Abstract
Direct monitoring of primary molecular-binding interactions without the need for secondary reactants would markedly simplify and expand applications of high-throughput label-free detection methods. A simple interferometric technique is presented that monitors the optical phase difference resulting from accumulated biomolecular mass. As an example, 50 spots for each of four proteins consisting of BSA, human serum albumin, rabbit IgG, and protein G were dynamically monitored as they captured corresponding antibodies. Dynamic measurements were made at 26 pg/mm2 SD per spot and with a detectable concentration of 19 ng/ml. The presented method is particularly relevant for protein microarray analysis because it is label-free, simple, sensitive, and easily scales to high-throughput.
Keywords: dynamic monitoring, optical biosensor, protein microarray, immunoassay, interference
Detecting biomolecular interactions is fundamental to our understanding of both cell physiology and disease progression. Antigen–antibody, receptor–ligand, virus–cell and protein–DNA binding interactions are monitored and quantified in biological and medical research by long established and extensively used solid-phase immunoassays such as enzyme-linked immunosorbent assays (ELISA) (1) and Western blotting (2). These techniques typically employ secondary probes that bind to captured analytes that are in turn detected with either fluorescent or enzyme-linked reagents, thereby converting the presence of target biomolecules to a measurable signal. Yet, the detection of analytes through secondary probes is intrinsically complex, requiring multiple layers of interacting components that provide specificity without interfering with one another. Monitoring the primary bimolecular interaction (i.e., binding of analyte to its cognate probe) would greatly simplify many immunoassays. Thus, label-free measurement of binding events is a potentially powerful tool, simpler and more efficient than secondary probe-based systems (3). Label-free detection has been demonstrated by using electrical, electromechanical, and optical detection methods (4). Especially notable is surface plasmon resonance (SPR), which has been the basis for such commercial biosensors as the Biacore instruments (GE Healthcare Life Sciences). SPR allows real-time, sensitive monitoring of an analyte binding to probes immobilized on a gold substrate. Although SPR has been the standard for label-free detection for single analytes, extension to large-format arrays has achieved limited success (5, 6). The importance of high-throughput platforms has been demonstrated by the success of gene arrays in the analysis of nucleic acids (7, 8). This success has further provided impetus for the proposal that protein–peptide arrays could achieve similar utility for the analysis of multiple binding events (9, 10).
In this article, we describe a label-free sensing technique based on the optical phase difference due to the accumulation of biological material on solid substrates. We demonstrate picogram per square millimeter sensitivity, real-time kinetics, and the binding of multiple analytes to immobilized probe molecules—all in a simple platform amenable to high-throughput screening.
Results
Detection Principle.
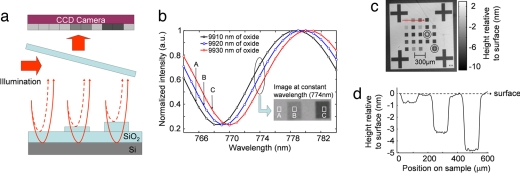
Interferometric measurements can provide enormous sensitivity and resolution of optical path length differences (OPD), with the best current systems achieving <10−10rad resolution (11). The basic working principle of our platform, the spectral reflectance imaging biosensor (SRIB), is the use of spectroscopy to quantify the interference signature of light reflected from a layered structure that allows the accurate measurement of the optical thickness of a designated transparent film (Fig. 1a). The layered substrate is a silicon wafer with a top layer of thermally grown silicon dioxide (SiO2) used as the solid support for biomolecular probes. The transparent film consists of the SiO2 layer and any additional material on its top surface. The magnitude of the total reflected light at a specific wavelength will depend on the OPD between the top surface of the transparent film and the buried reference surface at the SiO2–Si interface. Any binding on the top surface increases the OPD (oxide thickness plus biomolecule layer), manifests as a characteristic shift of the spectral reflectivity, and is also visible as an intensity difference at a specific wavelength (Fig. 1 b and Inset).
Fig. 1.
Basic principles of SRIB. (a) Simple theory of SRIB. The surface is illuminated with the laser, and the reflected light is imaged on a CCD. Any additional height on the surface will change the optical path difference between the top surface and buried oxide–silicon interface. (b) Data acquisition. The sample reflectance is recorded as an intensity image by a camera at each wavelength in 1-nm steps, forming a reflectivity-vs.-wavelength curve at each pixel of the CCD. Different reflectivity-vs.-wavelength curves are formed as a result of different optical path differences. A high-contrast image is shown as the Inset, at a wavelength of 774 nm, where the slope of the curve is highest. The thickness at each pixel is found by processing these recorded curves and displayed to show the surface profile. (c) System calibration. The SRIB system was calibrated by imaging 25 squares with varying average depths on the chip and comparing the results with the measurements of commercial profilometers. These samples were prepared by standard photolithography. Average etch depth varies from 7.5 to 0.1 nm. Concentric circles are used to find the average height of the square (see Methods). (d) Line cut of the image indicated by the red arrow shown on above image.
The optical characteristics of layered dielectrics are well understood, and we use conventional formulations (12) to extract the OPD from recorded spectral reflectivity data (see Methods). A thin protein layer causes a phase retardation of optical waves propagating in the direction perpendicular to the surface that is the product of refractive index and film thickness, assuming a uniform film. Experimental interpretations of the OPD for partial coverage have included both a constant refractive index accompanied by a change in thickness (13) or a constant thickness accompanied by a change in refractive index (14). These interpretations represent the two extreme approximations in modeling the OPD and would result in dramatic differences in case of label-free biosensors where an evanescent field interacts with surface-bound molecules such as in SPR. Because our fields near the surface are slowly varying, the correlation to the amount of biomolecules captured is nearly independent of which assumption is used, with a maximum error of 5% as shown in supporting information (SI) Fig. S1. Thus, we choose to interpret our OPD signal due to biomolecule binding as a change in thickness at a constant refractive index of 1.45, commonly used as the refractive index of a monolayer of protein (15, 16). This does not necessarily yield the actual thickness because the refractive index of the molecular layer may be different, but it does yield the actual surface-adsorbed mass density calculated from the measured thickness. It was shown by radiolabeling methods that in similar optical detection methods, 1 pm height change corresponds to ≈1 pg/mm2 of binding material on the surface (15) or to a ≈1 × 10−5 change in refractive index (17).
The experimental setup (Fig. S2) consists of a tunable laser made spatially incoherent by passing the beam through spinning ground-glass disks before illuminating the substrate (Fig. S3). The sample reflectance is recorded as an intensity image by a camera at each wavelength in 1-nm steps (Fig. 1 a and b). Different reflectivity-vs.-wavelength curves are formed as a result of different optical path differences (see Methods). To calibrate the measurements, a pattern of square regions of varying average depths at nanometer scale were etched into the silicon dioxide (Fig. 1c). SRIB measurements corresponded well to independently confirmed depths by atomic-force microscopy (AFM) and commercial white-light interferometry.
Label-Free Detection of Antibody–Antigen Interactions.
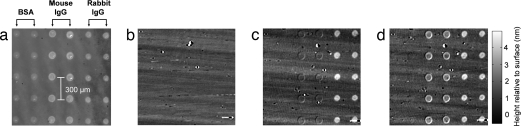
The selectivity of the SRIB platform was tested by measuring the binding of antibodies to their corresponding antigens spotted on the sample surface (Fig. 2). The surface was initially activated to permit the covalent attachment of the spotted probe molecules (see Methods). SRIB images after spotting easily discern the 10 spots each of BSA, rabbit IgG, and mouse IgG (Fig. 2a). The sample was initially blocked with BSA (1% in PBS containing 0.1% Tween-20 (PBST) and then first incubated with goat anti-rabbit IgG in PBST (5 μg/ml) for 30 min, followed by goat anti-mouse IgG (5 μg/ml). The chip was imaged with SRIB at each incubation step, and specific binding of the antibodies was detected (Fig. 2 c and d). Also note the marginal cross-reactive binding of anti-rabbit to the mouse IgG in Fig. 2d. No detectable binding at the control spots (BSA) was observed during either incubation.
Fig. 2.
Selective antibody detection. (a) Initial data acquired after washing the spotted sample. Initial heights of the spots were measured to be ≈0.5 nm for BSA spots, ≈1.2 nm for rabbit IgG spots, and ≈1 nm for mouse IgG spots, corresponding to 0.5 ng/mm2, 1.2 ng/mm2, and 1 ng/mm2 of material density on the surface, respectively. All subsequent data were referenced to the initial spotted sample to track the relative biomolecule accumulation. Height information of each image is scaled equally, which is shown with the gray-level scale bar. (b) After subtraction of the reference image. (c) Anti-rabbit incubation. Binding was detected as a 2.5 ng/mm2 incremental surface accumulation at the rabbit IgG spots after 5 μg/ml goat anti-rabbit IgG incubation. (d) Anti-mouse incubation. A 1.5 ng/mm2 incremental surface accumulation at the mouse IgG spots was detected after 5 μg/ml goat anti-mouse IgG incubation.
Dynamic Detection of Biomolecular Interactions.
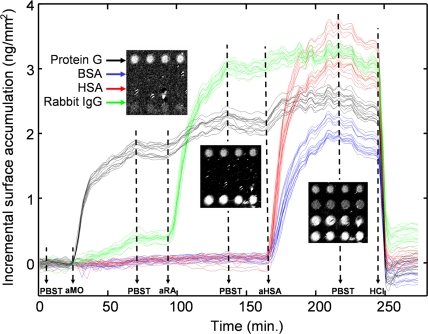
To measure the binding dynamics, samples were placed in a custom flow chamber sealed with a glass window that enabled optical measurements. Four different probes were spotted on an epoxysilanized chip, and their interactions with various analytes were monitored dynamically (Fig. 3). Fifty spots for each of BSA, human serum albumin (HSA), rabbit IgG, and protein G were spotted on the surface. Before antibody incubation, the surfaces of the flow chamber and sample were blocked by flowing BSA (1 mg/ml in PBST) in the chamber for 30 min at room temperature. SRIB images of the change in OPD were acquired over the entire chip in <1 min, easily sufficient to determine binding kinetics. Fig. 3 shows the time evolution of the experiment for 64 individual spots, with Inset images of 16 spots in four rows at different stages of the experiment. Following the first PBST flow for 20 min, goat anti-mouse IgG in PBST (10 μg/ml) was reacted for 50 min, and a 1.8 nm height increase was detected at the protein G spots. Note that ≈15% cross-reactivity was detected between anti-mouse IgG and the spotted rabbit IgG (Fig. 3a). Measurable dissociation was detected when the chip was washed with PBST alone, at ≈70 min. Subsequently, goat anti-rabbit IgG was introduced (10 μg/ml), and binding to the spotted rabbit IgG was observed. Not surprisingly, goat anti-rabbit IgG was also seen to associate with the immobilized protein G. Some dissociation of protein G and rabbit IgG was seen again during a second PBST wash (≈135 min). Next, the flow of rabbit anti-HSA IgG antibody (10 μg/ml) followed, and the binding of the polyclonal rabbit antibody was detected both to the HSA and BSA, with a faster initial rate of binding apparent for anti-HSA and HSA compared with that of BSA.
Fig. 3.
Dynamic binding kinetics of different antigens to their antibodies. Kinetics of 64 spots are plotted. The noise floor is 26 pm per spot. Sixteen of the spots are shown at different stages of the experiment as Insets. Solutions are introduced as indicated on the timeline: (PBST) PBS buffer with Tween 20, (aMO) goat anti-mouse IgG, (aRA) goat anti-rabbit IgG, (aHSA) rabbit anti-HSA, (HCl) 20 mM hydrochloric acid. A movie of the binding events is available (Movie S1). Fluctuations on the plots, between the 170th min and 250th min in the timeline, are caused by the laser intensity noises and can be reduced by adjustments and improvements in the optical setup.
Kinetic association and dissociation rates are readily calculated for the data generated from each spot (Fig. S4). The on rates (ka) averaged over 16 randomly placed spots are found to be ka = (1.66 ± 0.18) ×104 m−1s−1 for BSA and ka = (3.15 ± 0.15) ×104 m−1s−1 for HSA (±1 SD is given). It should be noted that these “on” and “off” rates are surface-derived rate values and often differ from rates in bulk solution. It is not surprising that the polyclonal rabbit anti-HSA antibody binds to both human and bovine albumin because a 76% sequence identity exists between HSA and BSA. Finally, hydrochloric acid [HCl 20 mM (pH 2.0), 10 min] was used to strip the bound analytes from the spotted probes. This allowed the efficient reuse of the chip (data not shown) with no noticeable loss of binding activity. A compressed-time movie of this experiment for all 200 spots is provided as Movie S1). The RMS baseline noise was 26 pm per spot during in-solution measurements, and averaging the height of 16 spots containing the same probe reduced the noise floor to <8 pm, corresponding to detection of >8 pg/mm2 bound target.
To exhibit the potential of SRIB as a diagnostics tool, simultaneous detection of antigens and antibodies was also demonstrated (Fig. S5). HSA antigens were first captured by spotted rabbit anti-HSA antibodies, and subsequently, these antigens captured the same type of antibody in the buffer during a second incubation. Cross-reactivity between anti-HSA and BSA was again detected as in the previous experiment.
Dilution Experiment to Determine the Sensitivity.
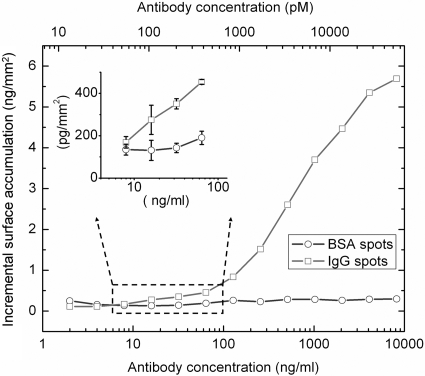
The sensitivity of the SRIB platform was tested by investigating the minimum-detectable concentration of antibody binding to spotted antigens in a separate experiment. The SiO2 surface was activated by epoxy silanization as described in Methods, and an array with three spots of BSA and three spots of rabbit IgG was applied to the chip. After blocking with BSA in PBST and washing, the chip was placed in the flow chamber and reacted with increasing concentrations of goat anti-rabbit IgG in PBST, allowing 45-min incubations at each concentration. After each incubation period, five SRIB images were collected for statistical analysis before adding the next antibody concentration solution. Fig. 4 displays the height change of a rabbit IgG spot and BSA spot as a function of antibody concentration. The noise floor in this experiment was measured as 50 pm on the negative control spots. Limit of detection was determined by dividing 1.96 times the standard deviation by the slope of the binding curve in its linear region. This gives a limit of detection at 19 ng/ml antibody concentration with a confidence limit of 95%, considering only the type I error. Recent experiments have shown a nearly 10-fold improvement in the noise floor.
Fig. 4.
Dilution curve for antibody–antigen interaction. Dilution curves for a pair of IgG–BSA spots are plotted. The sample was incubated in the solutions with increasing concentrations of antibody, each for 45 min., and imaged by SRIB. The antibody concentrations are shown for two different units: nanograms per milliliter and picomoles. Molarity was calculated by assuming the molecular mass of the antibody is 150 kDa. A concentration of 19 ng/ml is clearly detectable for an individual spot, corresponding to ≈120 pM, with a confidence limit of 95%. Because the error bars are very small compared with the height scale of the main plot, a portion of the curve is blown up. The error bars correspond to the standard deviation of measured height of each spot in five consecutive measurements.
Discussion
SRIB is an inexpensive and simple platform for label-free, high-throughput sensing. We have demonstrated the multiplexed and dynamic detection of protein–protein interactions over hundreds of spots simultaneously with high sensitivity. Label-free sensing array formats generally have a tradeoff among sensitivity, speed, and throughput. Sensitivity is inversely proportional to the square root of the product of sample area, integration time, and pixel number on the detector, assuming no noise contributions as the sample area increases and shot-noise limited detection. On the other hand, throughput is proportional to measurement area and can depend on time and hardware configuration.
As an example, SRIB dynamic experiments measured 200 spots simultaneously by using 500 × 500 pixels of a 0.35-megapixel camera. Averaging over ≈300 pixels yielded a noise floor of 26 pg/mm2 per spot, which was further reduced to 8 pg/mm2 when 16 spots were averaged. Throughput can always be traded for the required sensitivity by averaging over spots of the same probe or alternatively, by using a larger number of pixels per feature. Increasing throughput to thousands of spots can be achieved by a lower magnification or, if sensitivity is critical, by a large-format camera. For example, a 10-megapixel camera would yield approximately 1,000 spots with <10 pg/mm2 noise floor.
Dependence of sensitivity on averaging also exists in other label-free imaging methods including imaging SPR (17). Picometer noise floor was shown by several researchers (15, 18), but, when imaging a multiplexed array is required, the reported sensitivity drops. Averaging improves sensitivity by increasing the total number of detected photons and can reduce the effects of surface roughness. SRIB compares well by having both high sensitivity per unit area and the ability to measure a large area simultaneously.
SPR has been the leading technology among the label-free detection methods and has been used in the Biacore biosensors. Biacore measurements have recently expanded from four channels (Biacore T100) with imaging SPR technology, yet reported examples are still limited to a few hundred spots (17). Wavelength-dependent reflectance from layered surfaces has also been studied for optical detection of biomolecular interactions for many years (18–24). We argue that SRIB presents advantages when compared with these previous techniques. First, techniques that use broadband light sources and spectrometers are single-point detection methods that inherently require the scanning of either the substrate or the illumination and collection optics to form an image (19, 21). Such serial detection restricts applicability to low-throughput samples with limited dynamics because the imaging rate drops with increasing sample size. Another advantage is that, whereas the wavelength resolution of optical band-pass (24) and Fabry–Perot interference filters (18) is generally not <1 nm, SRIB samples the spectrum more accurately by scanning the wavelength of laser with a two-orders-of-magnitude-narrower line width.
Instead of a conventional glass slide as the reflecting substrate, a polished, thermally grown oxide layer was used because the uniformity and smoothness of these surfaces are markedly better. As an alternative to gold and glass surfaces, the chemical composition of the SiO2 surface is precisely known, so one can achieve repeatable surface functionalization that is critical because the sensitivities of all detection systems depend greatly on functional probe density and surface uniformity. SRIB shares the advantages of evanescent techniques like SPR in being insensitive to concentration changes away from the surface. But in contrast to SPR, the SRIB signal response is also independent of temperature and refractive index changes in the bulk solution [termed “bulk effect” (25)], allowing the possibility of thermal disassociation experiments. Similarly, the SRIB response is independent of the specific conformation of molecules on the surface. Regardless of how the molecules are oriented, the SRIB signal correlates to the surface accumulation with <5% maximum error, promising quantitative measurements. In addition to their optical and biomolecular advantages, silicon substrates are broadly compatible with most fabrication and processing technologies, so they can be used for lab-on-a-chip applications (7).
Proteomic microarrays have a wide range of applications, varying from parallel detection of antibodies for diagnosis of allergies and infectious diseases (26), to early detection of cancer (27), peptide analysis (28), identifying promoter sequences recognized by various transcription factors (29), and even to the detection of multiple biohazards (30). But it is recognized that, unlike gene-arrays, the production and use of protein arrays are still in their early stages, and there are numerous obstacles that need to be overcome (3). Even in a case where spotting thousands of different proteins is not feasible, high-throughput assays can still be useful for printing replicates on the same array, as has been done extensively in the case of DNA microarrays to markedly improve the reliability (31). As these experiments suggest, SRIB is one more step in the development of protein-array technology and is highly sensitive, easily implemented, and relatively cheap, with no fundamental limit for high throughput.
Materials and Methods
Optical Setup.
The reflection coefficient of a single oxide layer can be approximated by:
where r1 and r2 are the Fresnel reflection coefficients of the air–SiO2 (or buffer–SiO2) and SiO2–Si interfaces, respectively, and φ is the optical phase difference between the two reflections given by
Here, d is the SiO2 thickness, nox is the refractive index of SiO2, λ is the wavelength of the incident light, and θ is the angle of incidence. The angle and polarization dependence of the Fresnel reflection coefficients vanish for perpendicularly incident light (θ = 0°). In this work, molecular layers accumulating on the surface were assumed to have the same refractive index with SiO2, and surface accumulation was accompanied by a change in total transparent film thickness. Different optical thicknesses at different positions are apparent as shifted intensity vs. wavelength curves (Fig. 1b). Eq. 1 was fitted to the spectral reflectance data to find the OPD at each image pixel, interpreted as d − thickness, yielding a surface-profile image.
The optical configuration for SRIB is shown in Fig. S1. The measurement technique requires a temporally coherent light source (a laser), but spatial coherence causes unwanted artifacts due to diffraction and speckle. In our system, the illumination beam is passed through two ground-glass disks with at least one rotating, reducing the spatial coherence and eliminating artifacts in the image (Fig. S2). A fiber-coupled tunable diode laser (TLB6300 Velocity; NewFocus) is tuned in 1-nm steps from 764 to 784 nm, and a camera records an intensity image at each wavelength. Laser power out of the single-mode fiber is ≈1 mW. After changing the laser wavelength, it is necessary to wait ≈1 sec for laser power to be stabilized. Images were collected with ≈20 ms exposure time, and 25 images are collected and averaged for each wavelength to improve the signal-to-noise ratio. Because of the ground-glass diffusion, the beam is semicollimated when incident on the substrate. A Nikon 50-mm camera lens was used to image the reflection of the surface on the CCD (Rolera XR; QImaging), with a magnification ≈0.9. The pixel size of the camera is 13.7 μm, and 500 × 500 pixels are used to image approximately an area of 9 × 9 mm. Both the laser and the image grabber were controlled with National Instruments Labview software.
Because the sensing modality is based on optical thickness, smoothness of the layered substrate is crucial. Silicon samples were chemically and mechanically polished to <0.4 nm roughness as measured by AFM, followed by thermal growth of an oxide layer. Thermal oxide growth is self-limiting and highly uniform, virtually eliminating noise associated with variations in oxide thickness. The Si–SiO2 wafers were provided by Silicon Valley Microelectronics (SVM). The AFM and the white-light interferometer that were used to characterize the surface are Veeco Dimension 3100 and Zygo NewView 6000 Series, respectively.
Data Analysis.
Each pixel of the CCD collects an intensity-vs.-wavelength series of data points that are fitted to a curve of the governing reflection coefficient function R as described by Eq. 1. Every fitted curve then corresponds to a thickness that is mapped to an image. Each image is analyzed by using custom software developed in Matlab. The user selects circular regions of interest and denotes three concentric circles (Fig. 1c) that determine the “inner spot,” the middle ignored region (to reduce any edge effects) and the “background.” The thickness of a given spot above the surface is reported as the mean thickness of the inner spot minus the mean thickness of the background. During dynamic measurements, an initial reference image is subtracted from all subsequent images. The resulting “difference image” then indicates any increase/decrease that is observed upon the specific binding/dissociation of a target-probe complex. It is the resulting thickness increase that is reported in the dynamic data and sensitivity measurements.
Surface Preparation.
Unless stated otherwise, all reagents were of HPLC quality and purchased through Sigma–Aldrich. The surfaces were first cut into ≈15 × 15 mm squares and cleaned by sonication in acetone and rinsed in methanol, followed by a rinse with deionized (dI)H2O. The wafers were then placed in a solution of 10% NaOH for 10 min, rinsed in dIH2O, and dried under argon. The dried chips were epoxysilanized by immediately placing them in a solution of 3% (vol/vol) 3-(glycidoxypropyl)trimethoxysilane (United Chemical Technologies) in toluene for 3 min and then rinsing for 5 min in toluene. Samples were then dried under argon, covered, placed in a vacuum dessicator, and stored up to 2 months before use.
Protein Spotting.
The epoxysilanized chips were used directly and were spotted by using a desktop spotting unit (Odyssey Calligrapher minarrayer; Bio-Rad). For the initial selectivity experiment, BSA was spotted at 1 mg/ml concentration in PBS (pH 7.4; Fisher) containing 2.5% glycerol (to prevent the spot evaporation). Rabbit IgG purified from serum and monoclonal mouse IgG (I 5006 and I 56535, respectively; Sigma) were spotted at 400 μg/ml concentrations in PBS containing 1% glycerol.
For the dynamic-binding experiment, spotting concentrations were 4 mg/ml for BSA, 1 mg/ml for protein G (10–1200; Invitrogen), 2 mg/ml for rabbit-IgG, and 4 mg/ml for HSA (A9511; Sigma), all in PBS containing 2.5% glycerol. The wafers were allowed to incubate for at least 1 h in the Bio-Rad Calligrapher humidity chamber (55% humidity) before being stored overnight in the presence of dessicant. For the final sensitivity experiment, BSA and rabbit-IgG spotting concentrations were 4 mg/ml and 2 mg/ml, respectively.
Assay Procedure.
The spotted wafers were washed by rinsing three times in PBST and three times in PBS alone; each wash was ≈30 sec on a shaker at room temperature. The wafers were subsequently blocked by using 1%BSA (wt/vol) in PBST for 1 h. The same wash procedure was repeated with a final wash in dIH2O to remove any remaining salts, and the wafers were dried under argon. During the end-point experiment, binding to spotted rabbit and mouse IgG (the “antigen”) was detected upon incubating the chip with goat-anti rabbit IgG (R 1131; Sigma) and goat-anti mouse IgG from whole antiserum (M 5899; Sigma), respectively, at 5 μg/ml specific antibody concentrations in PBST (washing as before).
For the dynamic-binding experiments, the chip was secured into a custom flow cell after washing. The flow cell has a volume of ≈500 μl and has an antireflection coated front window. Solutions are driven through the flow cell by using a peristaltic pump at a rate of 400 μl/min. BSA (1%) in PBST is initially flowed through the system to block the associated tubing and the flow cell, followed by a flow of PBST for ≈15 min to remove any weakly bound probes before any tests are performed. Dynamic binding to protein G, rabbit IgG, HSA, and BSA was observed upon the addition of goat-anti mouse IgG (115-005-003; Jackson ImmunoResearch), goat-anti rabbit IgG, and rabbit-anti human albumin (Sigma, A3293), respectively, each analyte having the same concentration of 10 μg/ml. Measurement of the limit of sensitivity for the binding of goat anti-rabbit IgG to rabbit IgG was performed in the same flow chamber. The sample was incubated with antibody concentration of 2 ng/ml, 4 ng/ml, 8 ng/ml, 16 ng/ml, 32 ng/ml, 64 ng/ml, 128 ng/ml, 256 ng/ml, 512 ng/ml, 1024 ng/ml, 2.05 μg/ml, 4.1 μg/ml, and 8.2 μg/ml. After an incubation period of 45 min., five scans were taken successively to calculate the standard deviation of the measurement.
Supplementary Material
Acknowledgments.
We acknowledge Arthur Wu for help with the fabrication of artificial samples. This work was supported by National Institutes of Health Grant R21 GM074872-01A1 and Army Research Laboratory Grant W911NF-06-2-0040.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711421105/DCSupplemental.
References
- 1.Engvall E, Perlman P. Enzyme-linked immunosorbent assay (ELISA) Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 2.Gershoni JM, Palade GE. Protein blotting: Principles and applications. Anal Biochem. 1983;131:1–15. doi: 10.1016/0003-2697(83)90128-8. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P. A perspective on protein microarrays. Nat Biotechnol. 2002;20:225–229. doi: 10.1038/nbt0302-225. [DOI] [PubMed] [Google Scholar]
- 4.Ramachandran N, Larson DN, Stark PRH, Hainsworth E, LaBaer J. Emerging tools for real-time label-free detection of interactions on functional protein microarrays. FEBS J. 2005;272:5412–5425. doi: 10.1111/j.1742-4658.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 5.Homola J, Vaisocherova H, Dostalek J, Piliarik M. Multi-analyte surface Plasmon resonance biosensing. Methods. 2005;37:25–36. doi: 10.1016/j.ymeth.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Hong X, Kao F-J. Microsurface plasmon resonance biosensing based on gold-nanoparticle film. Appl Optics. 2004;43:2868–2873. doi: 10.1364/ao.43.002868. [DOI] [PubMed] [Google Scholar]
- 7.Wang J. From DNA biosensors to gene chips. Nucleic Acids Res. 2000;28:3011–3016. doi: 10.1093/nar/28.16.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 9.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 11.Ando M, et al. Stable operation of a 300-m laser interferometer with sufficient sensitivity to detect gravitational-wave events within our galaxy. Phys Rev Lett. 2001;86:3950–3954. doi: 10.1103/PhysRevLett.86.3950. [DOI] [PubMed] [Google Scholar]
- 12.Yeh P. Optical Waves in a Layered Media. New York: Wiley; 1988. [Google Scholar]
- 13.Schmitt HM, Brecht A, Piehler J, Gauglitz G. An integrated system for optical biomolecular interaction analysis. Biosens Bioelectron. 1997;12:809–816. [Google Scholar]
- 14.De Feijter JA, Benjamins J, Veer FA. Ellipsometry as a tool to study the adsorption behaviour of synthetic and bi-polymers at the air-water interface. Biopolymers. 1978;17:1759–1772. [Google Scholar]
- 15.Piehler J, Brecht A, Gauglitz G. Affinity detection of low molecular weight analytes. Anal Chem. 1996;68:139–143. doi: 10.1021/ac9504878. [DOI] [PubMed] [Google Scholar]
- 16.Prime KL, Whitesides GM. Self-assembled organic monolayers: Model systems for studying adsorption of proteins on surfaces. Science. 1991;252:1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 17.Shumaker-Parry JS, Campbell CT. Quantitative methods for spatially resolved adsorption/desorption measurements in real time by surface plasmon resonance microscopy. Anal Chem. 2004;76:907–917. doi: 10.1021/ac034962a. [DOI] [PubMed] [Google Scholar]
- 18.Nikitin PI, Gorshkov BG, Nikitin EP, Ksenevich TI. Picoscope, a new label-free biosensor. Sensor Actuat B. 2005;111–112:500–504. [Google Scholar]
- 19.Gao T, Lu J, Rothberg LJ. Biomolecular sensing using near-null single wavelength arrayed imaging reflectometry. Anal Chem. 2006;78:6622–6627. doi: 10.1021/ac0609226. [DOI] [PubMed] [Google Scholar]
- 20.Moiseev L, Ünlü MS, Swan AK, Goldberg BB, Cantor CR. DNA conformation on surfaces measured by fluorescence self-interference. Proc Natl Acad Sci USA. 2006;103:2623–2628. doi: 10.1073/pnas.0511214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu X, et al. Oblique-incidence reflectivity difference microscope for label-free high-throughput detection of biochemical reactions in a microarray format. Appl Optics. 2007;46:1890–1895. doi: 10.1364/ao.46.001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenison R, Yang S, Haeberli A, Polisky B. Interference-based detection of nucleic acid targets on optically coated silicon. Nat Biotechnol. 2001;19:62–65. doi: 10.1038/83530. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham B, Li P, Lin B, Pepper J. Colorimetric resonant reflection as a direct biochemical assay technique. Sensor Actuat B. 2002;81:316–328. [Google Scholar]
- 24.Gauglitz G. Multiple reflectance interference spectroscopy measurements made in parallel for binding studies. Rev Sci Ins. 2005;76 062224-1–062224-10. [Google Scholar]
- 25.Grassi JH, Georgiadis RM. Temperature-dependent refractive index determination from critical angle measurements: implications for quantitative SPR sensing. Anal Chem. 1999;71:4392–4396. doi: 10.1021/ac990125q. [DOI] [PubMed] [Google Scholar]
- 26.Mezzasoma L, et al. Antigen microarrays for serodiagnosis of infectious diseases. Clin Chem. 2002;48:121–130. [PubMed] [Google Scholar]
- 27.Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 28.Emili AQ, Cagney G. Large-scale functional analysis using peptide or protein arrays. Nat Biotechnol. 2000;18:393–397. doi: 10.1038/74442. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee S, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowe-Taitt CA, et al. Simultaneous detection of six biohazardous agents using a planar waveguide array biosensor. Biosens Bioelectron. 2000;15:579–589. doi: 10.1016/s0956-5663(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 31.Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.