Abstract
Two functions are proposed for the conserved family of UCS proteins: helping to fold myosin motor proteins and stimulating the motor function of folded myosins. We examined both functions in yeast. The fission yeast UCS protein (Rng3p) concentrates in nodes containing myosin-II (Myo2) and other proteins that condense into the cytokinetic contractile ring. Both the N-terminal (central) and C-terminal (UCS) domains of Rng3p can concentrate independently in contractile rings, but only full-length Rng3p supports contractile ring function in vivo. The presence of Rng3p in ATPase assays doubles the apparent affinity (KATPase) of both native Myo2 and recombinant heads of Myo2 for actin filaments. Rng3p promotes gliding of actin filaments by full-length Myo2 molecules, but not Myo2 heads alone. Myo2 isolated from mutant strains defective for Rng3p function is soluble and supports actin filament gliding. In budding yeast the single UCS protein (She4p) acts on both myosin-I isoforms (Myo3p and Myo5p) and one of two myosin-V isoforms (Myo4p). Myo5p turns over ≈10 times faster in she4Δ cells than wild-type cells, reducing the level of Myo5p in cells 10-fold and in cortical actin patches ≈4-fold. Nevertheless, Myo5p isolated from she4Δ cells has wild-type ATPase and motility activities. Thus, a fraction of this yeast myosin can fold de novo in the absence of UCS proteins, but UCS proteins promote myosin stability and interactions with actin.
Keywords: co-chaperone, motor proteins, protein stability
Proteins with Unc-45-/CRO1-/She4p-related (UCS) domains have been implicated in the folding, assembly and functions of myosin motor proteins in animals and fungi. In addition to a C-terminal UCS domain (1), all of these proteins have a central domain (conserved in animals, but not in yeast) and animal UCS proteins have an N-terminal tetratricopeptide (TPR) domain. Unc-45 from Caenorhabditis elegans, the first UCS protein to be characterized, is required for assembly of myosin-II in muscle (2) and cytokinesis (3). Loss of Unc-45 function results in reduced myosin-II levels in muscle because of ubiquitin-mediated degradation by proteasomes (4). Unc-45 binds Hsp90 (via its TPR domain) and the myosin-II motor (via its UCS domain) (5) and is proposed to act as a cochaperone ensuring the folding of myosin motors (1, 5, 6). The action of UCS proteins is not restricted to myosin-II. The budding yeast UCS protein, She4p, acts on type I myosins Myo3p and Myo5p, and type V myosin, Myo4p (7, 8). The fission yeast UCS protein, Rng3p (9), interacts with myosin-II (Myo2p) via its UCS domain (10). Purified Myo2 has actin-activated ATPase activity, but requires Rng3p to move actin filaments effectively in the presence of ATP (10). Rng3p is also required for fission yeast to assemble and constrict the contractile ring of actin filaments and Myo2p during cytokinesis (9).
Here, we report experiments with fission yeast Rng3p and budding yeast She4p to investigate the contributions of UCS proteins to de novo folding of myosins and motor activity of folded myosins. Both the N-terminal “central” and UCS domains of Rng3p are required for function in vivo and to enhance interaction of Myo2 with actin filaments in the presence of ATP. Deletion of the gene for the single budding yeast UCS protein, She4p, reduces cellular levels of the type I myosin Myo5p, owing to faster turn over, but Myo5p isolated from she4Δ cells is soluble and biochemically active. Thus, yeast UCS proteins contribute to both the stability and function of myosin motors.
Results
Rng3p Associates with Assembling and Mature Contractile Rings.
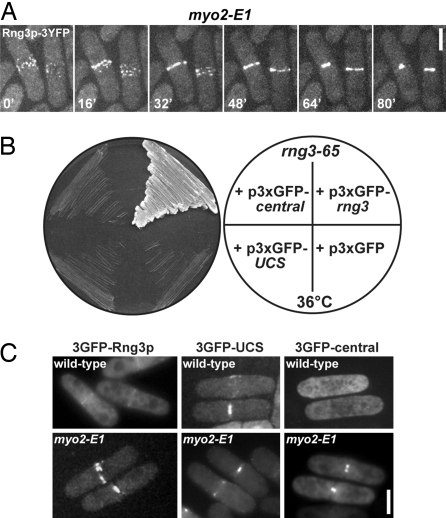
Rng3p-3YFP concentrated in precursors of the contractile ring (nodes) in temperature-sensitive myo2-E1 mutant cells grown at a permissive temperature of 23°C (Fig. 1A). These cells overexpress Rng3p (11) and concentrate it in contractile rings (9). In wild-type cells, Rng3p-3YFP is only detected some time after nodes have condensed into mature rings (10). Accumulation of Rng3p in mature contractile rings did not rely on the septation initiation network (SIN), which triggers ring constriction late in mitosis (12). Rng3p-3xYFP was found in unconstricted rings of arrested temperature-sensitive sid2-250 cells (data not shown) lacking a functional SIN at the restrictive temperature (13).
Fig. 1.
Rng3p central and UCS domains can independently target the contractile ring but both are needed for Rng3p function in vivo. (A) Spinning disk confocal fluorescence micrographs of live myo2-E1 cells expressing Rng3p-3YFP (MLP 683) at 23°C. Cells were mounted on 25% gelatin pads in EMM medium. A time-lapse series of two cells captured at 16-min intervals is shown. Stacks of 10 confocal z sections at intervals of 0.5 μm were collected (projected at maximum intensity) at each time point. (Scale bar: 5 μm.) (B) Viability of a rng3-65 strain (MLP 570) carrying p3xGFP-rng3 (positive control), empty vector (negative control), p3xGFP-UCS, and p3xGFP-central. Vectors contained the weak-strength 81nmt1 inducible promoter. Transformants were streaked on an EMM Ura− plate and grown at 36°C. (C) Epifluorescence images of live wild-type (MLP 479) (Upper) and myo2-E1 (TP 60) (Lower) cells containing plasmids expressing 3GFP-Rng3p (p3xGFP-rng3), 3GFP-UCS (p3xGFP-UCS), or 3GFP-central (p3xGFP-central). Cells were mounted in EMM Ura− medium and photographed after growth in EMM Ura− medium at 25°C. (Scale bar: 5 μm.)
Both the Central and UCS Domain of Rng3p Are Required for Function in Vivo.
To test the independent functions of the Rng3p N-terminal central and C-terminal UCS domains in vivo, we made GFP-fusion constructs of each domain. 3GFP-Rng3p rescued the temperature-sensitivity of rng3-65 cells and concentrated weakly in contractile rings of wild-type cells compared with its strong concentration in rings of myo2-E1 cells (Fig. 1 B and C). Whereas neither truncated form of Rng3p complemented the temperature-sensitivity of the rng3-65 strain (Fig. 1B), 3GFP-UCS concentrated prominently in nodes and contractile rings of both myo2-E1 and wild-type cells (Fig. 1C). 3GFP-central concentrated in highly constricted rings of myo2-E1 cells but not in wild-type cells (Fig. 1C). The average diameter of 3GFP-central rings was 1.1 μm (range 0.5–2 μm), much smaller than the maximum diameter (≈4.5 μm).
Purification of Functional Myo2 Motor Domains.
Acquiring isolated Myo2 motor domains was appealing, because they are expected to be soluble in physiological concentrations of salt like muscle myosin subfragment-1. This solubility was essential for the detailed characterization of muscle myosin ATPase kinetics (14) and crystal structures (15).
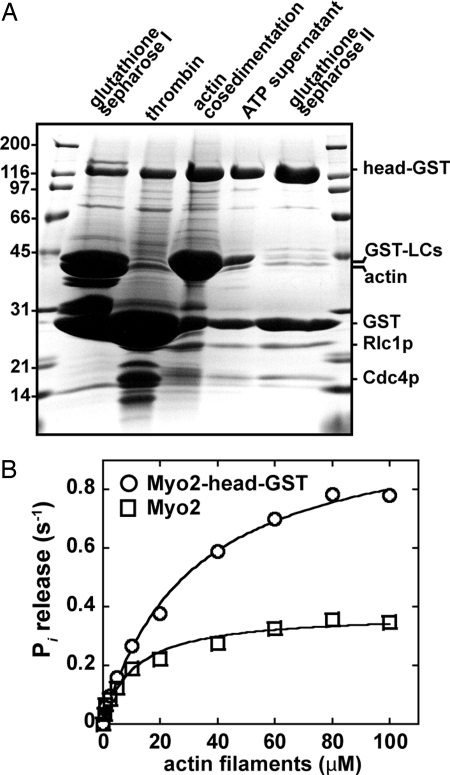
To purify a soluble form of Myo2 (Myo2-head-GST) from fission yeast we co-overexpressed a truncated form of Myo2p (amino acids 1–815, Myo2p motor plus light chain domain) with cleavable GST-tags on both light chains and a permanent C-terminal GST-tag on the heavy chain. Given the dimeric nature of GST, the soluble construct has two myosin heads, each with two light chains. GST facilitates attachment of the myosin heads to cover-slips for in vitro motility assays (16). Fig. 2A summarizes the purification, which yields ≈2 mg of purified Myo2-head-GST from 8-liter of culture. Actin filaments stimulated the steady-state Mg-ATPase activities of both Myo2 (full-length Myo2p plus light chains) and Myo2-head-GST (Fig. 2B). The actin-activated ATPase (VMAX ≈ 1 s−1; KATPase ≈ 30 μM) and K+ Ca2+ ATPase (≈4 s−1) activities of Myo2-head-GST were similar to those of purified Myo2 [Fig. 2B and supporting information (SI) Table S1].
Fig. 2.
Purification of active Myo2-head-GST. (A) Coomassie-stained SDS/PAGE gel summarizing the purification steps of head-GST. Outside lanes display molecular mass markers (kDa). Representative head-GST samples taken during the purification are indicated (left to right): head-GST affinity purified by enrichment of GST-tagged light chains/head on glutathione-Sepharose; thrombin-treated head (removes GST from light chains); head-GST pelleted by cosedimentation with actin filaments in the absence of ATP; soluble head-GST recovered by resuspending the actin-head-GST pellet in the presence of ATP followed by a second round of actin filament pelleting; purified head-GST after removal of residual actin by rebinding of head-GST to glutathione Sepharose. (B) Head-GST (circles) and Myo2 (squares) actin-activated Mg2+-ATPase activity shown as a function of polymerized actin concentration. Basal head-GST and Myo2 ATPase activities generated from control reactions (lacking actin filaments) were subtracted from all rates derived in the presence of polymerized actin (0.5–100 μM based on the concentration of polymerized actin subunits). All assays included myosin at a final concentration of 200 nM in 2 mM Tris·HCl (pH 7.2), 10 mM imidazole, 100 mM KCl, 0.1 mM CaCl2, 3 mM MgCl2, 2 mM ATP, and 1 mM DTT. Each curve was generated from average values obtained from five different datasets and fit to Michaelis–Menten kinetics, using KaleidaGraph software.
Effect of Rng3p on the Actin-Activated ATPase and Motility Activities of Myo2 and Myo2-Head.
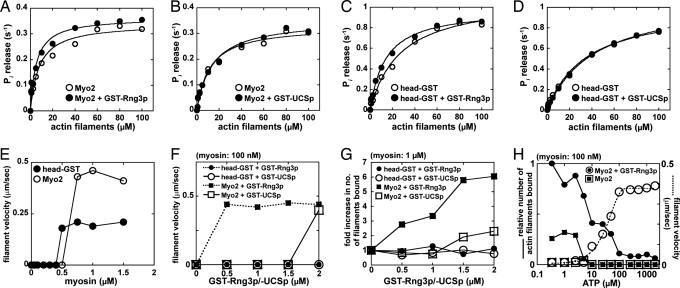
Full-length Rng3p, but not the UCS domain, slightly increased the apparent affinity of Myo2 (Fig. 3 A and B and Table S1) and Myo2-head-GST (Fig. 3 C and D and Table S1) for actin filaments, measured by a reproducible 1.5 to 2-fold reduction in KATPase values. Rng3p did not increase the maximum ATPase activity of either Myo2 or Myo2-head-GST (Table S1).
Fig. 3.
Full-length Rng3p and Myo2p are required for maximal motor activity in vitro. (A–D) Actin-activated Mg2+-ATPase activity shown as a function of polymerized actin concentration for Myo2 plus/minus GST-Rng3p (A), Myo2 plus/minus GST-UCS (B), Head-GST plus/minus GST-Rng3p (C), and Head-GST plus/minus GST-UCS (D). Basal ATPase activities were subtracted from all rates derived in the presence of polymerized actin (0.5–100 μM). All assays included myosin at a final concentration of 200 nM (±800 nM GST-Rng3p/-UCSp) in 2 mM Tris·HCl (pH 7.2), 10 mM imidazole, 100 mM KCl, 0.1 mM CaCl2, 3 mM MgCl2, 2 mM ATP, and 1 mM DTT. For each curve, three separate datasets were averaged and displayed as a single dataset that was fit to Michaelis–Menten kinetics, using KaleidaGraph software. Activities measured in the presence of GST-Rng3p (or GST-UCS) were always recorded in unison with corresponding measurements lacking GST-Rng3p or GST-UCS. (E) The ability of head-GST and Myo2 to support binding and in vitro motility of actin filaments in 1 mM ATP is plotted as a function of the myosin concentration applied to motility chambers. (F and G) The ability of head-GST and Myo2 to support in vitro motility is plotted as a function of GST-Rng3p or GST-UCS concentration. (F) Head-GST and Myo2 were delivered into chambers at a low concentration (100 nM) that does not support filament binding and motility. (G) Head-GST and Myo2 were delivered into chambers at a high concentration (1 μM) that supports filament motility. The number of filaments bound by myosin at the cover-slip surface was averaged by using four 130-μm2 frames taken from fluorescence micrographs from two independent experiments. Average filament numbers are plotted as relative values, with 1 being equal to the number of filaments bound in the absence of GST-Rng3p or GST-UCS. (H) The relative number of actin filaments (where 1 represents the highest recorded value) bound by Myo2 were plotted as a function of the ATP concentration in the running buffer (small filled squares, 50 nM Myo2 alone; small filled circles, 50 nM Myo2 plus 500 nM GST-Rng3p). Filament numbers were quantitated and averaged as described in G. The rate at which Myo2-bound actin filaments move in these assays is also displayed as a function of ATP concentration (large open squares, 50 nM Myo2 alone; large open circles, 50 nM Myo2 plus 500 nM GST-Rng3p).
ATP-dependent actin filament gliding in vitro depends on the presence of Rng3p when the slide is coated with low concentrations of Myo2, but not when the slide was coated with high concentrations of Myo2. At thresholds >750 nM Myo2 (or 500 nM Myo2-head-GST), actin filaments bound to the slides and moved without Rng3p (Fig. 3E). The gliding rates were 0.4 μm/sec for all concentrations of Myo2 and 0.2 μm/sec for all concentrations of Myo2-head-GST (Table S1).
In the presence of ATP Rng3p promoted binding of actin filaments to Myo2 on the slide surface (Fig. 3 F–H). This effect of Rng3p allowed relatively low concentrations of Myo2 (100 nM) to support gliding movements of actin filaments (Fig. 3F). Although gliding rates did not change, the number of actin filaments bound to slides coated with Myo2 increased with the concentration of Rng3p (Fig. 3 F and G). The ATP concentration was an important variable in experiments with low concentrations of Myo2 on the slide. At low ATP concentrations, Myo2 bound but did not move actin filaments (Fig. 3H). At physiological concentrations of ATP, Rng3p was required for Myo2 to support actin filament binding and motility (Fig. 3H).
The Rng3p UCS domain was less active than full-length Rng3p in activating Myo2 motility. Although 0.5 μM Rng3p effectively increased filament binding by low and high concentrations of Myo2, higher concentrations of Rng3p UCS domain (≈2 μM) were needed to activate motility (Fig. 3F) by enhancing the number of filaments bound by Myo2 (Fig. 3G). The effect of Rng3p UCS domain is likely nonspecific considering its failure to enhance Myo2 actin activated ATPase activity (Fig. 3B) and our observation that high concentrations of BSA (equivalent to Rng3p concentrations >10 μM) can enhance actin filament binding in motility assays with either Myo2 or Myo2-head. Given that the Rng3p UCS domain associates with Myo2p (10), the GST-UCS construct represents a negative control for motility experiments involving full-length Rng3p.
In contrast, neither full-length Rng3p nor the Rng3p UCS domain showed any sign of activating motility at low concentrations (100 nM) of Myo2-head-GST (Fig. 3F). Furthermore, with high concentrations of Myo2-head-GST (1 μM), neither Rng3p nor the Rng3p UCS domain increased the number of filaments bound to the surface (Fig. 3G).
Isolation of Functional Myo2 from a rng3 Mutant Strain.
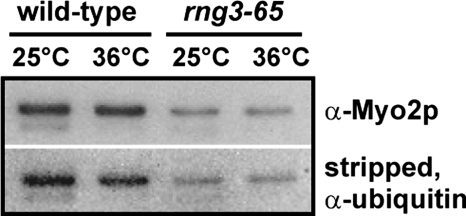
Our reconstitution experiments showing that Rng3p stimulates the motility activity of enzymatically active Myo2 do not rule out a role for Rng3p in the de novo folding of Myo2 in cells (1, 5, 6). An ideal approach to test whether Rng3p is required for Myo2 folding would be to characterize Myo2 from a rng3Δ strain. However, given that loss of rng3 is lethal, we induced overexpression of Myo2 in a rng3-65 temperature-sensitive strain grown under both permissive (25°C) and restrictive (36°C) conditions. At both temperatures, we recovered 3- to 4-fold less Myo2 from rng3-65 cells than from wild-type cells (Fig. 4). Overexpressed Myo2p heavy chains were ubiquitinated to the same degree in wild-type and rng3-65 cells (Fig. 4). Myo2 from rng3-65 cells (induced at 25 or 36°C) supported in vitro motility (Table S1), showing that cells can produce functional Myo2 when Rng3p function is compromised.
Fig. 4.
Isolation of soluble Myo2 from rng3-65 cells. Myo2p and its GST-tagged light chains were overexpressed and 1-step purified from wild-type (MLP 509) and rng3-65 (MLP 586) cells in which the myo2 promoter was replaced with the nmt41 (medium-strength) inducible promoter. Myo2 expression was induced for 22 h at 25°C followed by further induction for 4 h at 25°C or 36°C. Samples purified in one-step by affinity chromatography on glutathione-Sepharose were run on SDS/PAGE gels, transferred to nitrocellulose, and probed with anti-Myo2p tail antibodies to compare levels of Myo2p. The blot was stripped with 4% trichloroacetic acid and reprobed with anti-ubiquitin antibodies to compare levels of Myo2p ubiquitination. Samples were prepared identically and the volumes loaded on gels normalized based on the relative protein concentrations of soluble lysed cell extracts.
Effect of the Budding Yeast UCS Protein on the Cellular Levels and Activities of Myosin-I.
To characterize myosin in the complete absence of its respective UCS protein we turned to budding yeast, which has a single nonessential UCS gene, SHE4 (17). She4p is required for the function of type I myosins (Myo3p and Myo5p), and one type V (Myo4p) myosin (7, 8), but cells are viable (albeit temperature-sensitive) without She4p. For visualization and isolation we replaced the MYO5 gene with MYO5-GFP or MYO5-GST under the control of the native promoter in both wild-type and she4Δ cells (18).
The steady-state amount of Myo5-GST in cell extracts was ≈10-fold lower in she4Δ cells than wild-type cells (Fig. 5 A–C, E, and F) owing to much faster turnover of the newly synthesized protein (Fig. 6C. We carried out pulse–chase experiments in she4Δ cells and wild-type cells with the genomic promoter for MYO5 replaced with an inducible GAL1 promoter. We induced Myo5p-GST expression with a 1-h pulse of galactose. During a 90-min chase without galactose, wild-type cells produce a small amount of additional Myo5p-GST (20%), but Myo5p-GST declined rapidly in she4Δ cells with a half-life of ≈30 min (Fig. 6C). Because wild-type and she4Δ cells synthesized the same amount of Myo5-GST during the 1-h pulse, and because the steady-state level of Myo5-GST is 10-fold lower in she4Δ cells, the half life of Myo5p is >5 h in wild-type cells.
Fig. 5.
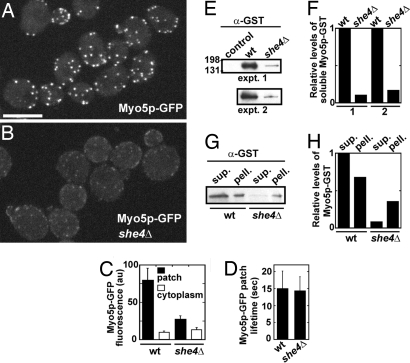
UCS protein She4p maintains the cellular levels of budding yeast Myo5p (myosin-I). Genomic integration at the MYO5 locus was used to generate MYO5-GFP and MYO5-GST strains. Spinning disk confocal fluorescence microscopy of live wild-type (MLY 701) and she4Δ (MLY 724) budding yeast cells expressing Myo5p-GFP was used to generate images, which were analyzed using Image J software. Cells were mounted on 25% gelatin pads in CSM medium. Still or time-lapse images were recorded in stacks of 12 confocal z sections (at intervals of 0.5 μm) and projected at maximum intensity. Relative levels of Myo5-GST recovered from wild-type (MLY 697) and she4Δ (MLY 720) cells were estimated by densitometry of Myo5-GST bands, using Image J software. Myo5p-GST bands were detected by using anti-GST antibodies after immunoblotting of SDS/PAGE gels containing glutathione Sepharose-enriched or fractionated cell lysate samples. (A and B) The subcellular localization of Myo5p-GFP (as patches) in wild-type and she4Δ cells. (Scale bar: 5 μm.) (C) Relative levels of Myo5p-GFP fluorescence detected in both cortical patches and cytoplasm of wild-type and she4Δ cells (n = 10). (D) Average lifetimes of Myo5p-GFP in cortical patches (n = 50) of wild-type and she4Δ cells. (E) Immunoblots indicating the levels of native Myo5p-GST enriched from control (JC 1284, a strain lacking integration of GST at the MYO5 locus), wild-type (MLY 697), and she4Δ (MLY 720) cells. An ≈160-kDa band reacted with α-GST antibodies reflecting the predicted size of Myo5p-GST (155 kDa). The experiment was performed in duplicate, using two alternative segregants corresponding to MLY 697 and 720. (F) Relative levels of Myo5p-GST detected from immunoblots shown in E. (G) Immunoblots indicating levels of native Myo5p-GST in wild-type and she4Δ cell extracts. Cells were lysed and spun to separate extracts into soluble supernatant (sup.) and insoluble pellet (pell.) samples. (H) Relative levels of Myo5p-GST detected from immunoblots in G.
Fig. 6.
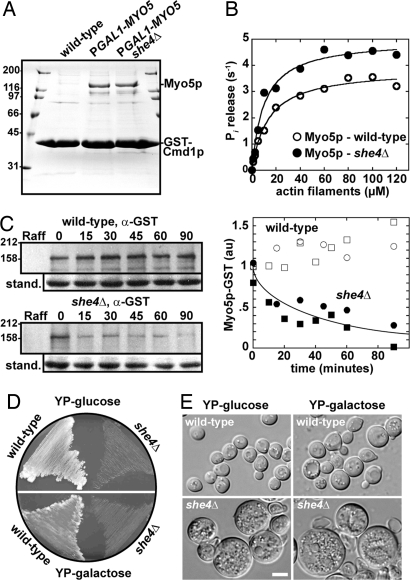
Purification and turnover of Myo5p in the absence of She4p. The genomic MYO5 promoter was replaced with the inducible GAL1 promoter. Myo5p was co-overexpressed with calmodulin (Cmd1p) in budding yeast (A and B) or alone as a Myo5p-GST fusion (C–E). (A) Coomassie-stained SDS/PAGE gel indicating the recovery of soluble Myo5p from wild-type (control cells lacking an integrated GAL1 promoter, Y 258), GAL1promoter-MYO5 (MLY 745), and she4Δ GAL1promoter-MYO5 (MLY 758) cells. All three strains contained a plasmid (pPGAL1-GST-CMD1-TRP1 or pPGAL1-GST-CMD1-URA3) expressing GST-Cmd1p from the galactose-inducible promoter. Protein was enriched by one-step purification on glutathione Sepharose. Outside gel lanes show molecular mass markers (kDa). (B) Actin-activated ATPase activity of Myo5p plotted as a function of polymerized actin concentration. Myo5p was purified in one step (as described in A) from wild-type and she4Δ cells. The concentration of Myo5p in these impure samples was estimated by using densitometry measurements of Coomassie-stained protein bands with rabbit skeletal muscle myosin as the standard. Basal Myo5p ATPase activities were subtracted from all rates derived in the presence of polymerized actin (1–120 μM). All assays included Myo5p at a final concentration of 50–75 nM in 2 mM Tris·HCl (pH 7.2), 10 mM imidazole, 100 mM KCl, 0.1 mM CaCl2, 3 mM MgCl2, 2 mM ATP, and 1 mM DTT. For each curve, two separate datasets were averaged and displayed as a single dataset fit to Michaelis–Menten kinetics, using KaleidaGraph software. (C) Pulse–chase experiment on the biosynthesis and turnover of Myo5p-GST. Wild-type (MLY 801, PGAL1-MYO5-GST) and she4Δ (MLY 802, she4Δ PGAL1-MYO5-GST) cells were grown in rich media containing 1% raffinose before induction of Myo5p-GST expression by addition of 2% galactose for 1 h. Cells were harvested and Myo5p-GST expression repressed by growth in rich media containing 2% glucose. Cell samples were taken before induction (Raff), after induction for 1 h (0) and at time points 15–90 min after initiating repression. Equal amounts of soluble protein in total cell extracts were separated by SDS/PAGE and analyzed by immunoblotting with antibodies to GST. The 155-kDa band is the size expected for Myo5p-GST. The graph shows the time course of the density of the Myo5p-GST bands from two experiments normalized for the intensity of the band in wild-type cells at chase time 0. Circles denote values derived from the blots on the left, squares represent data from an independent experiment. The curved line is an exponential with a half time of 26 min. (D) Wild-type (MLY 801, PGAL1-MYO5-GST) and she4Δ (MLY 802, she4Δ PGAL1-MYO5-GST) cells were grown at 36°C on rich media containing glucose (repressing conditions, Upper) and rich media containing galactose to induce overexpression of Myo5p-GST (Lower). (E) Representative DIC images of wild-type and temperature-sensitive she4Δ cells (taken from plates shown in D).
The concentration of Myo5p-GFP expressed from the native genomic promoter was much lower in the cytoplasm and cortical actin patches of she4Δ cells than wild-type cells (Fig. 5 A–C). Nevertheless, the lifetimes of Myo5p in patches were ≈15 sec in both she4Δ and wild-type cells (Fig. 5D).
Myo5p divided almost equally between the soluble and insoluble fractions of wild-type cells lysed in the presence of high salt and ATP (Fig. 5 G and H). Compared with wild-type cells, Myo5p from she4Δ cells was reduced ≈10-fold in the soluble fraction and ≈2-fold in the insoluble fraction (Fig. 5 G and H). Crude Myo5p-GST enriched from the soluble fraction of wild-type and she4Δ cells had similar ATPase activities (Table S2).
Myo5p purified from she4Δ cells (Fig. 6A) had slightly higher actin-activated ATPase activity (Fig. 6B and Table S3) and supported actin filament gliding at higher rates than Myo5p isolated from wild-type cells by Sun et al. (19) or ourselves (Table S3). We obtained sufficient Myo5p for these assays by overexpressing both the MYO5 heavy chain (from a genomic GAL1 promoter) and the Myo5p light chain calmodulin (20) fused to a GST tag (from a plasmid with the GAL1 promoter) in wild-type and she4Δ cells. Overexpression and 1-step purification of Myo5p/GST-Cmd1p from both wild-type and she4Δ backgrounds yielded soluble Myo5p (Fig. 6A). Given that fully functional Myo5p can be recovered from she4Δ cells, She4p is not essential for de novo folding of the Myo5p motor.
We tested whether increasing the rate of Myo5p synthesis can compensate for the rapid degradation of the protein in the absence of She4p. Overexpression of Myo5p failed to suppress the temperature-sensitivity of she4Δ cells or alter wild-type cells (Fig. 6 D and E).
Discussion
Role of UCS Proteins in Myosin Folding and Stability.
Because one can isolate Myo5p with robust ATPase and motor activity from budding yeast cells lacking UCS proteins, a UCS protein is not absolutely essential for this myosin-I to fold de novo in the cell. However, the steady-state concentration of Myo5p is 10-fold lower in cells lacking a UCS protein, similar to decreased levels of myosin-II in the body wall muscle of a C. elegans UNC-45 mutant strain (4), or in fission yeast cell extracts derived from a myo2-E1 rng3-65 double mutant (21). The reduced steady-state concentration of Myo5p is due to rapid degradation of newly synthesized Myo5p in the absence of She4p. Therefore, the budding yeast UCS protein She4p helps to maintain the steady-state levels of myosin in vivo, either by helping Myo5p to fold before it is degraded or inhibiting the degradation of folded Myo5p. The available data do not exclude either mechanism, although we note that a fraction of newly synthesized native Myo5p folds into a fully active motor protein in the absence of a UCS protein. She4p may protect Myo5p from rapid degradation by ubiquitin-mediated proteolysis given that Myo2p (Fig. 5) and C. elegans myosin-II (4) are ubiquitinated. Like Myo5p, the Myo2p motor unlikely requires a UCS protein to fold because Myo2p motors are soluble in budding yeast cells lacking She4p (22) or fission yeast cells defective in Rng3p function (Fig. 4).
If the sole function of She4p were simply to stabilize myosin levels in the cell, then increasing the rate of Myo5p synthesis by overexpressing its mRNA might offset rapid degradation and rescue the growth defects and temperature-sensitive endocytosis defects of she4Δ cells. However, overexpression of Myo5p failed to suppress the temperature-sensitivity of she4Δ cells (Fig. 6 D and E). This might be due to She4p being required to stabilize other proteins. However, suppressor mutations in MYO5 can rescue the growth defects of she4Δ cells (8), so the temperature-sensitive she4Δ phenotype largely reflects a loss of myosin-I function. The failure of Myo5p overexpression to compensate for its rapid degradation in she4Δ cells suggests that She4p supports Myo5p function in more than one way, such as promoting association of Myo5p with actin filaments. The phenotype of fission yeast cells with the temperature sensitive rng3-65 mutation is also difficult to reconcile with Rng3p being required only for folding Myo2. Given that very low levels of Myo2 activity suffice for cytokinesis (10) Myo2 would have to degrade very rapidly at the restrictive temperature in rng3-65 cells for the failure of cytokinesis to be explained by insufficient Myo2 alone.
Are UCS Proteins Required for Myosin Function?
Our new work and published work supports a second role for UCS proteins in myosin function in yeast cells. Biochemical experiments show that Rng3p promotes binding of Myo2 to actin filaments in the presence of ATP. At the cellular level, both UCS proteins studied here concentrate along with their myosin partners in transient structures that depend on myosin function. Rng3p colocalizes with Myo2p in the precursors of contractile rings (Fig. 1A) and mature contractile rings (9, 10), and She4p concentrates with Myo5p in cortical actin patches (7) that turn over in ≈15 sec. It seems unlikely that these transient structures are sites of myosin folding, so colocalization suggests that the UCS proteins act on folded myosins as they perform their cellular roles. Note, however, that in the whole cell and in the contractile ring, the number of Myo2 molecules exceeds molecules of Rng3p by about a factor of four (11), so some myosin-II proteins are not associated with Rng3p. Because Myo5p isolated from she4Δ cells has robust activity, we do not yet know whether She4p promotes the physiological activities of folded Myo5p.
Mechanism of Action of Rng3p.
Both the central and UCS domains of Rng3p are essential for cellular function. Despite their lack of independent function in vivo, the separate domains both accumulate in the contractile ring. Thus, function requires more than association of the UCS domain with Myo2. The central domain interacts directly with either Myo2p or Rng3p itself (e.g., by dimerization), and may recruit additional binding partners such as Hsp90 (21).
More Rng3p UCS domain concentrates in contractile rings than full-length Rng3p. The UCS domain concentrated in both precursor nodes and contractile rings, whereas Rng3p only concentrated in mature rings. This property may reflect a role for the central domain in regulating the association of the UCS domain with Myo2.
Full-length Rng3p, but not the UCS domain alone, promotes binding of Myo2 to actin filaments in physiological concentrations of ATP in both in vitro motility and ATPase assays (Fig. 3). This effect was most striking in motility assays with millimolar ATP, where Rng3p promoted actin filament binding and gliding motility by Myo2. The ability of Rng3p to promote motility does not reflect a role in preventing motor aggregation and loss of function, because low Myo2 concentrations bound actin filaments in the absence of ATP and Rng3p.
Rng3p likely interacts with the head of Myo2 but also seems to depend on the tail to promote motility. Rng3p promotes the actin-activated ATPase of Myo2-head-GST, but not the ability of Myo2-head-GST to support motility. We note that Rng3p reduced the KATPase for actin filaments from 10 to 5 μM for Myo2 and from 30 to 14 μM for Myo2-head-GST (Table S4). Thus, even with Rng3p, the apparent affinity of Myo2-head-GST for actin filaments may fall short of an affinity threshold required for motility. Alternatively, the ability of Myo2 to form filaments containing multiple motors may be needed for Rng3p to potentiate actin filament binding in motility assays. The latter possibility may explain why UCS proteins colocalize with myosin-II assembled in fission yeast contractile rings (9, 10) and C. elegans body wall muscle (4).
Methods
Yeast Strains, Growth Conditions, and Cell and Genetic Methods.
We used standard methods described in SI Methods to grow yeast and modify their genomes. Table S4 lists fission and budding yeast strains used in this study. Table S5 lists fission and budding yeast plasmids.
Microscopy.
We imaged yeast strains expressing GFP fusion proteins by spinning disk confocal or epi-fluorescence microscopy and processed the images with MetaMorph, UltraView RS, Image J, and PhotoShop software.
Protein Purification.
Details on protein purifications and biochemical assays are provided in the SI Methods. Briefly, full-length Myo2 (Myo2p/Cdc4p/Rlc1p) was purified as described in ref. 10. We purified GST-tagged myosin constructs from budding and fission yeast by affinity chromatography and additional steps. We purified actin from rabbit skeletal muscle for ATPase and in vitro motility assays (10). SI Methods provides additional details about gel electrophoresis and immunoblots (10). We used a pulse–chase experiment to determine the stability of Myo5p-GST in budding yeast. Adding 2% galactose to the medium for 1 h induced a pulse of expression, after which we observe the amount of Myo5p-GST in extracts on immunoblots over time. Tables S1, S2, and S3 summarize the biochemical activities of purified Myo2, Myo2-head-GST and Myo5p.
Supplementary Material
Acknowledgments.
We thank Mohan Balasubramanian (Tamasek Life Sciences Laboratory, Singapore) for the rng3-65 and myo2-E1 mutants, John Chant (Genentech, Inc., South San Francisco, CA) for the she4 null strain, Dan Gelperin (Yale University, New Haven, CT) for the pep4 strain, Dannel McCollum (University of Massachusetts Medical School, Worcester) for the sid2-250 mutant, Mitsuhiro Yanagida (Kyoto University, Kyoto) for the protease-deficient strain, Susan Forsburg (University of Southern California, Los Angeles) for the pDS473a and pDS572a plasmids, and Wei-Lih Lee (University of Massachusetts, Amherst, MA) and John Cooper (Washington University School of Medicine, St. Louis) for the triple GFP/YFP plasmids. We thank Zina Gorelik for technical assistance and members of the T.D.P. laboratory for helpful discussions. This work was supported by National Institutes of Health Grant GM-26132 (to T.D.P.) and a New Research Initiative Award from the University of Vermont College of Medicine (to M.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802874105/DCSupplemental.
References
- 1.Hutagalung AH, Landsverk ML, Price MG, Epstein HF. The UCS family of myosin chaperones. J Cell Sci. 2002;115:3983–3990. doi: 10.1242/jcs.00107. [DOI] [PubMed] [Google Scholar]
- 2.Barral JM, Bauer CC, Ortiz I, Epstein HF. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol. 1998;143:1215–1225. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kachur T, Ao W, Berger J, Pilgrim D. Maternal UNC-45 is involved in cytokinesis and colocalizes with non-muscle myosin in the early Caenorhabditis elegans embryo. J Cell Sci. 2004;117:5313–5321. doi: 10.1242/jcs.01389. [DOI] [PubMed] [Google Scholar]
- 4.Landsverk ML, et al. The UNC-45 chaperone mediates sarcomere assembly through myosin degradation in Caenorhabditis elegans. J Cell Biol. 2007;177:205–210. doi: 10.1083/jcb.200607084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- 6.Yu Q, Bernstein SI. UCS proteins: Managing the myosin motor. Curr Biol. 2003;13:R525–527. doi: 10.1016/s0960-9822(03)00447-0. [DOI] [PubMed] [Google Scholar]
- 7.Wesche S, Arnold M, Jansen RP. The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr Biol. 2003;13:715–724. doi: 10.1016/s0960-9822(03)00264-1. [DOI] [PubMed] [Google Scholar]
- 8.Toi H, Fujimura-Kamada K, Irie K, Takai Y, Todo S, Tanaka K. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:2237–2249. doi: 10.1091/mbc.E02-09-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong KC, Naqvi NI, Iino Y, Yamamoto M, Balasubramanian MK. Fission yeast Rng3p: An UCS-domain protein that mediates myosin II assembly during cytokinesis. J Cell Sci. 2000;113(Pt 13):2421–2432. doi: 10.1242/jcs.113.13.2421. [DOI] [PubMed] [Google Scholar]
- 10.Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 12.Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balasubramanian MK, et al. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 15.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 16.Anson M, Geeves MA, Kurzawa SE, Manstein DJ. Myosin motors with artificial lever arms. EMBO J. 1996;15:6069–6074. [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 18.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Geli MI, Wesp A, Riezman H. Distinct functions of calmodulin are required for the uptake step of receptor-mediated endocytosis in yeast: The type I myosin Myo5p is one of the calmodulin targets. EMBO J. 1998;17:635–647. doi: 10.1093/emboj/17.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra M, D'Souza VM, Chang KC, Huang Y, Balasubramanian MK. Hsp90 protein in fission yeast Swo1p and UCS protein Rng3p facilitate myosin II assembly and function. Eukaryot Cell. 2005;4:567–576. doi: 10.1128/EC.4.3.567-576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lord M, Laves E, Pollard TD. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol Biol Cell. 2005;16:5346–5355. doi: 10.1091/mbc.E05-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.