Abstract
Survival of Porphyromonas gingivalis in the constantly changing oral environment depends on its ability to alter gene expression. We demonstrate here that P. gingivalis activates superoxide dismutase expression in response to oxidative stress and represses expression of FimA, a subunit of major fimbriae. Coordinated expression of fimA and sod is regulated by the redox-sensing transcription factor OxyR. Mutations in the oxyR gene result in a decreased expression of sod and in an elevated expression of fimA. In addition, we provide evidence that regulation of expression of fimA and sod by OxyR is mediated by direct interaction of OxyR and the promoters of these two genes. These results suggest that OxyR plays an important role in regulation of expression of virulence genes in P. gingivalis.
Keywords: fimbriae, SOD, OxyR
Introduction
Porphyromonas gingivalis is a secondary colonizer of dental plaque, and is significantly more prevalent in both supra- and subgingival plaque samples from periodontitis subjects in comparison with samples from healthy subjects (Ximenez-Fyvie et al., 2000a). The organism is associated with several forms of periodontitis (Socransky et al., 2002). It has been well recognized that expression of fimA, encoding a subunit (FimA) of long (major) fimbriae, is required for colonization of P. gingivalis on a variety of oral surfaces (Lamont & Jenkinson, 1998). A P. gingivalis strain with fimA deficiency has a diminished capacity to adhere to human gingival fibroblasts and epithelial cells (Hamada et al., 1994). FimA also participates in coaggregation with various early plaque-forming bacteria, such as Actinomyces viscosus, Streptococcus gordonii, Streptococcus oralis and Treponema denticola (Goulbourne & Ellen, 1991; Lamont et al., 1993; Amano et al., 1997). Expression of the fimA gene is modulated in P. gingivalis in response to environmental cues, such as temperature and ion supply (Amano et al., 1994; Xie et al., 1997). We have reported earlier that fimA expression is repressed in the presence of another oral bacterium, Streptococcus cristatus (Xie et al., 2000). A two-component system (fimS/fimR), first reported by Hayashi et al. (2000), is found to be involved in the fimbriation of P. gingivalis. Sequence analysis demonstrates that FimS/FimR contain conserved sequences that have been suggested to be phosphorylation sites for transmitters and receivers. Disruption of fimS or fimR results in significant reduction in FimA production. Recently, Nishikawa et al. (2004) reported that FimR controls the expression of several genes clustered around the fimA locus, and that the recombinant FimR does not bind to the fimA gene but binds to the first gene in the fimA cluster, pg2130.
Pathogenicity of P. gingivalis involves the products of many genes. One important virulence factor is superoxide dismutase (SOD), which is required for modest aerotolerance of P. gingivalis. A sod-deficient mutant is more sensitive to atmospheric oxygen (Lynch & Kuramitsu, 1999). It is reported that P. gingivalis can alter its expression of FimA and SOD under the conditions observed in an inflamed periodontal pocket, such as an elevated temperature (Amano et al., 1994). Expression of the fimA gene is decreased in P. gingivalis grown at 39 °C, whereas expression of sod is increased, suggesting a possible coordinated regulation of fimA and sod. Recent studies have shown that expression of sod is positively regulated by a redox-sensing transcription activator, OxyR (Diaz et al., 2006; Ohara et al., 2006). It remains unknown whether P. gingivalis fimA is one of the OxyR regulons. We hypothesize that expression of fimA and sod may be coordinately regulated through OxyR. Here we report that P. gingivalis selectively up-regulates expression of sod and represses fimA expression under aerobic conditions, and that the oxyR-deficient mutant increases fimA expression and promotes biofilm formation.
Materials and methods
Bacterial strains and growth conditions
Porphyromonas gingivalis strains were grown from frozen stocks in Trypticase soy broth (TSB) or on TSB blood agar plates, supplemented with yeast extract (1 mg mL−1), hemin (5 μg mL−1), and menadione (1 μg mL−1), at 37 °C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). Escherichia coli DH5α was used as the host for plasmids. Escherichia coli strains were grown in Luria–Bertani broth at 37 °C. Antibiotics were used when appropriate, at the following concentrations: gentamicin (100 μg mL−1), erythromycin (10 μg mL−1), ampicillin (50 μg mL−1), kanamycin (50 μg mL−1), and tetracycline (5 μg mL−1).
Experimental oxidative stress conditions
Porphyromonas gingivalis 33 277 (2 × 106) were spotted on TSB blood agar plates and grown anaerobically for 48 h. The cells were then divided into two groups. One group was continuously incubated in the anaerobic chamber for another 3, 6, or 24 h. The other group was exposed to air at 37 °C for the same time periods.
RNA isolation and reverse transcription (RT)-PCR
Porphyromonas gingivalis strains were grown to late-exponential phase (OD600 nm, 1.0–1.2) in 5 mL of TSB. Bacteria were harvested by centrifugation at 10 000 g and mixed in Trizol Reagent (Invitrogen, Carlsbad, CA). The RNA in the supernatant was then purified using an RNeasy mini spin column (Qiagen, Valencia, CA). RNA samples were digested on the column with RNAse-free DNAse. The total RNA was tested using an Agilent 2100 Bioanalyzer to insure the quality of the samples. RT-PCR was conducted using SuperScript™ One-Step RT-PCR Systems (Invitrogen) with about 10 ng of total RNA as a template, and the conditions indicated by the supplier, using 26 PCR cycles. For each reaction, a negative control with Taq polymerase and without reverse transcriptase was included. The RT-PCR products were visualized by 1.2% agarose gel.
Construction of the oxyR mutant
An insertional oxyR mutant was generated using ligation-independent cloning (LIC) of PCR mediated mutagenesis (LIC-PCR) (Aslanidis & de Jong, 1990; Xie et al., 2007). A 2.1-kb ermF-ermAM cassette was introduced into the oxyR gene by three steps of PCR to yield a 3884-bp oxyR-erm-oxyR DNA fragment as described previously (Fletcher et al., 1995). This 3884-bp fragment was then introduced into P. gingivalis 33 277 by electroporation (Wu et al., 2007). The oxyR deficient mutants were constructed via a double crossover event that replaces the oxyR with the oxyR-erm-oxyR into the 33 277 chromosome. The mutants were selected on TSB plates containing erythromycin (5 μg mL−1). The insertional mutation was confirmed by PCR analysis, and the mutants were designated as P. gingivalis OxyRE.
Production of OxyR recombinant protein
The DNA fragment encoding OxyR was amplified by PCR with primers rOxyR-F and rOxyR-R, which produced a 927-bp PCR product (Table 1). The PCR products were then cloned into pCRII-TOPO (Invitrogen, Carlsbad, CA). The Recombinant OxyR (rOxyR) was expressed in E. coli using a pET protein expression system (Novagen, Madison, WI). The DNA fragment of oxyR was subcloned into the pET-30b downstream of a histidine tag. The rOxyR was expressed in E. coli BL21 (DE3) cells carrying the pET-30b/OxyR plasmid in the presence of isopropyl-β-d-thiogalactopyranoside and kanamycin. His-tagged rOxyR was purified with Ni2+-charged His-bind resin (Novagen, Madison, WI). The His-tag on the recombinant protein was cleaved with enterokinase and removed by His-bind resin. Enterokinase was then removed using Ekapture agarose. The rOxyR was used to generate rabbit anti-OxyR antibody (Lampire, Ottsville, PA).
Table 1.
Oligonucleotide primers used in this study
| Primer name | Primer sequences (5′–3′) | Applications |
|---|---|---|
| fimA-F | CGGAACGAATAACCCAGAGA | For RT-PCR of fimA |
| fimA-R | CTGACCAACGAGAACCCACT | |
| sod-F | AATTCCACCACGGTAAGCAC | For RT-PCR of sod |
| sod-R | GAGCCGAATTGTTTGTCGAT | |
| oxyRF | TCGCAAGCCAAGCAAATAC | For RT-PCR of oxyR |
| oxyRR | GACACGAGGCAGGAGATAGG | |
| pg1737F | ATGAATCCGATCCGCCACCAC | For RT-PCR of pg1737 |
| Pg1737R | GCCTCCCATCCCAAAGCACT | |
| pg0270(1)F | GGGCTGCGATGAAGAAAGAT | For creating the oxyR mutant |
| pg0270(2)R | GATTCTTCCGCGCATACACT | |
| pg0270(1)F2 | TCGAGAAGAGAATCCTGATGTG | |
| pg0270(2)R2 | TCCGAACAGCAAAAGAACAG | |
| pg0270(1)R-erm | GATGTTGCAAATACCGATGAGCATTCGAGCTGCTGTATATTC | |
| pg0270(2)F-erm | CCTCTAGAGTCGACCTGCAGACAGGGCAGCATTTGGCTTG | |
| erm-F | GCTCATCGGTATTTGCAACA | |
| erm-R | CTGCAGGTCGACTCTAGAGG | |
| rOxyR-F | ATGAATATACAGCAGCTCG | For production of OxyR recombinant protein |
| rOxyR-R | TCAAGCCAAATGCTGCCCT | |
| fimAProm-F | CGACGCTATATGCAAGACAA | For generating biotin-labeled fimA promoter region |
| fimAProm-R | Bio-TGTAACGGGTTCTGCCTCGT | |
| fimAProm-RC | TGTAACGGGTTCTGCCTCGT | For generating cold fimA promoter region |
| sodProm-F | Bio-CTTTTCGCCCGAAGGTTTTATC | For generating biotin-labeled sod promoter region |
| sodProm-R | AACGTCTGATTTTTATTGTAATTAAG | |
| sodProm-FC | CTTTTCGCCCGAAGGTTTTATC | For generating cold sod promoter region |
| mfaProm-F | CTCTCGCGAGGGTCAATATC | For generating biotin-labeled sod promoter region |
| mfaProm-R | Bio-CGTCTTACCGGCTTCCCTAT | |
| mfaProm-RC | CGTCTTACCGGCTTCCCTAT | For generating cold sod promoter region |
| 16S rRNAF | TGTAGATGACTGATGGTGAAA | For real time PCR |
| 16S rRNAR | ACTGTTAGCAACTACCGATGT |
Chromatin immunoprecipitation (ChIP) qPCR assay
A ChIP assay was conducted following the description by (Nishikawa et al., 2004). Briefly, formaldehyde was added to the P. gingivalis 33 277 culture (20 mL) to a final concentration of 1%. The crossing-link reaction was stopped by the addition of glycine (125 mM). Cells were resuspended in 2 mL of lysis buffer (20 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.5 mg mL−1 TLCK, 10 mg mL−1 lysozyme) for 10 min at room temperature, followed by the addition of an equal volume of 2 × immunoprecipitation (IP) buffer (0.1 M Tris-HCl pH 7.0, 0.3 M NaCl, 2% Triton X-100, 0.2% sodium deoxycholate). Cells were sonicated to fragment chromosomal DNA which was used as the ‘input’ fraction for the ChIP assay.
Anti-OxyR antibody (25 μL mL−1) was added to the input fraction and rotated overnight at 4 °C, and complexes were then incubated for 1 h with 20 μL bed volume of pre-equilibrated protein A Sepharose CL-4B beads (Sigma). After washing, antibody–protein–DNA complexes were treated with 30 μL elution buffer (1% sodium dodecylsulphate, TE pH 8.0, 10 mM dithiothreitol) for 30 min at 37 °C. The eluates were used as the output fraction for real-time PCR analyses. Preimmune rabbit serum was used as a control. Input and output fractions were treated with RNAse A (10 μg mL−1), and proteinase K (2 μg mL−1, Sigma). DNA samples were precipitated by ethanol and resuspended in 30 μL dH2O.
Real-time PCR analysis of ChIP samples was performed using the QuantiTect SYBR Green PCR Kit (Qiagen) on the iCycler MyiQ™ Real-Time PCR detection system (Bio-Rad Laboratories Inc.) according to the manufacturer's instructions. Primers were designed to amplify promoter sequences of fimA, sod, and pg1737. Primers are listed in Table 1. Amplification reactions consisted of an initial activation at 95 °C for 15 min, and 40 cycles of 94 °C for 15 s, 58 °C for 30 s, and 72 °C for 30 s. Data were collected during the extension step and were expressed in arbitrary fluorescence units per cycle. A melting curve was used at the end to confirm that there was only one peak and only one product. Enrichment values (fold) were calculated according to output/input (Nishikawa et al., 2004).
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed using the LightShift Chemiluminescent EMSA Kit (PIERCE, Rockford, IL) as described previously (Wu et al., 2007). Biotin labeled DNA fragments were generated using 5′ biotin incorporated primers (Table 1). Binding of rOxyR to DNA was carried out in a 20-μL reaction mixture containing 20 fmol biotin-labeled DNA, 10 mM Tris, pH 7.5, 50 mM KCl, 1 mM dithiothreitol, 10 ng μL−1 poly (dI-dC), 2% glycerol, 0.05% NP-40, 2 mM MgCl with various amounts of purified rOxyR protein (0, 2.5, 5 and 10 μg) at room temperature for 30 min. Samples were then loaded and run into a 5% nondenaturing polyacrylamide gel in 0.5 × Tris-Borate-EDTA buffer. The DNA and protein complexes were then transferred to a positively charged nylon membrane (380 mA, 30 min). The biotin end-labeled DNA was detected using the Streptavidin-Horseradish Peroxidase Conjugate and the Chemiluminescent Substrate. Each EMSA experiment was repeated at least three times.
Biofilm assay
Porphyromonas gingivalis were grown overnight in TSB, and cells were then resuspended in 4 mL fresh media to an OD600 nm of 0.1 in a six-well plate (COSTAR) coated with human whole saliva and grown for 14 h. After the unbound cells were removed, each well was washed with 4 mL phosphate-buffered saline (PBS) three times. The cells in the P. gingivalis biofilm were lysed with lysis solution (Solution A, Invitrogen). DNA was extracted using Easy-DNA kit (Invitrogen) and resuspended in 50 μL TE buffer. The cells in biofilms were enumerated using QuantiTect SYBR Green PCR Kit with P. gingivalis species-specific 16S rRNA gene primers (Tran & Rudney, 1999).
Standards used to determine P. gingivalis cell numbers were prepared using genomic DNAs from the wild-type strain 33 277. A fresh culture of P. gingivalis 33 277, grown in TSB, was serially diluted in PBS and plated on TSB plates to obtain CFU per milliliter at each dilution. DNA was also isolated from the dilutions and a qPCR assay was run, as described above, to determine cell number. Three trials were performed on three separate cultures.
Results
Coordinate expression of fimA and sod in P. gingivalis in response to oxidative stress
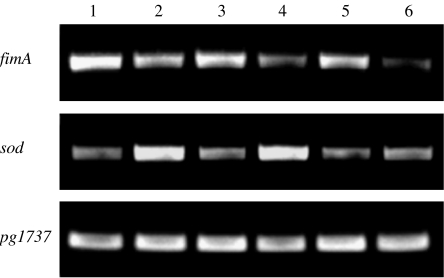
To determine whether expression of fimA and sod is coordinately regulated, we compared the mRNA level of these genes in response to oxidative stress using RT-PCR. As expected, expression of sod was increased in P. gingivalis 33 277 after the organism was exposed to air for 3 or 6 h (Fig. 1). An additional 24 h culture in an anaerobic chamber or in aerobic conditions had a notable affect on sod expression, although mRNA levels of sod were enhanced in P. gingivalis cells grown under oxidative stress, suggesting that expression of the sod is regulated by both oxidation stress and growth rate. In contrast to elevation of sod expression, expression of fimA was repressed when P. gingivalis was exposed to oxidative stress. In agreement with our earlier observation and in contrast to sod, expression of fimA was not affected by growth rate (Xie et al., 1997). As a control, expression of pg1737 encoding glycosidase was also examined, and no significant difference was detected in the mRNA levels when P. gingivalis was cultured either aerobically or anaerobically. The data demonstrate that expression of fimA and sod in P. gingivalis is coordinately regulated in response to oxidative stress.
Fig. 1.
Coordinate expression of fimA and sod in Porphyromonas gingivalis in response to oxidative stress. Porphyromonas gingivalis 33 277 was grown on TSB plates for 48 h anaerobically. The bacteria cells were continuously incubated in the anaerobic chamber for another 3, 6, or 24 h (lanes 1, 3, 5) or exposed to air for 3, 6, or 24 h (lanes 2, 4, 6). Expression of fimA and sod was determined using RT-PCR. Glycosidase (pg1737) expression served as a control.
Role of OxyR in coordinate regulation of sod and fimA
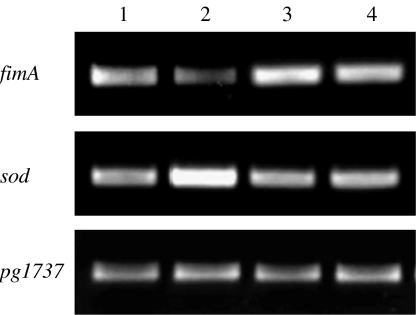
Previous studies have shown that expression of P. gingivalis sod is regulated by redox-sensing transcription activator OxyR (Diaz et al., 2006; Ohara et al., 2006). We postulated that OxyR may be also involved in fimA expression in P. gingivalis. To test the hypothesis, we constructed an oxyR mutant, P. gingivalis oxyRE, and expression of fimA and sod in the mutant was compared with that in wild-type 33 277. Higher expression level of fimA was observed in the oxyR mutant compared with the level in 33 277 (Fig. 2), suggesting that OxyR acts as a repressor of fimA in P. gingivalis. Comparable expression levels of fimA and sod were observed when the oxyR mutant was grown aerobically vs. anaerobically, clearly demonstrating that the oxyR mutant lost the ability to respond to oxidative stress. In addition, mutations in oxyR did not affect expression levels of sod when the P. gingivalis was grown anaerobically, suggesting that P. gingivalis OxyR only functions in its oxidized form resembling E. coli OxyR (Mongkolsuk & Helmann, 2002).
Fig. 2.
Expression of fimA and sod in wild-type strain 33 277 and in the oxyR mutant. Expression level of fimA and sod was determined by RT-PCR on RNA isolated from 33 277 grown anaerobically (lane 1), and aerobically (lane 2), or from the oxyR mutant grown anaerobically (lane 3), and aerobically (lane 4). Glycosidase (pg1737) expression served as a control.
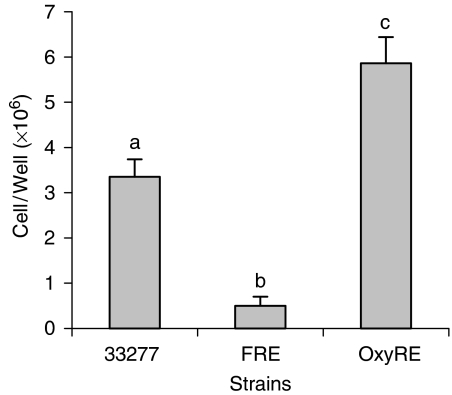
To determine if overexpressing FimA in the oxyR mutant has an impact on the binding ability of the strain, we compared biofilm formation among wild-type 33 277, the fimR mutant (FRE), and the oxyR mutant (OxyRE). We counted the number of bacteria in each biofilm using real-time PCR analysis. The number of bacteria in the FRE biofilms was sevenfold lower than in the wild-type strain. However, a twofold increase in bacteria was observed in biofilms of the oxyR mutant compared with wild-type 33 277 (Fig. 3). These findings demonstrate that OxyR influences the biofilm-forming potential of P. gingivalis.
Fig. 3.
Effect of OxyR and FimR on formation of Porphyromonas gingivalis biofilms. The ability of biofilm formation of wild-type strain 33 277, the fimR mutant (FRE), and the OxyR mutant (OxyRE) by quantization of cells bound on saliva coated 6 well plates using qPCR. The measurements were performed in triplicate; mean values are shown. Means with different letters are significantly different (P < 0.01; two-way anova and Student–Newman–Keuls Test).
Binding of OxyR to the fimA promoter region
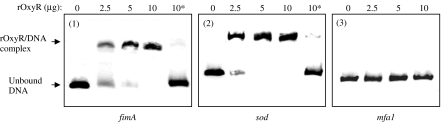
One possible mechanism for regulation of fimA expression by OxyR is the direct interaction of OxyR with the fimA promoter. To assess if the interaction occurs, we performed electrophoretic mobility shift assay. The promoter regions of fimA (positioned from −22 to −190) (Xie & Lamont, 1999) and sod (positioned from −1 to −210) were generated by PCR with the 5′ biotin-labeled primers (Table 1). The promoter region (positioned from +18 to −138) of mfa1, a gene encoding component of the short fimbriae of P. gingivalis, was used as a control. Recombinant OxyR (rOxyR) was expressed in a pET Expression System and purified from E. coli. As shown in Fig. 4, the DNA fragments of the sod and the fimA promoter regions were shifted in the presence of the rOxyR. As the concentration of rOxyR increased, the retarded protein–DNA complex became more evident, with a parallel loss of uncomplexed sod promoter DNA. Our data also suggested that the DNA binding affinity to the sod promoter region was higher than that to the fimA promoter region, at least in the context of our incubation conditions. Complete retardation of sod promoter-rOxyR complex was detected with as little as 5 μg rOxyR, whereas 10 μg rOxyR was required for a complete retardation of the fimA promoter fragment (Fig. 4). This binding activity was blocked by addition of cold probe. There was no DNA shift detected when rOxyR was incubated with the promoter region of mfa1. These data show that OxyR protein can bind specifically to the fimA and the sod promoter regions in vitro, acting as an activator of sod and as a repressor of fimA.
Fig. 4.
Interaction of rOxyR with the promoter region of fimA and sod. Electrophoretic mobility shift assays were performed in the presence or absence of rOxyR and 20 fmol biotin-labeled DNA. Increasing amounts of rOxyR were used in the assays. Asterisks indicate that 100-fold excess amounts of each specific competitor unlabeled probe were added to the reaction mixture with labeled probe.
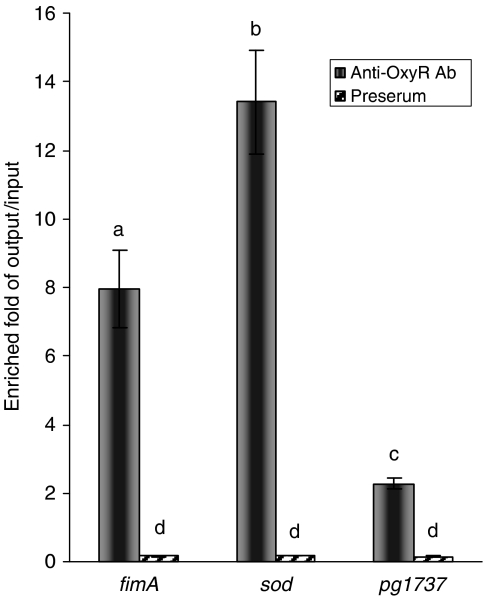
Using a ChIP qPCR assay, we further examined interactions between OxyR and the fimA promoter in vivo. Porphyromonas gingivalis 33 277 grown to late log phase was exposed to air for 3 h and then treated with formaldehyde to cross-link DNA-protein complexes. OxyR–DNA complexes were immunoprecipitated with anti-OxyR antibodies. A parallel sample using preimmune serum from the same rabbit was run as a background reference control. The immunoprecipitated DNA complexes were measured using real-time PCR analysis. As shown in Fig. 5, the promoter regions of fimA and sod were enriched eight- or 14-fold, respectively, by immunoprecipitation with anti-OxyR antibodies compared that to the twofold enrichment of the control gene, pg1737. Moreover, the enrichments of the promoter regions of fimA and sod were not observed in P. gingivalis 33 277 grown in aerobic conditions (data not shown). Although we could not rule out interaction of OxyR and the promoter region of sod in P. gingivalis grown under anaerobic conditions, it is likely that, at least, this interaction is enhanced when those bacteria were exposed to air. These results also indicate that OxyR binds to both sod and fimA promoter regions in vivo, although a relatively higher affinity is found for the DNA sequence of the sod promoter.
Fig. 5.
qPCR analysis of ChIP complex DNA. Immunoprecipitations were performed in the presence of anti-OxyR antibodies or preserum. The precipitated DNAs were amplified using promoter sequences of specific primers for fimA, sod, and pg1737 (as a negative control). Means with different letters are significantly different (P < 0.01; two-way anova and Student–Newman–Keuls Test).
Discussion
Porphyromonas gingivalis is a gram-negative and anaerobic bacterium commonly found in subgingival plaque, a causative agent of periodontitis (Ximenez-Fyvie et al., 2000b). Although the deep periodontal pocket and bacterial biofilms create safety niches for P. gingivalis, the organism can be exposed to oxidative stress conditions in the oral cavity, especially in its early stages of colonization. One mechanism used by P. gingivalis to protect itself from oxidative damage is to increase expression of oxidative stress related-genes. The results from recent two-dimensional electrophoresis and real-time PCR analyses demonstrate an elevated expression of SOD and alkyl hydroperoxide reductase (AhpC) at both transcriptional and translational levels in P. gingivalis after exposure to oxidative stress (Diaz et al., 2006; Ohara et al., 2006). The ability to selectively produce enzymes under regulation by oxygen switching transcriptional factors allows P. gingivalis to survive under aerobic conditions. The results of the present studies confirm elevation in sod gene expression in P. gingivalis in response to oxygen levels. In addition, we demonstrate here that the expression of FimA, a major component of the long fimbriae, is also regulated in response to oxidative stress. Expression of fimA is repressed in P. gingivalis under aerobic conditions, which is presumably necessary to conserve its energy against oxidative stress. Up-regulating expression of oxidative stress-related genes such as sod, and repressing expression of other nonessential cellular components under the aerobic condition such as fimA, may be critical strategies for the bacteria's survival in the oral cavity and in periodontal pockets with increased superoxide levels produced from neutrophils.
We have identified the OxyR as a transcription factor that coordinately regulates target genes in response to oxidative stress. The oxyR is widely distributed in most Gram-negative and some Gram-positive bacteria (Mongkolsuk & Helmann, 2002). OxyR is considered as a global regulator, since the OxyR regulon of E. coli includes genes involved in peroxide metabolism and protection, redox balance, and some important regulators such as the ferric uptake regulator, Fur (Zheng et al., 2001; Varghese et al., 2007). The molecular basis for OxyR regulation has been well characterized in E. coli. OxyR is a tetrameric DNA-binding protein that is activated under oxidative stress by forming a disulphide bond between two Cys residues (Zheng et al., 1998). Our genetic analysis of the P. gingivalis oxyR gene reveals that OxyR acts as a repressor of fimA in P. gingivalis, and that fimA is a member of the oxyR regulon. The mutation in oxyR results in an elevated expression of the fimA gene. The oxyR deficient mutants have lower expression levels of sod, which reduces the ability of the organism to adapt to oxidative stress. However, expression of sod in the wild-type strain and in the oxyR mutant is at similar levels when P. gingivalis cells were cultured in an anaerobic chamber. This finding suggests that, like E. coli, OxyR is activated under oxidative stress, and its activated form may have higher affinity to the promoter region of sod (Mongkolsuk & Helmann, 2002). Nevertheless, activation of OxyR is not required for repressing fimA transcription regulation, suggesting a constitutive repression of fimA expression by OxyR. It is reported that in E. coli, expression of Antigen 43, a self-recognizing surface adhesin, is repressed by OxyR (Haagmans & van der Woude, 2000). Similar to our observation in P. gingivalis, expression regulation of Ag43 is independent of the oxidation state of OxyR (Wallecha et al., 2003).
Porphyromonas gingivalis fimA is present in a single copy in the chromosome (Dickinson et al., 1988). A recent study has shown the presence of monocistronic and polycistronic mRNA of fimA (Park et al., 2007). Although the full extent of polycistronic mRNA of fimA has not been determined, it contains, at least, the fimA gene (pg2132) and its two immediately upstream genes (pg2130 and pg2131). It appears that fimA transcription involves a promoter upstream of pg2130 and an internal promoter immediately upstream of fimA. FimR, a transcriptional activator of fimA, can bind to promoter upstream of pg2130, but not to the fimA promoter region (Nishikawa et al., 2004). Therefore, a cascade regulation of fimA expression by FimR is proposed. Activation of fimA expression by FimR is through PG2130. We show here a novel regulatory mechanism of fimA expression involving OxyR. Unlike FimR, OxyR recognizes a different DNA motif in the internal promoter immediately upstream of fimA. The results reveal a complex regulatory system in expression of P. gingivalis fimA. Although several adhesive molecules are identified in P. gingivalis (Lamont & Jenkinson, 2000), FimA plays an important role in P. gingivalis colonization. Further understanding of regulation of FimA production may present attractive targets for inhibition of P. gingivalis biofilm formation.
Acknowledgments
This work was supported by Public Health Service grant DE014699 (HX) from the National Institute of Dental and Craniofacial Research.
Statement
OnlineOpen articles are made available in accordance with the terms of the Creative Commons Deed, Attribution 2.5 (further details available from http://www.creativecommons.org), which allows Open Access dissemination of the article, but does not permit commercial exploitation or the creation of derivative works.
Supplementary material
The following material is available for this article online:
Semi-quantitation RT-PCR products in Fig. 1.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1574-6968.2008.01116.x (This link will take you to the article abstract).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Amano A, Sharma A, Sojar HT, Kuramitsu HK, Genco RJ. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect Immun. 1994;62:4682–4685. doi: 10.1128/iai.62.10.4682-4685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano A, Fujiwara T, Nagata H, Kuboniwa M, Sharma A, Sojar HT, Genco RJ, Hamada S, Shizukuishi S. Prophyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. 1997;76:852–857. doi: 10.1177/00220345970760040601. [DOI] [PubMed] [Google Scholar]
- Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, Kolenbrander PE. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J Bacteriol. 2006;188:2454–2462. doi: 10.1128/JB.188.7.2454-2462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DP, Kubiniec MA, Yoshimura F, Genco RJ. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J Bacteriol. 1988;170:1658–1665. doi: 10.1128/jb.170.4.1658-1665.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, Macrina FL. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulbourne PA, Ellen RP. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J Bacteriol. 1991;173:5266–5274. doi: 10.1128/jb.173.17.5266-5274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans W, van der Woude M. Phase variation of Ag43 in Escherichia coli dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol. 2000;35:877–887. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- Hamada N, Watanabe K, Sasakawa C, Yoshikawa M, Yoshimura F, Umemoto T. Construction and characterization of a fimA mutant of Porphyromonas gingivalis. Infect Immun. 1994;62:1696–1704. doi: 10.1128/iai.62.5.1696-1704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi J, Nishikawa K, Hirano R, Noguchi T, Yoshimura F. Identification of a two-component signal transduction system involved in fimbriation of Porphyromonas gingivalis. Microbiol Immunol. 2000;44:279–282. doi: 10.1111/j.1348-0421.2000.tb02496.x. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol Immunol. 2000;15:341–349. doi: 10.1034/j.1399-302x.2000.150601.x. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Bevan CA, Gil S, Persson RE, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8:272–276. doi: 10.1111/j.1399-302x.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Lynch MC, Kuramitsu HK. Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect Immun. 1999;67:3367–3375. doi: 10.1128/iai.67.7.3367-3375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S, Helmann JD. Regulation of inducible peroxide stress responses. Mol Microbiol. 2002;45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Yoshimura F, Duncan MJ. A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol Microbiol. 2004;54:546–560. doi: 10.1111/j.1365-2958.2004.04291.x. [DOI] [PubMed] [Google Scholar]
- Ohara N, Kikuchi Y, Shoji M, Naito M, Nakayama K. Superoxide dismutase-encoding gene of the obligate anaerobe Porphyromonas gingivalis is regulated by the redox-sensing transcription activator OxyR. Microbiology. 2006;152:955–966. doi: 10.1099/mic.0.28537-0. [DOI] [PubMed] [Google Scholar]
- Park Y, Xie H, Lamont RJ. Transcriptional organization of the Porphyromonas gingivalis fimA locus. FEMS Microbiol Lett. 2007;273:103–108. doi: 10.1111/j.1574-6968.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Smith C, Haffajee AD. Subgingival microbial profiles in refractory periodontal disease. J Clin Periodontol. 2002;29:260–268. doi: 10.1034/j.1600-051x.2002.290313.x. [DOI] [PubMed] [Google Scholar]
- Tran SD, Rudney JD. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. J Clin Microbiol. 1999;37:3504–3508. doi: 10.1128/jcm.37.11.3504-3508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese S, Wu A, Park S, Imlay KR, Imlay JA. Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol Microbiol. 2007;64:822–830. doi: 10.1111/j.1365-2958.2007.05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallecha A, Correnti J, Munster V, van der Woude M. Phase variation of Ag43 is independent of the oxidation state of OxyR. J Bacteriol. 2003;185:2203–2209. doi: 10.1128/JB.185.7.2203-2209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lin X, Xie H. Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system. FEMS Microbiol Lett. 2007;271:214–221. doi: 10.1111/j.1574-6968.2007.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Lamont RJ. Promoter architecture of the Porphyromonas gingivalis fimbrillin gene. Infect Immun. 1999;67:3227–3235. doi: 10.1128/iai.67.7.3227-3235.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Cai S, Lamont RJ. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect Immun. 1997;65:2265–2271. doi: 10.1128/iai.65.6.2265-2271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Cook GS, Costerton JW, Bruce G, Rose TM, Lamont RJ. Intergeneric communication in dental plaque biofilms. J Bacteriol. 2000;182:7067–7069. doi: 10.1128/jb.182.24.7067-7069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Lin X, Wang BY, Wu J, Lamont RJ. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology. 2007;153:3228–3234. doi: 10.1099/mic.0.2007/009050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol. 2000a;27:722–732. doi: 10.1034/j.1600-051x.2000.027010722.x. [DOI] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000b;27:648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Semi-quantitation RT-PCR products in Fig. 1.