Abstract
The Archaea, and the viruses that infect them, are the least well understood of all of the three domains of life. They often grow in extreme conditions such as hypersaline lakes and sulfuric hot springs. Only rare glimpses have been gained into the structures of archaeal viruses. Here, we report the subnanometer resolution structure of a recently isolated, hypersalinic, membrane-containing, euryarchaeal virus, SH1, in which different viral proteins can be localized. The results indicate that SH1 has a complex capsid formed from single β-barrels, an important missing link in hypotheses on viral capsid protein evolution. Unusual, symmetry-mismatched spikes seem to play a role in host adsorption. They are connected to highly organized membrane proteins providing a platform for capsid assembly and potential machinery for host infection.
Keywords: archaeal virus, electron cryomicroscopy, infection, symmetry mismatch, capsid protein evolution
Because of the high mutation rates in viruses, amino acid sequence-analysis methods generally only reveal the relationships between closely related viruses. Instead, viral evolutionary links can be studied by using conserved structural information; for example, a tentative viral lineage has been constructed by analysis of the structures of the major capsid proteins (MCPs) of adenoviruses (infect mammals and birds), Paramecium bursaria Chlorella virus type 1 (infects unicellular algae) (1), and bacteriophages PRD1 (infects Gram-negative bacteria) and Bam35 (infects Gram-positive bacteria). The MCPs of all these viruses are trimers of double β-barrels in which the axis of the barrel is oriented normal to the capsid shell (2–4). Could this tentative viral lineage be expanded further? The pool of structural information on viruses that infect eukaryotic and bacterial hosts is relatively large and constantly growing, but our knowledge about viruses that infect Archaea is still very limited. The only three-dimensional structure available at 27-Å resolution is that of the Sulfolobus turreted icosahedral virus (STIV) (5), which infects Sulfolobus solfataricus in the archaeal kingdom Crenarchaeota. As the name suggests, the STIV capsid is icosahedral and has elaborate turret-like structures at the fivefold vertices (5). Its glycosylated MCP (6) also has a double β-barrel fold (7), suggesting a common ancestry with the PRD1-adenovirus lineage.
SH1 is a virus that infects the halophilic organism Haloarcula hispanica in the archaeal kingdom of the Euryarchaeota (8). Its linear dsDNA genome of 30,898 bp has 56 identified ORFs (9). It has been shown to have several lipid moieties and 11 structural protein species. The proteins have been classified into capsid proteins and lipid core proteins (found with the membrane vesicle enclosing the nucleic acid) by biochemical analysis of virion dissociation products (10). The most abundant proteins are the capsid-associated proteins VP3, VP4, and VP7 and the lipid core protein VP12. The viral membrane is composed of neutral lipids and three major archaeal phospholipids: phosphatidylglycerol, phosphatidylglycerophosphate methyl ester, and phosphatidylglycerosulfate. The lipids are selectively acquired from the host during viral assembly (9).
Here, we use electron cryomicroscopy (cryo-EM) and image reconstruction to study the overall structure of SH1 and the structure of the symmetry-mismatched spikes (11–14). We demonstrate that the spikes are composed of proteins VP3 and VP6 and that the simultaneous removal of VP2 with VP3 and VP6 drastically affects infectivity and particle integrity.
Results
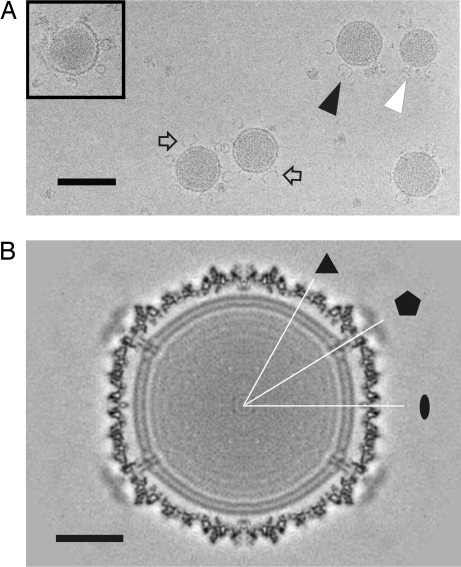
The natural habitat of SH1 and its host is a hypersaline (>3 M NaCl) salt lake, and the stability of the virus depends on high ionic strength (8, 10). This dependency creates a considerable challenge to cryo-EM of the virus, because the background noise attributable to the presence of 1 M NaCl reduces the contrast significantly. Because the virus is also large (≈115 nm from spike to spike; Fig. 1), the layer of vitrified water in which the particles are embedded is relatively thick, which increases the noise further. Two types of virus particles can be seen in the electron micrographs: (i) spherical virions (Fig. 1A, black arrowhead) with the protein shell intact and bifurcated spikes extending from the surface and (ii) spherical lipid core particles, off of which the protein shell has peeled (Fig. 1A, white arrowhead). These data were used for three-dimensional image reconstruction (Fig. 1; Table 1), first by using icosahedral symmetry to resolve the capsid (11, 12) and then by relaxing the symmetry to resolve the spikes (13, 14). At 9.6-Å resolution, the icosahedral reconstruction of the virion (Fig. 1B) shows clearly the protein capsid and the underlying lipid bilayer. The dimensions of the icosahedral capsid are 78.5 nm edge to edge, 78.0 nm facet to facet, and 79.5 nm vertex to vertex.
Fig. 1.
Cryo-EM of SH1. (A) Micrograph at 1.8-μm underfocus showing virions (black arrowhead), bifurcated spikes (arrow), and lipid core particles (white arrowhead). (Inset) Lipid cores are released when the intact virions uncoat. (B) Central section of the icosahedral reconstruction viewed down an icosahedral twofold axis of symmetry. Twofold (ellipse), threefold (triangle), and fivefold (pentagon) symmetry axes are indicated. (Scale bars: A, 100 nm; B, 20 nm.)
Table 1.
Statistics of SH1 reconstructions
| SH1 | Vertex | SH1-VP36 | SH1-VP236 | |
|---|---|---|---|---|
| Specific infectivity, plaque-forming units per mg of protein | 7.4 × 1012 | Not performed | 6.9 × 1011 | 1.2 × 109 |
| No. of particles used | 2,558 | 10,196* | 985 | 2,015 |
| No. of micrographs used | 116 | 169 | 58 | 39 |
| Underfocus, μm | 0.8–2.1 | 0.7–2.5 | 0.7–3.5 | 0.7–2.1 |
| Resolution, ņ | 9.6 | 34.0 | 10.5 | 10.0 |
*Vertices extracted from virus images and used in final reconstruction.
†Estimate based on the FSC 0.5 criterion (15).
The membrane of SH1 follows the shape of the capsid and contains icosahedrally ordered proteins. Under the fivefold vertices there are clear transmembrane structures. The membrane is highly curved at the site of the fivefold transmembrane complex, and there is a visible dent in the underlying layer at the same location (Fig. 1B). The average thickness of the membrane, measuring between the peaks of the density in a radial profile, is 2.4 nm. Despite major differences in archaeal and bacterial virus lipids, this membrane spacing is similar to that in the bacteriophage membranes of PRD1, PM2, and Bam35 (3, 16, 17), reflecting the similarity in the average lipid chain length (9). There are two additional ordered layers beneath the membrane, which we assume to be DNA. The average DNA-packaging density is ≈0.45 bp·nm−3 if the DNA occupies the entire volume enclosed by the membrane.
SH1 Has a Previously Undescribed Capsid Organization.
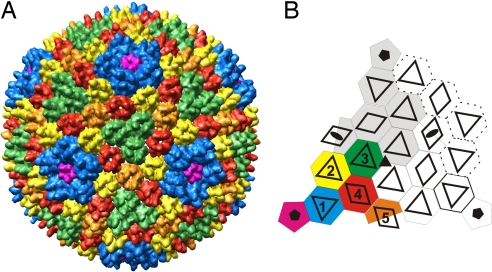
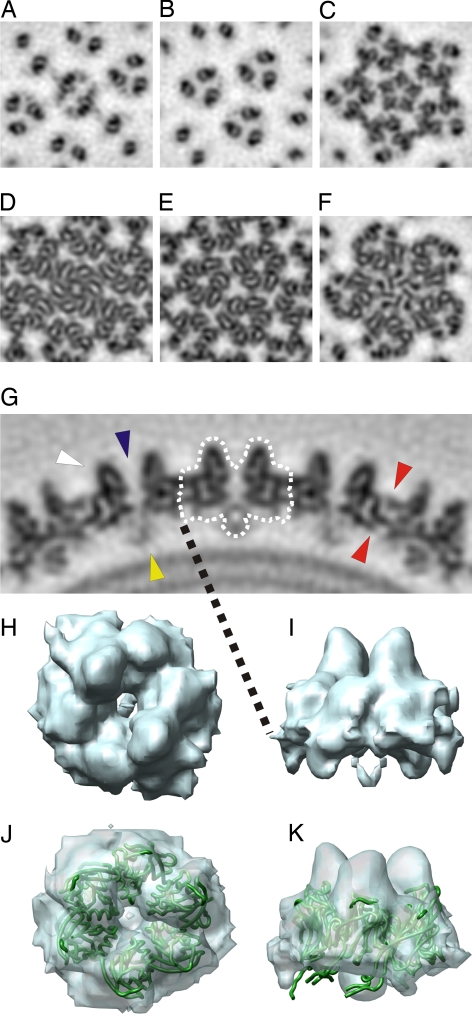
The protein capsid is arranged in a T = 28 lattice, which is skewed to the right (T = 28 dextro; Fig. 2A) (18). We determined the absolute hand by using tilt experiments (16, 19, 20). There are five icosahedrally independent capsomers (capsomers 1–5) in the asymmetric unit (Fig. 2B). One of the capsomers is located at the twofold axis of symmetry and belongs to two adjacent asymmetric units (Fig. 2). There are two distinct capsomer types (Figs. 2 and 3). Type II capsomers have two protruding towers (Figs. 2 and 3 A, H, and I), whereas type III capsomers have three towers (Figs. 2 and 3 B, J, and K). In the notation of Fig. 2, capsomers 4 and 5 are of type II, and capsomers 1, 2, and 3 are of type III. The capsomers adjacent to the vertices are rotated 60° relative to the other type III capsomers in the facet (Fig. 2B). These capsomers interact with pentamers at the capsid vertices (Figs. 3 C and F and 4). We manually segmented and averaged the capsomers (Figs. 2A and 3 H–K). Type II capsomers are slightly smaller (average mass, 178 kDa) than type III capsomers (average mass, 195 kDa). The bases of both type II and III capsomers appear hexameric, but in type II capsomers the base is slightly skewed (Fig. 3 D and E). Each of the six capsomer subunits in the base makes a bridge-like connection to an adjacent capsomer subunit on the surface of the capsid (Fig. 3G, red arrowheads). In sections through the capsid it is evident that there is a similar connection also at a lower radius (Fig. 3G, red arrowheads). The capsomer bases appear to be formed of six similarly sized β-barrels normal to the capsid surface with towers composed of separate β-barrel domains (Fig. 3G, white arrowhead). This observation was supported by the good fit of the small β-barrel from the PRD1 MCP [Protein Data Bank (PDB) ID code 1HX6] (21) into the averaged type III capsomer density (Fig. 3 J and K).
Fig. 2.
Architecture of the capsid. (A) Isosurface representation of the segmented capsid colored according to icosahedrally independent capsomer type viewed down a threefold symmetry axis. (B) Schematic diagram of a facet of a T = 28 dextro capsid, with one asymmetric unit colored. Transparent triangles and rhombs indicate tower positions in each capsomer. Adjacent facet capsomers are shown with dotted lines. Symmetry axes are indicated with black ellipses (twofold), a triangle (threefold), and pentagons (fivefold).
Fig. 3.
Capsomer types. (A–F) Planar cross-sections through the capsomer tips (A–C) and bases (D–F) along twofold (A and D), threefold (B and E), and fivefold (C and F) symmetry axes. (G) A central section along a plane that bisects the type II capsomers. A capsomer is outlined by a dotted white line. A tower barrel (white arrowhead), a channel through the center of the capsomer (blue arrowhead), the globular density connecting the capsomer to the membrane (yellow arrowhead), and some intercapsomer connections (red arrowheads) are indicated. (H and I) Isosurface representation of the averaged type II capsomer from the top (H) and the side (I). (J and K) Isosurface representation of the averaged type III capsomer (blue) fitted with the x-ray structure of the small β-barrel of the PRD1 MCP (PDB ID code 1HX6; green ribbon) viewed from the top (J) and the side (K).
Fig. 4.
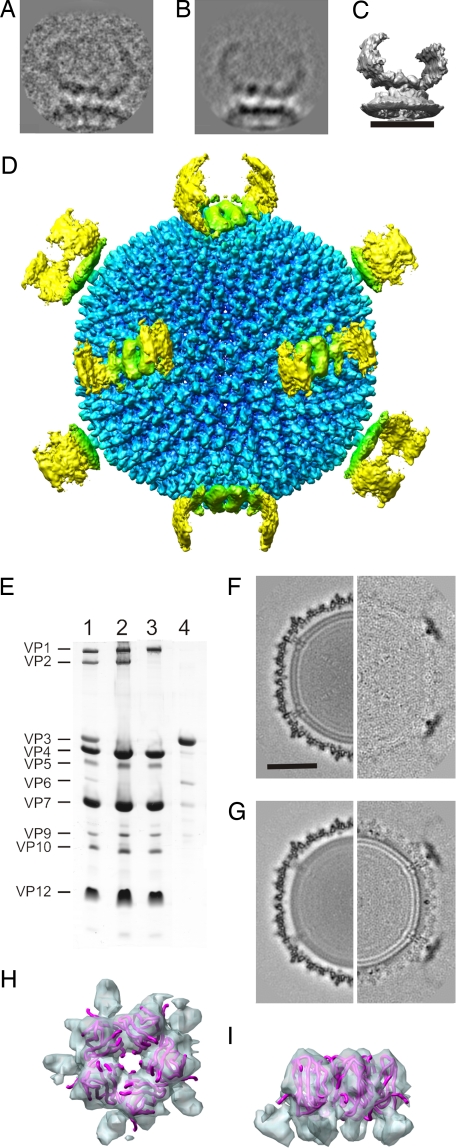
Analysis of the spike. (A) Example classum used in the vertex reconstruction. (B) Reprojection of the reconstruction in the same view as the example classum in A. (C) Surface rendering of the vertex reconstruction. (Scale bar, 20 nm.) (D) Radially depth-cued isosurface representation of an SH1 montage made from the icosahedral and vertex reconstructions along a twofold axis of symmetry. (E) Coomassie blue-stained Tricine-SDS/PAGE analysis of SH1 (lane 1), SH1-VP36 particles (lane 2), SH1-VP236 particles (lane 3), and the released material from the top of the gradient during production of SH1-VP36 particles (lane 4). (F) Central sections of the SH1-VP36 reconstruction (Left) and the difference map (Right) between SH1 and SH1-VP36. (Scale bar, 25 nm.) (G) Central sections of the SH1-VP236 reconstruction (Left) and the difference map (Right) between SH1 and SH1-VP236. (H and I) Isosurface representation of the pentamer (blue) fitted with the x-ray structure of the β-barrels of the pentameric P31 of PRD1 (PDB ID code 1W8X; pink ribbon) viewed from the top (H) and the side (I).
The membrane is effectively covered by the tight interdigitation of the capsomer bases (Fig. 2A). Although the capsomers appear to have a hole running through them from the outside of the capsid to the inside (Fig. 3G, blue arrowhead), the base of the capsomer is actually closed by a globular density that connects the capsomer to the membrane (Figs. 3G, yellow arrowhead, and 5A). These connections do not affect the membrane curvature significantly.
Fig. 5.
Radial cross-sections of the membrane. (A) Peripheral membrane proteins (radius, 33 nm). (B) Transmembrane complex in between the layers (radius, 31 nm). Protein is colored black.
The Spikes Contain Proteins VP3 and VP6.
The horn-like spikes at the fivefold vertices seen in the micrographs (Fig. 1A, arrow) are poorly represented in the icosahedral virion reconstruction as a result of low occupancy and inappropriate averaging (Fig. 1B). We used three approaches to interpret this structure: an asymmetric in situ vertex reconstruction (13, 14), disassembly of the spikes from the virion, and difference imaging of the resulting particles to identify the constituent proteins. For the asymmetric in situ reconstruction, 60,252 spike subimages were extracted from the virion images (Fig. 4A), of which 17% could be used in the reconstruction (Table 1). At 34-Å resolution we can see that the spike has twofold symmetry and that it is apparently built of 10 parallel tubular domains (Fig. 4 C and D). The mass of the spike is ≈1.3 MDa.
Given the prominent nature of the spikes, we hypothesized that they could be involved in receptor binding. Hence, we carried out dissociation experiments on purified virions to find conditions under which subviral particles with diminished infectivity could be isolated and studied further by cryo-EM. Having observed that calcium ions stabilized the virions, dissociation studies were carried out in calcium-free buffers. It was possible to remove proteins VP3 and VP6 by prolonged incubation in sulfate-containing buffer (Fig. 4E). In the resulting particles (SH1-VP36) in which ≈10% of VP3 was left, the specific infectivity decreased by 1 order of magnitude (Table 1). The released VP3 and VP6 copurified in a size-exclusion column and appeared as isolated spikes in electron cryomicrographs [see supporting information (SI) Fig. S1]. Incubation of virions in SH1 buffer at 37°C (Fig. 4E) resulted in particles that lacked VP2 as well as VP3 and VP6 (SH1-VP236). The specific infectivity of SH1-VP236 was reduced by 3 orders of magnitude (Table 1). Size-exclusion chromatography of the released material indicated that VP2 did not copurify with VP3 and VP6 (data not shown).
Difference imaging between the icosahedral reconstructions of the virus and the subviral particles (Table 1 and Fig. 4 F and G) as well as image processing of the vertices from SH1-VP236 and SH1-VP36 images (data not shown) revealed that the spikes were missing in both instances. Thus, multiple copies of VP3 and VP6 form the horn-like spikes.
In addition to the protruding spike structure, there is a pentameric protein embedded in the capsid shell (Figs. 2 and 3) and a clear transmembrane domain (Figs. 1 and 4). The pentamer (Fig. 3 C and F) appears to consist of five small β-barrels with an estimated mass of 85 kDa. In support of this, the PRD1 pentamer atomic model (68.7 kDa; PDB ID code 1W8X) (22) fitted inside (Fig. 4 H and I). The SH1 pentamer is attached to the capsid via five arms of density radiating out from the fivefold axis (Fig. 3F). It remained in place after the dissociation experiments; thus, VP2, VP3, and VP6 most probably do not contribute to it.
Under the pentamer there is a shaft that traverses the lipid membrane (Figs. 1 and 5). The shaft is surrounded by five additional groups of transmembrane proteins, most evident in a section between the leaflets (Fig. 5B). The outer leaflet contains well ordered, punctate patterns of densities spreading out from the fivefold axes (Fig. 5). Removal of VP3 and VP6 did not change the transmembrane structure under the fivefold vertex (Fig. 4F), whereas removal of VP2 in addition to the VP3 and VP6 disordered the membrane, the DNA, and the membrane proteins (Fig. 4G).
Discussion
SH1 resides in a hypersaline environment and, as a consequence, is relatively difficult to study. However, searching in such an environment may reveal unexpected virus structures. Indeed, SH1 has an unusual capsid architecture (T = 28 dextro) with two different capsomer types: the type II capsomers at and adjacent to the twofold axes and type III capsomers at all other locations (Fig. 2). Here, we address how our current knowledge of the protein composition and life cycle of SH1 (8–10) can help us to explain the virion structure.
SH1 has 15 potential structural protein species seen by SDS/PAGE, 11 of which have been identified by using protein chemistry (9). The most abundant capsid proteins are VP3 (37.5 kDa), VP4 (25.7 kDa), and VP7 (20.0 kDa). We demonstrated that VP3 and VP6 are major components of the horn-like spikes. Hence, VP4 and VP7 are the MCPs, as has been proposed earlier (10). Although posttranslational modifications, similar to those observed in STIV (6), may complicate the picture, here, we consider two simple ways to explain the observed capsomer masses and symmetries with the two candidate capsid proteins, VP4 (denoted A) and VP7 (denoted B): (i) type II and III capsomers are homodimers and homotrimers of the same protein ([A2, A3], [B2, B3], [A2, B3], or [B2, A3]) or (ii) type II and III capsomers are heteromeric ([A6B2, A6B3] or [B6A2, B6A3]). Possibility i can be ruled out on the basis of the estimated capsomer masses. The fact that the relative average masses of the type II and III capsomers (178 and 195 kDa, respectively; Fig. 3 H–K) differ so little rules out the possibility that the capsomers consist of two and three copies of the same protein. Furthermore, dimers and trimers of VP4 (25.7 kDa) and VP7 (20 kDa) would be too small to explain the capsomer masses. Hence, it is unlikely that the type III capsomer has the double β-barrel fold found in members of the PRD1-adenovirus lineage, which would require an MCP in the range of 65 kDa. In addition, the T = 28 lattice precludes the use of a trimer for the type II capsomer on the twofold axis of symmetry (23). To explain possibility ii, we assume that the bases of both type II and III capsomers consists of one and the same protein that can take a round hexameric and a skewed hexameric form. The capsid proteins of HK97 and herpes simplex virus 1 (HSV-1) are examples of such proteins: in the smaller procapsid state the capsid proteins are in the skewed conformation, and the unskewed conformation corresponds to the mature capsid (20, 24). In the case of SH1, we suggest that the towers on the capsomers are built by a decorating protein that may also stabilize the capsomer. The conformation of the base affects how many copies of the decorating protein can be accommodated: the skewed conformation (type II) of the base protein can only accommodate two copies, and the round conformation (type III) can only accommodate three copies. Conformation-dependent binding has been observed previously in, for instance, HSV-1, in which a single MCP, VP5, makes the pentameric and hexameric capsomers, and the minor protein, VP26, only decorates the hexamers (25). For SH1 we favor the explanation that the capsomer base is a hexamer of VP7, and VP4 is a decorating protein that makes the towers. This hypothesis predicts capsomer sizes of 171 kDa (type II) and 197 kDa (type III), the difference being one copy of VP4 (26 kDa). These predicted values are close to the values that we observe. Hence, we predict a copy number of 1,620 for VP7, 720 for VP4, and 60 for the pentamer. Likely candidates for the pentamer are either VP7 in a third conformation, which promotes the assembly of VP3 and VP6, or VP9, because it is a minor capsid protein of the appropriate mass (16.5 kDa).
In complex viruses, the structural components need to assemble in a regular fashion to form the complete virion. Capsid assembly is regulated by the use of pathways in which smaller subassemblies (e.g., capsomers) are first assembled and then incorporated into the growing capsid. In SH1, the membrane vesicle is selectively enriched in particular lipid moieties (9). The numerous and well-ordered membrane proteins that we see in the virion may selectively interact with lipids of a particular acyl chain length (26) already in the host cytoplasmic membrane to create a virion-specific patch. Further assembly can be then nucleated at this site by specific interactions between the virus' helical transmembrane proteins and the capsid proteins as in PM2 (N. Abrescia, J. Grimes, H.M.K., R. Assenberg, G. Sutton, S.J.B, J. Bamford, D.H.B., and D. Stuart, unpublished data). Lateral interaction of the capsid proteins can promote further growth of the capsid on the membrane, stabilized by divalent cations (8). The current literature supports the hypothesis that the genome is then transported into this lipid-containing procapsid by using a packaging ATPase (8, 27).
The phospholipids present in SH1 are all negatively charged. If the DNA is directly in contact with the membrane, then these charges need to be neutralized. Recent molecular dynamics simulations have indicated that in an anionic palmitoyloleoylphosphatidylglycerol bilayer, Na+ ions can form ion bridges between the lipids, neutralizing these charges (28). Hence, in SH1, it is likely that the high salinity of the native environment helps to stabilize not only protein–protein interactions but also the membrane and the membrane–DNA interactions and, thus, indirectly affects infection.
Archaeal cells are surrounded by a proteinaceous layer called the S-layer. There is currently very little understanding of how viral DNA penetrates this layer or the membrane. The relative complexity of the SH1 spike suggests that this entry may be accomplished in multiple stages. The large, twofold symmetric spike appears similar to the terminal structure seen in electron micrographs of the archaeal virus Acidianus filamentous virus 1 (29) [but not to the turret-like spikes of STIV (5)]. It has been suggested that the purpose of the terminal structure in Acidianus filamentous virus 1 is to increase the probability of host adsorption. In SH1, we removed the horn-like spikes by removing proteins VP3 and VP6 and saw a directly proportionate decrease in infectivity. Although it is possible that this treatment causes changes in capsid stability or release of minor viral proteins such as nucleases affecting the titer, we propose that the spikes contribute to host adsorption. The pillars of organized transmembrane proteins and the increased separation of the membrane leaflets seen under the pentamers look like a potential prefusion complex. Major disruption of the membrane, including this complex, by the removal of VP2, VP3, and VP6 caused a significant reduction in infectivity (Fig. 4; Table 1). Thus, the large ordered transmembrane shaft and the surrounding peripheral proteins are probably involved in genome translocation into the host cell.
One of the main incentives for this study was to extend our knowledge of virus evolution. The simplest self-assembling icosahedral shells are formed from 60 subunits (18). Examples have been described in which the building blocks are single β-barrel proteins lying either tangential (as found in parvoviruses) (30) or normal (similar to the sTALL-1 from the tumor necrosis factor cytokine family that forms virus-like particles) to the surface (31, 32). These structures could then have given rise to more complex capsids with larger T numbers as protein interactions were modified (33) and additional assembly components were introduced. Thus, SH1 can be seen as a molecular fossil, because it is the first example of a complex capsid formed from these single, vertical β-barrels. Thus, we now have highly suggestive data indicating that the β-barrel fold is very widespread, found in viruses infecting both branches of the Archaea as well as the Bacteria and Eukarya. It is possible that the double β-barrel fold seen, for instance, in the archaeal virus STIV (7) has evolved via gene duplication or gene fusion from the single β-barrel fold in the context of a virus with both pentameric and hexameric capsomers consisting of single vertical barrels (1, 4, 23). This hypothesis is supported by the occurrence of single vertical β-barrels found at the vertices of SH1 (Fig. 4) and the double β-barrel viruses [e.g., the adenovirus penton (34), PRD1 P31 (22), and Bam35 (3)]. The double β-barrel fold is probably more stable than a dimer of two β-barrels. Its major advantage is that as the number of capsomers increases, the number of capsomer components is dramatically reduced by using trimers instead of hexamers, hence reducing the potential number of errors in assembly (35, 36).
In conclusion, this study has given us insights into three main processes relevant to life: the formation of large assemblies, the interaction of proteins with membranes, and virus protein evolution through the examination of molecular fossils.
Materials and Methods
SH1 was grown on H. hispanica and purified as described (8, 10) by using sucrose rate zonal centrifugation in 18% artificial salt water (37). Purified SH1 virions were resedimented in a linear 5% to 20% sucrose rate zonal gradient in SH1 buffer [40 mM Tris·HCl (pH 7.2)/1 M NaCl/40 mM MgCl2; 82,392 × g, 70 min, 22°C], supplemented with 5 mM CaCl2, and concentrated to 4 mg·ml−1 by ultrafiltration. Protein concentration was measured by using the Bradford assay (38). The same virion batch was used for the production of subviral particles as described below.
For production of SH1-VP36 or SH1-VP236 particles, purified virions (1 mg·ml−1) were incubated in either 40 mM Tris·HCl (pH 7.2)/0.75 M Na2SO4/40 mM MgSO4 (42 h, 22°C) or SH1 buffer (20 h, 37°C), respectively, purified by 5% to 20% sucrose rate zonal centrifugation (82,392 × g, 70 min for SH1-VP236 and 90 min for SH1-VP36, 20°C) and concentrated by ultrafiltration. Protein composition was analyzed by Tricine-SDS/PAGE analysis (39). VP3 release was assessed from Coomassie blue-stained gels by using TINA 2.09c software as described previously (10).
For spike isolation, the virions were purified by sucrose rate zonal and CsCl isopycnic centrifugation (10) before dissociation as described above. Material from the top of the gradient was analyzed by size-exclusion chromatography [Tricorn Superdex 200 10/300 GL (GE Healthcare)] and cryo-EM in SH1 buffer/5 mM CaCl2.
For structural analysis, 3-μl droplets of the freshly purified virus, subviral particles, or isolated spikes (in SH1 buffer supplemented with 5 mM CaCl2) were pipetted onto Quantifoil grids, vitrified, imaged, and processed as described previously (14). Initial orientations were found by using a model-based approach (40) with PM2 (16) as the initial model. The orientations were further refined with POR and P3DR (11, 12). A full contrast transfer function (CTF) correction was applied when calculating the reconstructions in P3DR. Fourier shell correlation of 0.5 between two independent reconstructions (see Fig. S2 and Table 1) was used to estimate the resolution (15). Reconstruction statistics are listed in Table 1. The absolute hand of SH1 was determined by using tilt experiments, as described previously, by using HK97 proheads (kind gift of R. Duda, University of Pittsburgh, Pittsburgh, PA) as a reference particle (16, 19). The icosahedral reconstructions were filtered to 10.5-Å resolution before difference imaging.
The density map was manually segmented in EMAN (41). The type III capsomer mass was estimated by averaging the three different capsomers (1, 2, and 3 in Fig. 2). The rotations and translations between the capsomers were determined by fitting a pseudoatomic model of the capsomer into the reconstruction by using CoLoRes (42). The same procedure was applied to average the type II capsomers (4 and 5 in Fig. 2). The mass estimates were based on the volume of the segmented density at 1.2 SDs above the mean by using a protein density of 1.35 g·ml−1, calculated in EMAN (41). The PRD1 MCP (PDB ID code 1HX6) was fitted into the averaged type III capsomer by using CoLoRes (42), and the PRD1 P31 (PDB ID code 1W8X) was fitted into the pentamer by using Chimera (43)
The vertex reconstruction was carried out as described previously (13, 14). Vertex subimages were extracted from the virion data by using the icosahedral orientations. The data were then aligned and grouped into 30 classes with respect to the out-of plane angle (β) in 6° intervals. Multivariate statistical analysis in IMAGIC (44) was then applied to classify the data within β-groups up to 24° out of the plane (thus not overlapping the capsid) by rotation around the fivefold axis to determine the γ-angle). The data were first classified into 10 classes (corresponding to 36° intervals in the γ-angle). From the resulting class averages, 15 clear side views were selected and assigned γ-angles. An initial reconstruction was calculated by using these angles, assuming twofold symmetry. The data were then reclassified into 60 γ-classes, corresponding to 6° intervals, and a model-based iterative process implemented in IMAGIC was used to find the best orientations for the class averages. As a control, vertices were also classified from the SH1-VP36 and the SH1-VP236 images and did not show any averages in which spikes were evident.
Molecular graphics images were produced by using the University of California San Francisco Chimera package (43).
Supplementary Material
Acknowledgments.
We thank B. Löflund, T. Kemppinen, and S. Korhonen for excellent technical support; R. Tuma, J. Huiskonen, and R. Burnett for constructive comments; and the Electron Microscopy Unit (Institute of Biotechnology, Helsinki University) for providing the facilities. This work was supported by Academy of Finland Centre of Excellence Program in Virus Research Grant 1213467 (2006-2011) (to S.J.B. and D.H.B.), the European Science Foundation EUROCORES Program Euro-SCOPE from the European Commission's Sixth Framework Program (Contract ERAS-CT-2003-980409), and Academy of Finland Grants 1112244 (to S.J.B.) and 1210253 (D.H.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The reconstructions reported in this paper have been deposited in the Electron Microscopy Data Bank at the European Bioinformatics Institute with accession codes EMD-1351 (SH1-VP36), EMD-1352 (SH1-VP236), EMD-1353 (SH1 virion), and EMD-1498 (SH1 spike).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801758105/DCSupplemental.
References
- 1.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 3.Laurinmäki PA, Huiskonen JT, Bamford DH, Butcher SJ. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure (London) 2005;13:1819–1828. doi: 10.1016/j.str.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice G, et al. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci USA. 2004;101:7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maaty WS, et al. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol. 2006;80:7625–7635. doi: 10.1128/JVI.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khayat R, et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci USA. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter K, et al. SH1: A novel, spherical halovirus isolated from an Australian hypersaline lake. Virology. 2005;335:22–33. doi: 10.1016/j.virol.2005.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Bamford DH, et al. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J Virol. 2005;79:9097–9107. doi: 10.1128/JVI.79.14.9097-9107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kivelä HM, et al. Quantitative dissociation of archaeal virus SH1 reveals distinct capsid proteins and a lipid core. Virology. 2006;356:4–11. doi: 10.1016/j.virol.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 11.Ji Y, et al. A model-based parallel origin and orientation refinement algorithm for cryoTEM and its application to the study of virus structures. J Struct Biol. 2006;154:1–19. doi: 10.1016/j.jsb.2005.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marinescu DC, Ji Y. A computational framework for the 3D structure determination of viruses with unknown symmetry. J Parallel Distrib Comput. 2003;63:738–758. [Google Scholar]
- 13.Briggs JA, et al. Classification and three-dimensional reconstruction of unevenly distributed or symmetry mismatched features of icosahedral particles. J Struct Biol. 2005;150:332–339. doi: 10.1016/j.jsb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Huiskonen JT, Manole V, Butcher SJ. Tale of two spikes in bacteriophage PRD1. Proc Natl Acad Sci USA. 2007;104:6666–6671. doi: 10.1073/pnas.0608625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harauz G, van Heel M. Similarity measures between images: Exact filters for general geometry of 3D reconstructions. Optik. 1986;73:146–156. [Google Scholar]
- 16.Huiskonen JT, Kivelä HM, Bamford DH, Butcher SJ. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat Struct Mol Biol. 2004;11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 17.Huiskonen JT, Butcher SJ. Membrane-containing viruses with icosahedrally symmetric capsids. Curr Opin Struct Biol. 2007;17:229–236. doi: 10.1016/j.sbi.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 19.Cheng N, et al. Handedness of the herpes simplex virus capsid and procapsid. J Virol. 2002;76:7855–7859. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lata R, et al. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell. 2000;100:253–263. doi: 10.1016/s0092-8674(00)81563-9. [DOI] [PubMed] [Google Scholar]
- 21.Benson SD, Bamford JK, Bamford DH, Burnett RM. The x-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 Å resolution. Acta Crystallogr D Biol Crystallogr. 2002;58:39–59. doi: 10.1107/s0907444901017279. [DOI] [PubMed] [Google Scholar]
- 22.Abrescia NG, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 23.Simpson AA, Nandhagopal N, Van Etten JL, Rossmann MG. Structural analyses of Phycodnaviridae and Iridoviridae. Acta Crystallogr D Biol Crystallogr. 2003;59:2053–2059. doi: 10.1107/s090744490302225x. [DOI] [PubMed] [Google Scholar]
- 24.Trus BL, et al. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhou ZH, et al. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 26.Jensen MO, Mouritsen OG. Lipids do influence protein function: The hydrophobic matching hypothesis revisited. Biochim Biophys Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Strömsten NJ, Bamford DH, Bamford JK. In vitro DNA packaging of bacteriophage PRD1: a common mechanism for internal-membrane viruses. J Mol Biol. 2005;348:617–629. doi: 10.1016/j.jmb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Zhao W, et al. Atomic-scale structure and electrostatics of anionic palmitoyloleoylphosphatidylglycerol lipid bilayers with Na+ counterions. Biophys J. 2007;92:1114–1124. doi: 10.1529/biophysj.106.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bettstetter M, Peng X, Garrett RA, Prangishvili D. AFV1, a novel virus infecting hyperthermophilic archaea of the genus Acidianus. Virology. 2003;315:68–79. doi: 10.1016/s0042-6822(03)00481-1. [DOI] [PubMed] [Google Scholar]
- 30.Simpson AA, Chipman PR, Baker TS, Tijssen P, Rossmann MG. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure (London) 1998;6:1355–1367. doi: 10.1016/s0969-2126(98)00136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, et al. Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands. Cell. 2002;108:383–394. doi: 10.1016/s0092-8674(02)00631-1. [DOI] [PubMed] [Google Scholar]
- 32.Merckel MC, et al. The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture. Mol Cell. 2005;18:161–170. doi: 10.1016/j.molcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zubieta C, Schoehn G, Chroboczek J, Cusack S. The structure of the human adenovirus 2 penton. Mol Cell. 2005;17:121–135. doi: 10.1016/j.molcel.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 35.Burnett RM. The structure of the adenovirus capsid. II. The packing symmetry of hexon and its implications for viral architecture. J Mol Biol. 1985;185:125–143. doi: 10.1016/0022-2836(85)90187-1. [DOI] [PubMed] [Google Scholar]
- 36.Predki PF, Regan L. Redesigning the topology of a four-helix-bundle protein: monomeric Rop. Biochemistry. 1995;34:9834–9839. doi: 10.1021/bi00031a003. [DOI] [PubMed] [Google Scholar]
- 37.Nuttall SD, Dyall-Smith ML. HF1 and HF2: novel bacteriophages of halophilic archaea. Virology. 1993;197:678–684. doi: 10.1006/viro.1993.1643. [DOI] [PubMed] [Google Scholar]
- 38.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 39.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 40.Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryo-electron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 41.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 42.Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.van Heel M, et al. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.