primary hepatocytes (not shown). Based on these observations, we suggest that Elovl-5 is responsible for the elongation of C20 PUFAs to C22 PUFAs in primary hepatocytes. The finding that Elovl-5 mRNA is expressed in a broad array of tissues (Fig. 1) (13), regulated during postnatal development (Fig. 5), by dietary fat and the PPARα agonist WY14,643 (Fig. 2) suggests that changes in Elovl-5 expression and activity may be important for the maintenance of cellular long-chain fatty acids. Whether changes in Elovl-5 expression and activity affect fatty acid-regulated nuclear receptor function is currently under investigation.
It is unclear whether changes in elongase activity during postnatal development are attributable solely to changes in elongase expression. Elongases are condensing enzymes that interact with 3-keto acyl-CoA reductase, a dehydratase, and trans-2,3-enoyl-CoA reductase to elongate fatty acids (6–8). The rate of fatty acid elongation is generally viewed as being determined by the activity of the elongase (condensing enzyme) and not the reductases or dehydratase. However, the low elongase activity in fetal liver may be the result of deficient expression of the reductases and dehydratase as well as elongases. Additional studies will be required to examine the expression of these enzymes in fetal liver.
The developmental and nutritional studies have allowed us to assess the role of key transcription factors in the control of elongase expression. During the perinatal and postnatal periods, two distinct patterns of Elovl and desaturase expression become apparent. These patterns of regulation can be linked to changes in SREBP-1c nuclear content or PPARα activation by the high-fat milk diet. Both SREBP-1c and PPARα play major roles in whole-body and hepatic lipid metabolism (43, 47). Changes in SREBP-1c nuclear content closely parallel changes in FAS, Δ9 desaturase, and Elovl-6 expression (Fig. 5). SREBP-1c is expressed in fetal livers at levels near adult values. However, during the suckling period, SREBP-1c is absent from the nucleus. FAS, Δ9 desaturase, and Elovl-6 increase after weaning when SREBP-1c nuclear content increases. SREBP-1c nuclear abundance has been linked, directly or indirectly, to the transcriptional control of each of these genes (11, 43, 48). The regulation of SREBP-1c nuclear content during postnatal development is mediated by changes in the proteolytic conversion of the SREBP-1c precursor (~125 kDa) to the mature form (~65 kDa) and its transport to the nucleus (35). Ingestion of the high-fat milk diet (~65% calories as fat) begins at parturition. The ingestion of the high-fat milk diet, coupled with low blood insulin levels (44, 49), likely contributes to this regulatory scheme.
The expression of Elovl-5 does not parallel changes in SREBP-1c nuclear content during postnatal development (Fig. 5). In fact, the induction of Elovl-5 mRNA parallels the induction of the PPARα-regulated transcripts, CYP4A, and Δ5 and Δ6 desaturase. In addition, treatment of primary hepatocytes with T1317, an LXR agonist, significantly increases SREBP-1c nuclear content as well as several SREBP-1c-regulated transcripts (33). T1317 only modestly induced Elovl-5 mRNA (<30%) in primary hepatocytes (not shown). Unlike Elovl-6 (10, 12), Elovl-5 does not appear to be a target for either SREBP-1c or LXRα.
Although the postnatal induction of Elovl-5 and Δ5 and Δ6 desaturase closely parallels the induction of PPARα-regulated transcripts [e.g., CYP4A (Fig. 6)] (34, 50), the role that PPARα plays in this regulatory scheme is less clear. PPARα agonists, such as WY14,643 (Fig. 2), and fibrates (31) induce hepatic elongase activity toward a broad range of fatty acids. Our studies show that WY14,643 induced Elovl-1, -5, and -6 mRNAs (Fig. 2). However, studies with primary hepatocytes indicated that although PPARα target genes, such as CYP4A and acyl CoA oxidase, are well induced in primary hepatocytes (5, 34), transcripts encoding elongases (e.g., Elovl-5) are only marginally (<25%) induced (not shown). The lack of a significant WY14,643 effect on Elovl-5 (or other elongases) in primary hepatocytes suggests that these enzymes are not a direct target of PPARα. Clearly, many factors change during postnatal development [e.g., insulin, glucagon glucocorticoids, leptin, T3 (44, 51, 52)] that could affect hepatic elongase expression. These and other factors are being evaluated for their contribution to elongase expression.
The finding of two distinct regulatory patterns for elongase and desaturase expression during postnatal development likely has physiological significance for the synthesis and/or maintenance of tissue levels of 20:4, n-6 and 22: 6, n-3. Analysis of hepatic fatty acid profiles in fetal, 10 day old, and 30 day old liver showed that the mol% of 20:4, n-6 and 22:6, n-3 remained essentially unchanged, at 15–20 mol% (not shown). Blood of pregnant female rats fed a Harlan-Teklad chow diet contained levels of 20:4, n-6 and 22:6,n-3 at 10 and 5 mol%, respectively (not shown). Because both elongase and desaturase expression was low in fetal liver, the high tissue content of 20:4,n-6 and 22:6,n-3 is likely derived from maternal blood. At parturition, neonates rely on maternal milk and endogenous pathways to sustain the production of n3 and n6 PUFAs. Rat milk is deficient in 20:4,n-6 and 22:6,n-3 (i.e., 0.5 and 0.1 mol%, respectively). Neonates grow very rapidly, and the only mechanism available to sustain the constant level of hepatic 20:4,n-6 and 22:6,n-3 in the absence of an extrahepatic source of C20–22 PUFA is to induce PUFA synthesis. The capacity of the neonatal liver to synthesize 20:4,n-6 and 22:6,n-3 from dietary precursors (18:2,n-6 and 18:3,n-3) is achieved, at least in part, by inducing mRNAs encoding Δ5 and Δ6 desaturase and Elovl-5 (Figs. 6, 7).
Fetal liver has high blood levels of 20:4,n-6 and 22:6,n-3, low elongase activity, and low levels of mRNAs encoding Elovl-5 and Δ5 and Δ6 desaturase. Adult animals fed fish oil (a rich source of 20:5,n-3, 22:5,n-3, and 22:6,n-3) had low 20:4,n-6 and 20:5,n-3 elongation activity (Fig. 5) and suppressed expression of mRNAs encoding Elovl-5 and Δ5 and Δ6 desaturase (Fig. 2). Thus, changes in Elovl-5 and Δ5 and Δ6 desaturase expression respond to changes in dietary PUFA composition and tissue requirements for C20–22 PUFAs. Elovl-2 has also been implicated in PUFA synthesis (18, 19). Yet, Elovl-2 mRNA remained unresponsive during postnatal development after fasting/refeeding and fish oil treatment. Based on these findings, Elovl-5, and not Elovl-2, appears to play the major role, along with Δ5 and Δ6 desaturase, in the adaptive response of PUFA synthesis after changes in dietary lipid composition. This response is mediated, at least in part, through the regulation of the cellular abundance of the mRNAs encoding these enzymes. Additional studies will be needed to further define the roles that Elovl-2 and Elovl-5 play in the control of hepatic PUFA synthesis. In this regard, raising rats on n3 PUFA-deficient diets leads to changes in blood and tissue lipid profiles as well as to diminished cognitive function (53). Whether similar effects occur in null mutations of specific elongases or desaturases remains to be determined.
Feeding rats fish oil suppresses lipogenic gene expression (33, 54) and Elovl-5 mRNA (≥50%) (Fig. 2). Primary hepatocytes treated with 20:5,n-3 showed ~50% reduction in Elovl-5 mRNA (not shown). Many of the effects of fish oil on hepatic gene expression can be attributed to the suppression of nuclear levels of SREBP-1c or the activation of PPARα (2). Our findings suggest that neither factor plays a direct role in regulating Elovl-5 expression in primary hepatocytes. Hepatic L-pyruvate kinase gene transcription is suppressed by 20:5,n-3 through mechanisms that are independent of PPARα and SREBP-1c (34, 54). The target for 20:5,n-3 action on L-pyruvate kinase is a region that binds a carbohydrate regulatory element binding protein (ChREBP), MAX-like factor (Mlx), and Hepatic nuclear factor (HNF)-4 (55, 56). Elovl-5 mRNA is unresponsive to changes in glucose treatment of primary hepatocytes, suggesting that ChREBP and Mlx are not involved in Elovl-5 expression (not shown). The role of HNF-4 in Elovl-5 expression is under investigation.
Finally, the finding that WY14,643 increased hepatic and plasma levels of mead acid (20:3,n-9) in olive oil-fed rats was a surprise (Fig. 4). Mead acid is an elongation and desaturation (Δ5 and Δ6 desaturase) product of vaccenic acid (18:1,n-7) and oleic acid (18:1,n-9) (41). The effect of dietary n3 and n6 PUFAs on tissue and plasma levels of 20:3,n-9 has been well described (42). It is generally accepted that much of the PUFA-mediated suppression of 20:3,n-9 content in cells is attributable to competition among PUFAs for Δ6 desaturase (n3 > n6 > n9) (57). The increased tissue content of C20–22 PUFAs, coupled with the decline in both elongation and desaturation capacity (Figs. 2–4), likely accounts for the low levels of 20:3,n-9 in fish oil-fed animals. Contrary to the effects of fish oil, WY14,643 significantly increased hepatic and plasma levels of 20:3,n-9. The level of 18:2,n-6 in the olive oil diet (3.3 mol%) is sufficient to prevent EFAD. However, ingestion of an 18:1,n-9-enriched diet, coupled with the induction of hepatic Elovl-1, Elovl-5, and Elovl-6 plus Δ5 and Δ6 desaturases by WY14,643, is apparently sufficient to increase 20:3,n-9 synthesis and its accumulation in tissues and plasma. The consequence of increased 20:3,n-9 production is an increase in the hepatic 20:3,n-9/20:4,n-6 to 0.62, a value consistent with EFAD (42). Clearly, the increased expression of key enzymes involved in PUFA synthesis, plus ingestion of an 18:1,n-9-enriched diet, is sufficient to shift the tissue balance of 20:3,n-9 and 20:4,n-6.
Conclusions
We have examined the nutritional and developmental regulation of rat hepatic Elovl expression and correlated changes in elongase expression with elongase enzymatic activity. Of the four elongase subtypes expressed in rat liver, Elovl-5 is the most abundant elongase transcript. Rat hepatic Elovl-5 expression is regulated at the pretranslational level by dietary n3 PUFAs and PPARα agonist and during postnatal development. These same factors regulate hepatic Elovl activity. Changes in the expression of the elongases (and their activity) and desaturases are sufficient to induce changes in tissue and plasma levels of specific fatty acids (e.g., 20:3,n-9). Unlike Δ5, Δ6, and Δ9 desaturases, the elongases do not display a uniform response to fasting-refeeding, fish oil, or PPARα agonist. In this regard, changes in nuclear SREBP-1c levels correlate with changes in Elovl-6 expression but not other elongases. Although PPARα agonists induce all desaturases and three elongases (Elovl-1, -5, and -6), the mechanism for this control remains unresolved, although it likely represents an adaptive response to changes in hepatic/whole-body metabolism induced by WY14,643 (58). Clearly, additional studies will be required to define the molecular basis for the regulation of expression of Elovl-5 and other elongases by PPARα and dietary and developmental factors.
Acknowledgments
This research was supported by National Institute of Diabetes and Digestive Diseases Grant 43220, U.S. Department of Agriculture Grant 2003-35200-13400, and the Michigan Experimental Agriculture Station. The authors thank Dr. L. Karl Olson for critical review of the manuscript.
Abbreviations
- ChREBP
carbohydrate regulatory element binding protein
- CYP
cytochrome P450
- DNL
de novo lipogenesis
- EFAD
essential fatty acid deficiency
- Elovl
fatty acid elongase
- HiCHO
high-carbohydrate
- HNF
hepatic nuclear factor
- LXR
liver X receptor α
- MLX
MAX-like factor
- PPAR
peroxisome proliferator-activated receptor
- SREBP
sterol regulatory element binding protein
References
- 1.Jump DB, Clarke SD. Regulation of gene expression by dietary fat. Annu Rev Nutr. 1999;19:63–90. doi: 10.1146/annurev.nutr.19.1.63. [DOI] [PubMed] [Google Scholar]
- 2.Jump D. Fatty acid regulation of gene transcription. Crit Rev Clin Lab Sci. 2004;41:41–78. doi: 10.1080/10408360490278341. [DOI] [PubMed] [Google Scholar]
- 3.Hillgartner FB, Salati LM, Goodridge AG. Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- 4.Pawar A, Xu J, Jerks E, Mangelsdorf DJ, Jump DB. Fatty acid regulation of liver X receptors (LXR) and peroxisome proliferator-activated receptor alpha (PPARalpha) in HEK293 cells. J Biol Chem. 2002;277:39243–39250. doi: 10.1074/jbc.M206170200. [DOI] [PubMed] [Google Scholar]
- 5.Pawar A, Jump DB. Unsaturated fatty acid regulation of PPAR-alpha in rat primary hepatocytes. J Biol Chem. 2003;278:35931–35939. doi: 10.1074/jbc.M306238200. [DOI] [PubMed] [Google Scholar]
- 6.Leonard AE, Pereira SL, Sprecher H, Huang YS. Elongation of long-chain fatty acids. Prog Lipid Res. 2004;43:36–54. doi: 10.1016/s0163-7827(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 7.Moon YA, Horton JD. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J Biol Chem. 2003;278:7335–7343. doi: 10.1074/jbc.M211684200. [DOI] [PubMed] [Google Scholar]
- 8.Prasad MR, Nagi MN, Ghesquier D, Cook L, Cinti DL. Evidence for multiple condensing enzymes in rat hepatic microsomes catalyzing the condensation of saturated, monounsaturated and polyunsaturated acyl coenzyme A. J Biol Chem. 1986;261:8213–8217. [PubMed] [Google Scholar]
- 9.Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A. Role of a new mammalian gene family in the biosynthesis of very long fatty acids and sphingolipids. J Cell Biol. 2000;149:707–717. doi: 10.1083/jcb.149.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon YA, Shah NA, Mohapatra S, Warrington JA, Horton JD. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J Biol Chem. 2001;276:45358–45366. doi: 10.1074/jbc.M108413200. [DOI] [PubMed] [Google Scholar]
- 11.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuzaka T, Shimano H, Yahagi N, Yoshikawa T, Amemiya-Kudo M, Hasty AH, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Osuga J, Takahashi A, Yato S, Sone H, Ishibashi S, Yamada N. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J Lipid Res. 2002;43:911–920. [PubMed] [Google Scholar]
- 13.Inagaki K, Aki T, Fukuda Y, Kawamoto S, Shigeta S, Ono K, Suzuki O. Identification and expression of a rat fatty acid elongase involved in the biosynthesis of C18 fatty acids. Biosci Biotechnol Biochem. 2002;66:613–621. doi: 10.1271/bbb.66.613. [DOI] [PubMed] [Google Scholar]
- 14.Westerberg R, Tvrdik P, Unden A-B, Mansson J-E, Norlen L, Jakobsson A, Holleran WH, Elias PM, Asadi A, Flodby P, Toftgard R, Capecchi MR, Jacobsson A. Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem. 2004;279:5621–5629. doi: 10.1074/jbc.M310529200. [DOI] [PubMed] [Google Scholar]
- 15.Lagali PS, Liu J, Ambasudhan R, Kakuk LE, Bernstein SL, Seigel GM, Wong PW, Ayyagari R. Evolutionarily conserved ELOVL4 gene expression in the vertebrate retina. Invest Ophthalmol Vis Sci. 2003;44:2841–2850. doi: 10.1167/iovs.02-0991. [DOI] [PubMed] [Google Scholar]
- 16.Tvrdik P, Asadi A, Kozak LP, Nedergaard J, Cannon B, Jacobsson A. Cig30, a mouse member of a novel membrane protein gene family, is involved in the recruitment of brown adipose tissue. J Biol Chem. 1997;272:31738–31746. doi: 10.1074/jbc.272.50.31738. [DOI] [PubMed] [Google Scholar]
- 17.Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, Li Y, Metzker ML, Allikmets R, Zack DJ, Kakuk LE, Lagali PS, Wong PW, MacDonald IM, Sieving PA, Figueroa DJ, Austin CP, Gould RJ, Ayyagari R, Petrukhin K. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27:89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- 18.Leonard AE, Bobik EG, Dorado J, Kroeger PE, Chuang LT, Thurmond JM, Parker-Parnes JM, Das T, Huang YS, Mukerji P. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of PUFA. Biochem J. 2000;350:765–770. [PMC free article] [PubMed] [Google Scholar]
- 19.Leonard AE, Kelder B, Bobik EG, Chuang LT, Lewis CJ, Kopchick JJ, Mukerji P, Huang YS. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids. 2002;37:733–740. doi: 10.1007/s11745-002-0955-6. [DOI] [PubMed] [Google Scholar]
- 20.Fan YY, Chapkin RS. Importance of dietary gamma-linolenic acid in human health and nutrition. J Nutr. 1998;128:1411–1414. doi: 10.1093/jn/128.9.1411. [DOI] [PubMed] [Google Scholar]
- 21.Alava MA, Iturralde M, Gonzalez B, Pineiro A. Fatty acid desaturation: effect of alphafetoprotein on alpha-linolenic acid conversion by fetal rat hepatocytes. Prostaglandins Leukot Essent Fatty Acids. 1999;60:209–215. doi: 10.1054/plef.1999.0026. [DOI] [PubMed] [Google Scholar]
- 22.Cinci G, Guerranti R, Pagani R, Carlucci F, Terzuoli L, Rosi F, Marinello E. Fatty acid composition of phospholipids, triglycerides and cholesterol in serum of castrated and estradiol treated rats. Life Sci. 2000;66:1647–1654. doi: 10.1016/s0024-3205(00)00484-7. [DOI] [PubMed] [Google Scholar]
- 23.Clore JN, Li L, Rizzo WB. Effects of fructose and troglitazone on phospholipid fatty acid composition in rat skeletal muscle. Lipids. 2000;35:1281–1287. doi: 10.1007/s11745-000-0644-5. [DOI] [PubMed] [Google Scholar]
- 24.de Antueno RJ, Knickle LC, Smith H, Elliot ML, Allen SJ, Nwaka S, Winther MD. Activity of human Delta5 and Delta6 desaturases on multiple n-3 and n-6 polyunsaturated fatty acids. FEBS Lett. 2001;509:77–80. doi: 10.1016/s0014-5793(01)03135-0. [DOI] [PubMed] [Google Scholar]
- 25.Asadi A, Jorgensen J, Jacobsson A. Elovl1 and p55Cdc genes are localized in a tail-to-tail array and are co-expressed in proliferating cells. J Biol Chem. 2002;277:18494–18500. doi: 10.1074/jbc.M111503200. [DOI] [PubMed] [Google Scholar]
- 26.Keyes SR, Cinti DL. Biochemical properties of cyto-chrome b5-dependent microsomal fatty acid elongation and identification of products. J Biol Chem. 1980;255:11357–11364. [PubMed] [Google Scholar]
- 27.Nagi MN, Cook L, Gherquier D, Cinti DL. Site of inhibition of rat liver microsomal fatty acid chain elongation system by Dec-2-ynoyl coenzyme A. J Biol Chem. 1986;261:13598–13605. [PubMed] [Google Scholar]
- 28.Walker KA, Harwood JL. Evidence for separate elongation enzymes for very-long-chain-fatty-acid synthesis in potato (Solanum tuberosum) Biochem J. 1986;237:41–46. doi: 10.1042/bj2370041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suneja SK, Osei P, Cook L, Nagi MN, Cinti DL. Enzyme site-specific changes in hepatic microsomal fatty acid chain elongation in streptozotocin-induced diabetic rats. Biochim Biophys Acta. 1990;1042:81–85. doi: 10.1016/0005-2760(90)90059-7. [DOI] [PubMed] [Google Scholar]
- 30.Chang JH, Lunt DK, Smith SB. Fatty acid composition and fatty acid elongase and stearoyl-CoA desaturase activities in tissues of steers fed high oleate sunflower seed. J Nutr. 1992;122:2074–2080. doi: 10.1093/jn/122.11.2074. [DOI] [PubMed] [Google Scholar]
- 31.Alegret M, Cerqueda E, Ferrando R, Vazquez M, Sanchez RM, Adzet T, Merlos M, Laguna JC. Selective modification of rat hepatic microsomal fatty acid chain elongation and desaturation by fibrates: relationship with peroxisome proliferation. Br J Pharmacol. 1995;114:1351–1358. doi: 10.1111/j.1476-5381.1995.tb13355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprecher H. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim Biophys Acta. 2000;1486:219–231. doi: 10.1016/s1388-1981(00)00077-9. [DOI] [PubMed] [Google Scholar]
- 33.Pawar A, Botolin D, Mangelsdorf DJ, Jump DB. The role of liver X receptor-alpha (LXR-alpha) in the fatty acid regulation of hepatic gene expression. J Biol Chem. 2003;278:40736–40743. doi: 10.1074/jbc.M307973200. [DOI] [PubMed] [Google Scholar]
- 34.Pan DA, Mater MK, Thelen AP, Peters JM, Gonzalez FJ, Jump DB. Evidence against the peroxisome proliferator-activated receptor alpha (PPARalpha) as the mediator for poly-unsaturated fatty acid suppression of hepatic L-pyruvate kinase gene transcription. J Lipid Res. 2000;41:742–751. [PubMed] [Google Scholar]
- 35.Botolin D, Jump DB. Selective proteolytic processing of rat hepatic sterol regulatory element binding protein-1 (SREBP-1) and SREBP-2 during postnatal development. J Biol Chem. 2003;278:6959–6962. doi: 10.1074/jbc.M212846200. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura MT, Nara TY. Structure, function and dietary regulation of delta6, delta5 and delta9 desaturases. Annu Rev Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- 37.Jump DB, Clarke SD, MacDougald O, Thelen A. Polyunsaturated fatty acids inhibit S14 gene transcription in rat liver and cultured hepatocytes. Proc Natl Acad Sci USA. 1993;90:8454–8458. doi: 10.1073/pnas.90.18.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura MT, Cho HP, Clarke SD. Regulation of hepatic delta-6 desaturase expression and its role in the polyunsaturated fatty acid inhibition of fatty acid synthase gene expression in mice. J Nutr. 2000;130:1561–1565. doi: 10.1093/jn/130.6.1561. [DOI] [PubMed] [Google Scholar]
- 39.Matsuzaka T, Shimano H, Yahagi N, Amemiya-Kudo M, Yo-shikawa T, Hasty AH, Tamura Y, Osuga J, Okazaki H, Iizuka Y, Takahashi A, Sone H, Gotoda T, Ishibashi S, Yamada N. Dual regulation of mouse Delta(5)- and Delta(6)-desaturase gene expression by SREBP-1 and PPARalpha. J Lipid Res. 2002;43:107–114. [PubMed] [Google Scholar]
- 40.Miller CW, Ntambi JM. Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proc Natl Acad Sci USA. 1996;93:9443–9448. doi: 10.1073/pnas.93.18.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fulco AJ, Mead JF. Metabolism of essential fatty acids. VIII Origin of 5,8,11-eicosatrienoic acid in the fat-deficient rat. J Biol Chem. 1959;234:1411–1416. [PubMed] [Google Scholar]
- 42.Holman RT. The ratio of trienoic:tetraenoic acids in tissue lipids as a measure of essential fatty acid requirement. J Nutr. 1960;70:405–410. doi: 10.1093/jn/70.3.405. [DOI] [PubMed] [Google Scholar]
- 43.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girard J, Ferre P, Pegorier J-P, Duee P-H. Adaptations of glucose and fatty acid metabolism during perinatal period and suckling-weaning transition. Physiol Rev. 1992;72:507–562. doi: 10.1152/physrev.1992.72.2.507. [DOI] [PubMed] [Google Scholar]
- 45.Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor-alpha, beta and gamma during rat embryonic development. Endocrinology. 1998;139:2748–2754. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- 46.Cook WS, Jain S, Jia Y, Cao W-Q, Yeldandi AV, Reddy JK, Rao MS. Peroxisome proliferator-activated receptor alpha-responsive genes induced in the newborn, but not perinatal liver of peroxisomal fatty acyl CoA oxidase null mice. Exp Cell Res. 2001;268:70–76. doi: 10.1006/excr.2001.5266. [DOI] [PubMed] [Google Scholar]
- 47.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 48.Bennett MK, Lopez JM, Sanchez HB, Osborne TF. Sterol regulation of fatty acid synthase promoter. Coordinate feedback regulation of two major lipid pathways. J Biol Chem. 1995;270:25578–25583. doi: 10.1074/jbc.270.43.25578. [DOI] [PubMed] [Google Scholar]
- 49.Girard J. Metabolic adaptations to change of nutrition at birth. Biol Neonate. 1990;58(Suppl 1):3–15. doi: 10.1159/000243294. [DOI] [PubMed] [Google Scholar]
- 50.Ren B, Thelen AP, Peters JM, Gonzalez FJ, Jump DB. Polyunsaturated fatty acid suppression of hepatic fatty acid synthase and S14 gene expression does not require peroxisome proliferator-activated receptor alpha. J Biol Chem. 1997;272:26827–26832. doi: 10.1074/jbc.272.43.26827. [DOI] [PubMed] [Google Scholar]
- 51.Girard J, Perdereau D, Foufelle F, Prip-Buus C, Ferre P. Regulation of lipogenic enzyme gene expression by nutrients and hormones. FASEB J. 1994;8:36–42. doi: 10.1096/fasebj.8.1.7905448. [DOI] [PubMed] [Google Scholar]
- 52.Ahima RS, Prabakaran D, Flier JS. Postnatal leptin surge and regulation of circadian rhythm of leptin by feeding. Implications for energy homeostasis and neuroendocrine function. J Clin Invest. 1998;101:1020–1027. doi: 10.1172/JCI1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catalan J, Moriguchi T, Slotnick B, Murthy M, Greiner RS, Salem N., Jr Cognitive deficits in docosahexaenoic acid-deficient rats. Behav Neurosci. 2002;116:1022–1031. doi: 10.1037//0735-7044.116.6.1022. [DOI] [PubMed] [Google Scholar]
- 54.Mater MK, Thelen AP, Pan DA, Jump DB. Sterol response element-binding protein 1c (SREBP1c) is involved in the polyunsaturated fatty acid suppression of hepatic S14 gene transcription. J Biol Chem. 1999;274:32725–32732. doi: 10.1074/jbc.274.46.32725. [DOI] [PubMed] [Google Scholar]
- 55.Liimatta M, Towle HC, Clarke S, Jump DB. Dietary polyunsaturated fatty acids interfere with the insulin/glucose activation of L-type pyruvate kinase gene transcription. Mol Endocrinol. 1994;8:1147–1153. doi: 10.1210/mend.8.9.7838147. [DOI] [PubMed] [Google Scholar]
- 56.Stoeckman AK, Ma L, Towle HC. Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. J Biol Chem. 2004;279:15662–15669. doi: 10.1074/jbc.M311301200. [DOI] [PubMed] [Google Scholar]
- 57.Johnson PV. Dietary fat, eicosanoids and immunity. Adv Lipid Res. 1985;21:103–141. doi: 10.1016/b978-0-12-024921-3.50010-1. [DOI] [PubMed] [Google Scholar]
- 58.Reddy JK, Mannaerts GP. Peroxisomal lipid metabolism. Annu Rev Nutr. 1994;14:343–370. doi: 10.1146/annurev.nu.14.070194.002015. [DOI] [PubMed] [Google Scholar]


