Abstract
Carnitine is essential for the transfer of long-chain fatty acids across the mitochondrial membrane for subsequent beta-oxidation. A defect in the high affinity carnitine transporter OCTN2 causes autosomal recessive primary carnitine deficiency that can present with hypoketotic hypoglycemia, mainly in infancy, or cardiomyopathy. Heterozygotes for primary carnitine deficiency can have mildly reduced plasma carnitine levels and can develop benign cardiac hypertrophy. In animal models, heterozygotes for this disease have a higher incidence of cardiomyopathy with aging. This study tested whether heterozygosity for primary carnitine deficiency was associated with cardiomyopathy. The frequency of mutations in the SLC22A5 gene encoding the OCTN2 carnitine transporter was determined in 324 patients with cardiomyopathy and compared to that described in the normal population. Missense variations identified in normal controls and patients with cardiomyopathy were expressed in Chinese Hamster Ovary cells to confirm a functional effect. Exons 2–10 of the SLC22A5 gene were amplified by PCR in the presence of LCGreen I™ and analyzed by dye-binding/high-resolution thermal denaturation. Exon 1 of the gene was sequenced in all patients. Heterozygosity for a few variants (L144F, T264M, I312V, E317K and R488H) was found in 6/324 patients with cardiomyopathy. Expression of these variants in CHO cells indicated that T264M decreased, E317K increased, while L144F, I312V, and R488H did not significantly affect carnitine transport. Expression in CHO cells of all the variants identified in a normal population indicated that only 2 had a functional effect (L17F and Y449D), while L144F, V481I, V481F, M530V, and P549S did not change significantly carnitine transport. The frequency of variants affecting carnitine transport was 2/324 patients with cardiomyopathy (0.61%) not significantly different from frequency of 3/270 (1.11%) in the general population. These results indicate that heterozygosity for primary carnitine deficiency is not more frequent in patients with unselected types of cardiomyopathy and is unlikely to be an important cause of cardiomyopathy in humans.
Keywords: SLC22A5, OCTN2, carnitine transporter, cardiomyopathy, carnitine
Primary carnitine deficiency (OMIM# 212140) is an autosomal recessive disorder of the carnitine cycle that results in defective fatty acid oxidation [1, 2]. It has a frequency of ranging from 1:40,000 to 1:120,000 newborns in different parts of the world [3–5] and is possibly the second most frequent disorder of fatty acid oxidation after medium chain acyl CoA dehydrogenase deficiency. About 0.5–1% of the population carries one abnormal allele for this condition [3], although the precise frequency of carriers in the USA is still unclear.
Primary carnitine deficiency is caused by defective activity of the novel organic cation/carnitine transporter OCTN2 [6, 7], resulting in urinary carnitine wasting, low serum carnitine levels, and decreased intracellular carnitine accumulation [8]. Carnitine is essential for the transfer of long-chain fatty acids from the cytosol to mitochondria for subsequent beta oxidation and its lack impairs the ability to use fat as fuel during periods of fasting or stress. This can result in an acute metabolic decompensation, most often early in life, with hepatic encephalopathy, hypoketotic hypoglycemia, Reye syndrome, and sudden infant death, or in a more insidious presentation, which may be very early in childhood but is more often of later onset, with cardiomyopathy [9]. The gene for primary carnitine deficiency, SLC22A5 (MIM# 603377) encoding OCTN2, maps to chromosome 5q31 and different mutations have been identified in patients with primary carnitine deficiency (summarized in [9]).
Heterozygotes for primary carnitine deficiency have mildly reduced plasma carnitine levels [3, 8]. Over time, heterozygotes develop benign cardiac hypertrophy and it is unclear whether they have a higher incidence of cardiomyopathy or heart disease [3]. In one of our original families, a maternal aunt of the proband died of an unusual type of cardiomyopathy non responsive to conventional management [8].
The jvs (juvenile visceral steatosis) mouse is a model for primary carnitine deficiency [10]. Affected mice accumulate fat in visceral tissue and die of hypoglycemia or cardiomyopathy as patients with primary carnitine deficiency [10, 11]. Heterozygous mice have increased deposits of triglycerides and cholesterol and a higher rate of cardiomyopathy with aging [11, 12]. The presence of additional risk factors, such as hypertension, can increase the risk for cardiomyopathy in heterozygous jvs−/+ mice [13]. It is unclear whether heterozygosity for primary carnitine deficiency is a risk factor for cardiomyopathy in humans.
In this paper, we evaluate the SLC22A5 gene encoding the OCTN2 carnitine transporter in patients with cardiomyopathy. Our results do not support a major role of the carnitine transporter gene in human cardiomyopathy.
MATERIALS AND METHODS
The protocol for DNA studies was approved by the University of Colorado and by the University of Utah Institutional Review Board. Informed consent was obtained from all patients or their parents prior to DNA studies. GenBank sequence AB016625.1 was used as reference for the gene, NM_003060.2 was used as the reference sequence for the cDNA. Genomic DNA was obtained from 324 patients with cardiomyopathy part of the cardiomyopathy registry [14, 15]. About 10% of these patients had hypertrophic cardiomyopathy, while the rest had dilated cardiomyopathy.
Exon 1 of the SCL22A5 carnitine transporter gene was amplified by PCR and sequenced [16]. Exons 2–10 of the SCL22A5 carnitine transporter gene were amplified by PCR in the presence of the fluorescent dye LCGreen™ I and analyzed in a High Resolution Melter HR-1 (Idaho Technology Inc., Salt Lake City, UT) [17]. Each 96-well plate included a DNA sample with a known mutation in the exon screened (positive control) and a blank sample (no DNA) to control for contamination or nonspecific amplification. Samples generating abnormal melting curves (expected in heteroduplexes) were sequenced to identify putative mutations. The mutations identified were confirmed by amplifying and sequencing an independent PCR product.
The mammalian expression vector with the green fluorescent protein fused to the C-terminus of OCTN2 was generated as previously described [18]. Mutations were introduced in the OCTN2 cDNA by site-directed mutagenesis using the Quick Change system (Stratagene, La Jolla, CA, USA, http://www.stratagene.com) following the manufacturer’s instructions. The final clones were sequenced to confirm the presence of the mutation and the absence of PCR artifacts. The clones were transfected into CHO cells using lipofectamine [18, 19]. Cells were selected for 2 weeks in 0.8 mg/ml of G418 and then used for the transport assay. Expression of the transgene was verified by the presence of green fluorescent protein detected by confocal microscopy.
Chinese hamster ovary (CHO) cells were grown in Ham F12 medium supplemented with 6% fetal bovine serum. Carnitine (0.5 µM) transport was measured at 37 °C for 1 h in CHO cells as described previously [8, 16, 20–22]. Nonsaturable carnitine transport was measured in the presence of 2 mM cold carnitine and was subtracted from total transport to obtain saturable transport. Data are expressed as means ± SD and compared using analysis of variance.
RESULTS
Direct DNA sequencing of exon 1 of the SLC22A5 gene did not identify any significant variation in the DNA of patients with cardiomyopathy. Screening of the remaining exons (2–10) of the SLC22A5 gene by high-resolution melting analysis in 324 patients with cardiomyopathy identified 38 variants out of 2,916 exons screened. An example of the melting curve of selected exons is shown in Fig. 1. DNA sequencing identified polymorphisms not affecting the amino acid residues and described in the normal population [23] in the DNA of 31 patients and variants of unknown significance in 7 patients (Table I). The L144F variant has been reported in normal controls, while T264M, E317K, and I312V have not yet been reported. R488H has been reported in association with A142S in a patient with primary carnitine deficiency [16].
Fig. 1. Melting profile of normal and abnormal exons of the carnitine transporter gene in patients with cardiomyopathy.
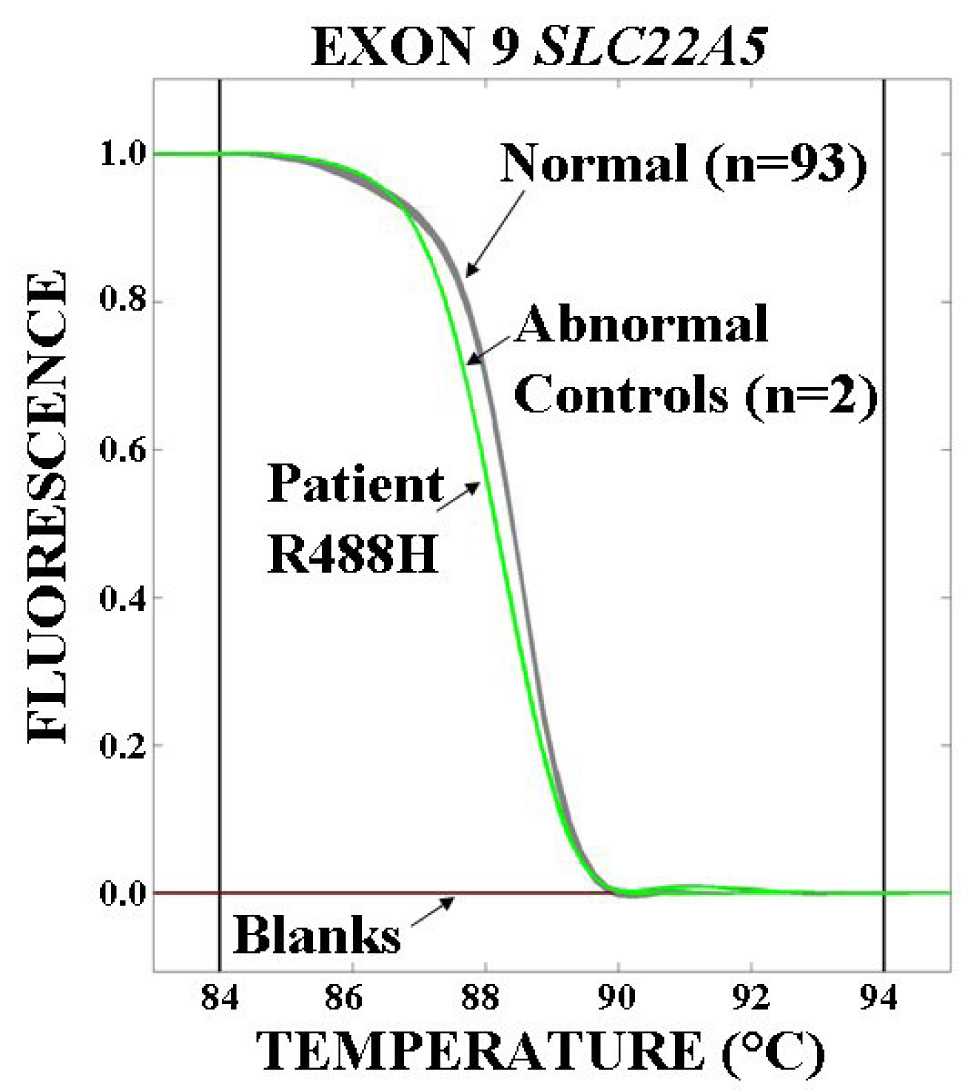
Exons 2–10 of the SLC22A5 gene encoding the OCTN2 carnitine transporter were amplified by PCR in the presence of the fluorescent dye LCGreen™ I and analyzed in a High Resolution Melter. Exon 9 is shown in the figure. Two blanks (flat lines) and two samples with known mutations (curves to the left with lower melting temperatures) were included in each run. Samples not grouping with the controls (number of normal control samples indicated in parentheses) were sequenced and variations in sequences were confirmed in an independent PCR product. In the example shown, all the variants (including the patient) were heterozygous for the same base pair change creating the same substitution (R488H) and appear as a single curve.
Table I.
Variations in the SLC22A5 gene in patients with cardiomyopathy.
| Code | Exon | Variation | Amino acid |
|---|---|---|---|
| 04-0906 | 2 | c.430 C>T | L144F |
| 04-0243 | 4 | c.791 C>T | T264M |
| 02-0212 | 5 | c.949 G>A | E317K |
| 03-1663 | 5 | c.934 A>G | I312V |
| 03-1661 | 5 | c.934 A>G | I312V |
| 02-1034 | 9 | c.1463 G>A | R488H |
| 02-2695 | 9 | c.1463 G>A | R488H |
To determine whether these variations had a functional effect, they were recreated in a mammalian expression vector and expressed in CHO cells. Carnitine transport was then measured and compared to the wild-type OCTN2 (Fig. 2A). T264 M significantly decreased while E317K significantly increased carnitine transport (Fig. 2B). Both patients carrying these mutations had dilated cardiomyopathy.
Fig. 2. Carnitine transport by CHO cells expressing missense mutations identified in patients with cardiomyopathy (A) and in normal controls (B).
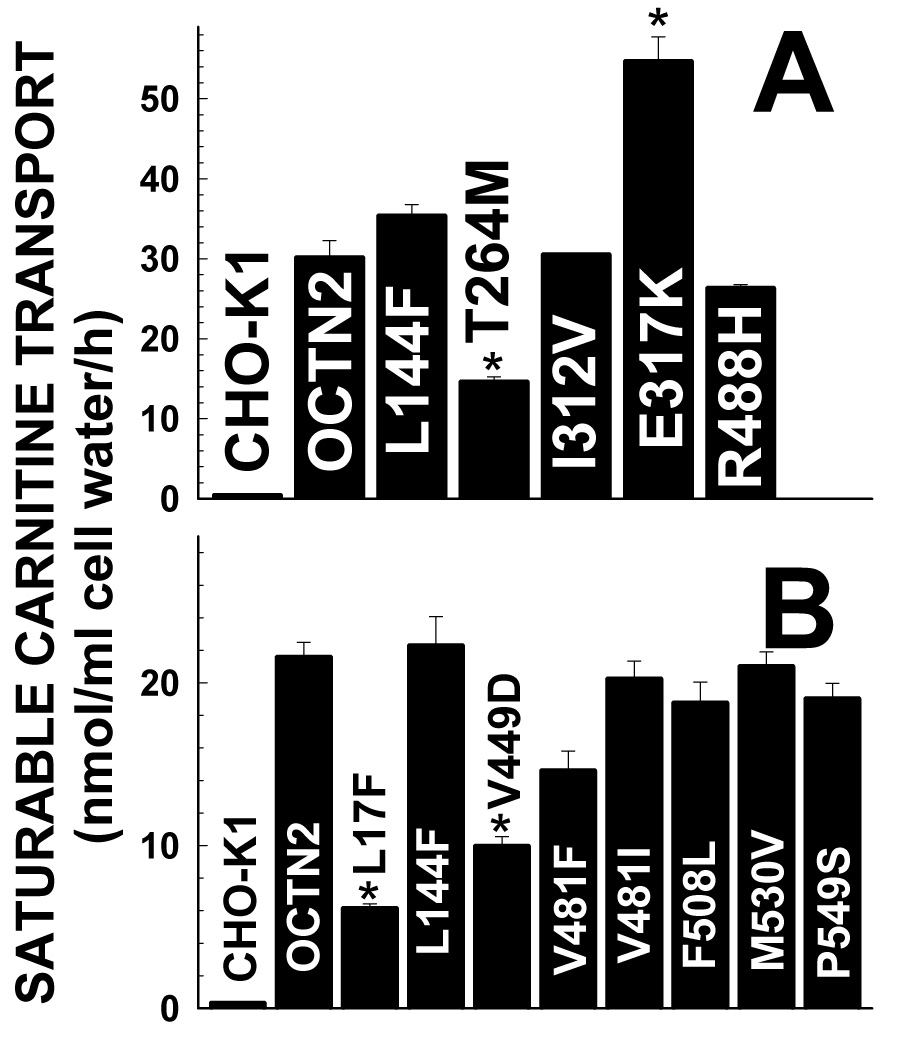
Carnitine (0.5 µM) transport was measured for 1 h at 37°C. Data are means ± SD of 6 observations. *p<0.01 versus wild-type OCTN2 using analysis of variance.
Variations in the SLC22A5 gene in the general population were recently reported [23]. It is unclear how many of these variations are normal polymorphisms or might represent mutations. We evaluated in our expression system the missense substitutions identified in normal controls (Fig. 2B). Only the L17F and Y449D substitutions significantly impaired carnitine transport (Fig. 2B), while L144F, V481F, V481I, F508L, M530V, and P549S did not significantly affect carnitine transport.
The frequency of variations in SLC22A5 gene significantly affecting carnitine transport (when the mutations are expressed in CHO cells) was 3/270 (1.11%) in the normal control population. In our patients with cardiomyopathy, the frequency of mutations was 2/324 (0.61%), not significantly different from that of the general population with odds ratio of 0.55 (95%CI, 0.09 to 3.33).
In one of our families, the sister of our proband 04-0243 (Table I) with the T264M mutation also had cardiomyopathy. DNA sequencing in family members indicated that the proband, but not her similarly affected sister carried the T264M mutation. By contrast, the unaffected mother of the proband carried the T264M mutation and had a normal echocardiogram in her mid seventies. These results indicate that variations in the SLC22A5 gene are an unlikely predisposition factor for cardiomyopathy in this family.
DISCUSSION
Primary carnitine deficiency impairs the accumulation of carnitine within organs and tissues [9]. In the heart, carnitine is essential for normal fatty acid beta-oxidation and even partial deficiency could lead to organ dysfunction. Heterozygous animal models of primary carnitine deficiency have a higher rate of cardiomyopathy with aging [11, 12]. Humans heterozygous for primary carnitine deficiency have mildly reduced plasma carnitine levels due to increased urinary losses [8] and might develop cardiac hypertrophy over time [3]. In patients with primary carnitine deficiency dilated cardiomyopathy is more frequently found [9] while cardiac hypertrophy can be seen in heterozygotes for this condition [3]. To test whether heterozygosity for primary carnitine deficiency is a risk factor for cardiomyopathy in humans, we studied the SLC22A5 gene encoding the OCTN2 carnitine transporter in 324 patients with cardiomyopathy (dilated and hypertrophic) using a combination of high-resolution melting [17] and direct sequencing (Fig. 1). High-resolution melting is exquisitely sensitive to heterozygous sequence changes [17]. Only a limited number of variations were identified in patients with cardiomyopathy (Table I) and most variations did not have any significant functional effect on carnitine transport (Fig. 2A). Only two changes, T264M and E317K significantly modified carnitine transport. T264M impaired carnitine transport, while E317K enhanced functional activity (Fig. 2A). All mutations reported to date in patients with classic primary carnitine deficiency impair carnitine transport [9]. It is not know whether a gain of function could result in any type of pathology. Since the changes in carnitine transport induced by E317K were significant, we conservatively assumed that it could be associated with a phenotype. With this assumption, only 2/324 patients with cardiomyopathy were heterozygous for mutations in the carnitine transporter gene. This compares with a rate of 0.5–1% in the general population estimated from the 1:40,000–1:120,000 prevalence of primary carnitine deficiency [3–5] and with a frequency of 1.11% obtained from the analysis of 270 normal controls [23] and our expression studies of the changes identified (Fig. 2B). These results agree with previous expression studies in HEK 293 cells [23] and in CHO cells [19]. Statistical analysis indicated that heterozygosity for primary carnitine deficiency was not a risk factor for cardiomyopathy (odds ratio=0.55 with a 95% confidence interval of 0.09 to 3.33, i.e. overlapping with the control group).
Cardiomyopathy, most often dilated, is one of the cardinal manifestations of the recessive disorder primary carnitine deficiency in children [9, 20]. It less clear whether cardiomyopathy can be observed in adults with primary carnitine deficiency. With the advent of expanded newborn screening, we have recently identified a numbers of adult mothers with this condition [24]. Despite very low plasma carnitine levels, low carnitine transport by fibroblasts of affected mothers (<10% of normal), and the demonstration of homozygosity or compound heterozygosity for mutations in the SLC22A5 gene, none of the mothers had cardiomyopathy when studied by echocardiogram or electrocardiogram [24]. It is worth noting, however, that one of the mothers reported had severe arrhythmia requiring placement of a pace-maker [24]. It is possible that the phenotype of this disease changes as people escape childhood presentation. This would explain the lack of association described here between cardiomyopathy and heterozygous variations in the carnitine transporter gene, but would not exclude the possibility that this gene could act as a modifier in other types of heart disease. It is also possible that given the large number of causes, genetic or environmental, for cardiomyopathy [25], lack of carnitine could be responsible only of a very limited number of cases, not identified in the current study.
ACKNOWLEDGEMENTS
We wish to thank Idaho Technology Inc. for allowing the use of the HR-1 High Resolution Melter and for valuable help in optimizing the screening assay. We also thank the Familial Cardiomyopathy Study subjects for their participation. This study was supported by Grant-in-Aid 0455086Y from the American Heart Association and in part by grants DK 53824 and 1K23Hl67915-01A1 from the National Institutes of Health.
Funding: Supported by Grant-in-Aid 0455086Y from the American Heart Association and in part by grants R01 DK 53824 and 1K23Hl67915-01A1 from the National Institutes of Health
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Roe C, Ding J. Mitochondrial fatty acid oxidation disorders. In: Scriver C, Beaudet A, Sly W, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 2297–2326. [Google Scholar]
- 2.Scaglia F, Longo N. Primary and secondary alterations of neonatal carnitine metabolism. Semin Perinatol. 1999;23:152–161. doi: 10.1016/s0146-0005(99)80047-0. [DOI] [PubMed] [Google Scholar]
- 3.Koizumi A, Nozaki J, Ohura T, Kayo T, Wada Y, Nezu J, Ohashi R, Tamai I, Shoji Y, Takada G, Kibira S, Matsuishi T, Tsuji A. Genetic epidemiology of the carnitine transporter OCTN2 gene in a Japanese population and phenotypic characterization in Japanese pedigrees with primary systemic carnitine deficiency. Hum Mol Genet. 1999;8:2247–2254. doi: 10.1093/hmg/8.12.2247. [DOI] [PubMed] [Google Scholar]
- 4.Wilcken B, Wiley V, Sim KG, Carpenter K. Carnitine transporter defect diagnosed by newborn screening with electrospray tandem mass spectrometry. J Pediatr. 2001;138:581–584. doi: 10.1067/mpd.2001.111813. [DOI] [PubMed] [Google Scholar]
- 5.Wilcken B, Wiley V, Hammond J, Carpenter K. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N Engl J Med. 2003;348:2304–2312. doi: 10.1056/NEJMoa025225. [DOI] [PubMed] [Google Scholar]
- 6.Tamai I, Ohashi R, Nezu J, Yabuuchi H, Oku A, Shimane M, Sai Y, Tsuji A. Molecular and functional identification of sodium ion-dependent, high affinity human carnitine transporter OCTN2. J Biol Chem. 1998;273:20378–20382. doi: 10.1074/jbc.273.32.20378. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Prasad PD, Leibach FH, Ganapathy V. cDNA sequence, transport function, and genomic organization of human OCTN2, a new member of the organic cation transporter family. Biochem Biophys Res Commun. 1998;246:589–595. doi: 10.1006/bbrc.1998.8669. [DOI] [PubMed] [Google Scholar]
- 8.Scaglia F, Wang Y, Singh RH, Dembure PP, Pasquali M, Fernhoff PM, Longo N. Defective urinary carnitine transport in heterozygotes for primary carnitine deficiency. Genet Med. 1998;1:34–39. doi: 10.1097/00125817-199811000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Longo N, Amat di San Filippo C, Pasquali M. Disorders of carnitine transport and the carnitine cycle. Am J Med Genet C Semin Med Genet. 2006;142:77–85. doi: 10.1002/ajmg.c.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuwajima M, Kono N, Horiuchi M, Imamura Y, Ono A, Inui Y, Kawata S, Koizumi T, Hayakawa J, Saheki T, et al. Animal model of systemic carnitine deficiency: analysis in C3H-H-2 degrees strain of mouse associated with juvenile visceral steatosis. Biochem Biophys Res Commun. 1991;174:1090–1094. doi: 10.1016/0006-291x(91)91532-h. [DOI] [PubMed] [Google Scholar]
- 11.Lahjouji K, Elimrani I, Wu J, Mitchell GA, Qureshi IA. A heterozygote phenotype is present in the jvs +/− mutant mouse livers. Mol Genet Metab. 2002;76:76–80. doi: 10.1016/s1096-7192(02)00017-3. [DOI] [PubMed] [Google Scholar]
- 12.Xiaofei E, Wada Y, Dakeishi M, Hirasawa F, Murata K, Masuda H, Sugiyama T, Nikaido H, Koizumi A. Age-associated cardiomyopathy in heterozygous carrier mice of a pathological mutation of carnitine transporter gene, OCTN2. J Gerontol A Biol Sci Med Sci. 2002;57:B270–B278. doi: 10.1093/gerona/57.7.b270. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi R, Asai T, Murakami H, Murakami R, Tsuzuki M, Numaguchi Y, Matsui H, Murohara T, Okumura K. Pressure overload-induced cardiomyopathy in heterozygous carrier mice of carnitine transporter gene mutation. Hypertension. 2007;50:497–502. doi: 10.1161/HYPERTENSIONAHA.107.088609. [DOI] [PubMed] [Google Scholar]
- 14.Mestroni L, Maisch B, McKenna WJ, Schwartz K, Charron P, Rocco C, Tesson F, Richter A, Wilke A, Komajda M. Guidelines for the study of familial dilated cardiomyopathies. Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy. Eur Heart J. 1999;20:93–102. doi: 10.1053/euhj.1998.1145. [DOI] [PubMed] [Google Scholar]
- 15.Ku L, Feiger J, Taylor M, Mestroni L. Cardiology patient page. Familial dilated cardiomyopathy. Circulation. 2003;108:e118–e121. doi: 10.1161/01.CIR.0000097493.70422.50. [DOI] [PubMed] [Google Scholar]
- 16.Amat di San Filippo C, Pasquali M, Longo N. Pharmacological rescue of carnitine transport in primary carnitine deficiency. Hum Mutat. 2006;27:513–523. doi: 10.1002/humu.20314. [DOI] [PubMed] [Google Scholar]
- 17.Dobrowolski SF, McKinney JT, Amat di San Filippo C, Giak Sim K, Wilcken B, Longo N. Validation of dye-binding/high-resolution thermal denaturation for the identification of mutations in the SLC22A5 gene. Hum Mutat. 2005;25:306–313. doi: 10.1002/humu.20137. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Meadows TA, Longo N. Abnormal sodium stimulation of carnitine transport in primary carnitine deficiency. J Biol Chem. 2000;275:20782–20786. doi: 10.1074/jbc.M000194200. [DOI] [PubMed] [Google Scholar]
- 19.Amat Di San Filippo C, Longo N. Tyrosine Residues Affecting Sodium Stimulation of Carnitine Transport in the OCTN2 Carnitine/Organic Cation Transporter. J Biol Chem. 2004;279:7247–7253. doi: 10.1074/jbc.M309171200. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Kelly MA, Cowan TM, Longo N. A missense mutation in the OCTN2 gene associated with residual carnitine transport activity. Hum Mutat. 2000;15:238–245. doi: 10.1002/(SICI)1098-1004(200003)15:3<238::AID-HUMU4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Taroni F, Garavaglia B, Longo N. Functional analysis of mutations in the OCTN2 transporter causing primary carnitine deficiency: lack of genotype-phenotype correlation. Hum Mutat. 2000;16:401–407. doi: 10.1002/1098-1004(200011)16:5<401::AID-HUMU4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Korman SH, Ye J, Gargus JJ, Gutman A, Taroni F, Garavaglia B, Longo N. Phenotype and genotype variation in primary carnitine deficiency. Genet Med. 2001;3:387–392. doi: 10.1097/00125817-200111000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Urban TJ, Gallagher RC, Brown C, Castro RA, Lagpacan LL, Brett CM, Taylor TR, Carlson EJ, Ferrin TE, Burchard EG, Packman S, Giacomini KM. Functional genetic diversity in the high-affinity carnitine transporter OCTN2 (SLC22A5) Molecular pharmacology. 2006;70:1602–1611. doi: 10.1124/mol.106.028126. [DOI] [PubMed] [Google Scholar]
- 24.Schimmenti LA, Crombez EA, Schwahn BC, Heese BA, Wood TC, Schroer RJ, Bentler K, Cederbaum S, Sarafoglou K, McCann M, Rinaldo P, Matern D, di San Filippo CA, Pasquali M, Berry SA, Longo N. Expanded newborn screening identifies maternal primary carnitine deficiency. Mol Genet Metab. 2006;90(4):441–445. doi: 10.1016/j.ymgme.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Wynne J, Braunwald E. Cardiomyopathy and myocarditis. In: Kasper D, Braunwald E, Fauci A, Hauser S, Longo D, Jameson J, editors. Harrison's Princples of Internal Medicine. New York: McGraw-Hill; 2005. pp. 1408–1414. [Google Scholar]


