Abstract
There is substantial evidence that cerebrospinal fluid (CSF) levels of both Aβ42 and tau/ptau are promising biomarkers for Alzheimer's disease (AD). We show that both Aβ and tau exhibit more than 10-fold interindividual variation in CSF levels suggesting that these biomarkers may also be effectively used as endophenotypes for genetic studies of AD. To test the role of common variation in the gene encoding microtubule associated protein tau (MAPT) in influencing CSF tau/ptau levels, we genotyped 21 MAPT single nucleotide polymorphisms (SNPs) in 313 individuals and tested for association with CSF tau/ptau levels. We identified alleles of several SNPs that show association with increased CSF tau/ptau levels. When CSF Aβ42 levels were used to stratify the sample into those with and without likely Aβ deposition in the brain the association was only observed in individuals with evidence of Aβ deposition. This association was replicated in an independent CSF series. When these SNPs were evaluated in a late-onset AD case control series the alleles associated with higher CSF tau/ptau were associated with an earlier age at onset but had no effect on risk for AD. In vivo gene expression studies show that these alleles are associated with increased MAPT mRNA levels in individuals with evidence of brain Aβ deposition. This endophenotype-based approach provides evidence for a gene (MAPT SNPs)–physiological environment (Aβ deposition) interaction that places changes in CSF tau after Aβ deposition and suggest that this interaction predisposes for the development of tauopathy and accelerated disease progression.
Keywords: Alzheimer's disease, genetics
Neurofibrillary tangles (NFT) composed of hyperphosphorylated tau protein are a feature of many neurodegenerative diseases, collectively known as tauopathies. Alzheimer's disease (AD) is characterized by the presence of NFT and amyloid-beta (Aβ) plaques. Association studies between the tau gene (MAPT) and AD have produced inconsistent results (1, 2). Failure to replicate reported associations is common and frequently due to inadequate power resulting from small sample size and genetic and environmental heterogeneity. Using endophenotypes, the issue of heterogeneity in clinical diagnoses is avoided. Quantitative endophenotypes may provide increased power for genetic studies, because they are measurable in many or all individuals and provide a biological model of disease and the possible effects of the associated genetic variation. Cerebrospinal fluid (CSF) Aβ and tau levels have emerged as promising endophenotypes for AD (3). Our recent studies show that levels of CSF 42 amino acid Aβ (Aβ42) and tau are correlated with AD status, disease severity [as measured by clinical dementia rating (CDR)] and Aβ deposition (4–6). CSF tau levels increase in a number of neurodegenerative disorders and are thought to reflect neuronal damage (7). However, elevated levels of soluble phosphorylated tau appear to be specific to AD (8). Leveraging the strengths of an endophenotype-based approach, we genotyped 21 SNPs in MAPT in a series of 313 individuals with measured CSF biomarkers.
Results
CSF tau and ptau181 levels approximated a normal distribution after log–log transformation. Tau levels showed 10-fold variation between individuals (mean = 375 pg/ml, range = 88–1358 pg/ml). Ptau181 levels showed similar levels of interindividual variation (mean = 63 pg/ml, range = 24–241 pg/ml). Our analyses showed significant association between CDR and both tau and ptau181 levels (P < 0.0001 for both tau and ptau181) and association between the apoliprotein E epsilon 4 allele (APOE ε4) and ptau181 (P = 0.013) with a similar but not significant (P = 0.090) trend in tau. Age also showed significant association with tau and ptau181 levels, but this effect was not independent of the CDR effect. We observed no significant association between gender and CSF tau or ptau181 levels.
We genotyped 21 SNPs in the MAPT region, including five SNPs that are commonly genotyped in disease association studies and four other SNPs from previous studies (9, 10) [supporting information (SI) Fig. S1]. The 21 SNPs that we genotyped tag ≈92% of the MAPT SNPs in the HapMap database with an r2> 0.80 (Fig. S2). Four SNPs showed association with CSF ptau181 levels that survived the false discovery rate (FDR) filter: rs16940758 (intron 2; P = 0.0068), rs3785883 (intron 4; P = 0.0061), rs2435211 (intron 5; P = 0.0057), and rs2471738 (intron 8; P = 0.0098; Table 1). The nonparametric Kruskall–Wallis test also yielded significant association for these SNPs. There is strong linkage disequilibrium (LD) between rs2471738 and rs2435211 (D′ = 1, r2 = 0.57) and between rs3785883 and rs16940758 (D′ = 1, r2 = 0.86) but little LD between the two pairs (r2 < 0.15). In each case, the minor allele is associated with increased CSF tau and/or ptau181.
Table 1.
Association of SNPs in MAPT with CSF tau and ptau181 levels
| SNP | MAF | Position | tau | ptau181 |
|---|---|---|---|---|
| rs7210728 | 0.35 | 41324209 | 0.90 (0.0011) | 0.54 (0.0056) |
| rs1560310† | 0.22 | 41334330 | 0.33 (−0.0088) | 0.68 (−0.0041) |
| rs1467967 | 0.34 | 41342006 | 0.66 (0.0036) | 0.95 (−0.00053) |
| rs3785880 | 0.39 | 41349204 | 0.88 (−0.0012) | 0.84 (−0.0018) |
| rs242556 | 0.25 | 41358078 | 0.72 (0.0031) | 0.86 (0.0017) |
| rs12947764 | 0.24 | 41360270 | 0.35 (0.0083) | 0.096 (0.017) |
| rs1001945 | 0.43 | 41361649 | 0.86 (−0.0014) | 0.42 (−0.0070) |
| rs242557 | 0.32 | 41375573 | 0.56 (0.0049) | 0.42 (0.0073) |
| rs878918 | 0.31 | 41387462 | 0.63 (0.0041) | 0.67 (0.0041) |
| rs16940758 | 0.21 | 41396462 | 0.069 (−0.020) | 0.0068 (−0.034) |
| rs3785883 | 0.22 | 41410269 | 0.078 (−0.020) | 0.0061 (−0.035) |
| rs754593 | 0.45 | 41410532 | 0.062 (−0.015) | 0.042 (−0.018) |
| rs2435211 | 0.28 | 41419081 | 0.025 (−0.045) | 0.0057 (−0.054) |
| rs8079215 | 0.29 | 41420688 | 0.42 (0.0068) | 0.25 (0.011) |
| rs2258689 | 0.19 | 41423219 | 0.93 (−0.0028) | 0.49 (−0.024) |
| rs2435200 | 0.42 | 41427688 | 0.19 (0.0098) | 0.23 (0.010) |
| rs2471738 | 0.19 | 41431900 | 0.025 (−0.078) | 0.0098 (−0.10) |
| rs2435203 | 0.14 | 41439134 | 0.94 (0.0022) | 0.84 (−0.0069) |
| rs7521 | 0.45 | 41461242 | 0.50 (0.0051) | 0.43 (0.0067) |
| rs1078997 | 0.16 | 41465690 | 0.73 (0.012) | 0.78 (0.011) |
| rs2696588 | 0.47 | 41578201 | 0.59 (0.0041) | 0.61 (0.0044) |
For each SNP the rs number, minor allele frequency (MAF), position on chromosome 17, and P values with estimates in parentheses for association with tau and ptau181. Values in boldface indicate P values <0.05.
†Major H1/H2 haplotypes.
In a subset of the CSF sample, individuals with CSF Aβ42 levels less than ≈500 pg/ml have been shown to have evidence of Aβ deposition as detected by positron emission tomography, using Pittsburgh compound B (PET PIB), whereas individuals with CSF Aβ42 levels greater than ≈500 pg/ml have negative PET PIB scans (4, 5). To evaluate the association in the likely presence or absence of Aβ deposition we used CSF Aβ42 levels greater or less than 500 pg/ml to stratify the sample approximating a neuropathological definition of cases and controls. For all four SNPs, we observed significant association in the low Aβ42 stratum but not in the high Aβ42 stratum (Table 2). We also observed significant P values for genotype by Aβ42 interaction terms for both tau and ptau181 (tau vs. rs3785883*Aβ42, P = 0.0011; ptau181 vs. rs3785883*Aβ42 P = 0.0033)
Table 2.
P values in the different strata for association of selected SNPs in MAPT with CSF tau and ptau181 levels
| Test | MAF | Stratum | n | Tau | Ptau181 |
|---|---|---|---|---|---|
| rs2471738† | 0.19 | Total sample | 304 | 0.025 | 0.0098 |
| CSF Aβ42 > 500 pg/ml | 168 | 0.45 | 0.76 | ||
| CSF Aβ42 > 500 pg/ml | 136 | 0.021 | 0.024 | ||
| rs16940758 | 0.21 | Total sample | 304 | 0.070 | 0.0068 |
| CSF Aβ42 > 500 pg/ml | 168 | 0.82 | 0.23 | ||
| CSF Aβ42 < 500 pg/ml | 136 | 0.0098 | 0.0026 | ||
| rs2435211† | 0.28 | Total sample | 304 | 0.025 | 0.0057 |
| CSF Aβ42 > 500 pg/ml | 168 | 0.97 | 0.53 | ||
| CSF Aβ42 < 500 pg/ml | 136 | 0.008 | 0.020 | ||
| rs3785883 | 0.22 | Total sample | 304 | 0.070 | 0.0061 |
| CSF Aβ42 > 500 pg/ml | 168 | 0.65 | 0.71 | ||
| CSF Aβ42 < 500 pg/ml | 136 | 0.0044 | 0.0011 |
Based on these findings, we genotyped the higher frequency SNP from each high LD pair (rs2435211 and rs3785883) in a sample of 49 Italian AD cases for whom CSF tau and ptau181 had been measured and performed a one-tailed test for association. In the 49 cases, rs2435211 showed significant association with tau (P = 0.013) but not ptau181 (P = 0.11). Rs3785883 showed significant association with both tau (P = 0.0087) and ptau181 (P = 0.023). Consistent with our initial finding, the minor alleles of these SNPs are associated with increased CSF tau and ptau181 levels.
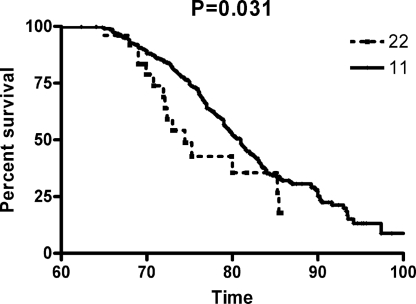
The observation that CSF tau levels are associated with MAPT SNPs only in the presence of Aβ deposition suggests that, rather than affecting risk for AD, these SNPs would influence age at onset (AAO) of clinical symptoms. Specifically, we hypothesized that the minor allele of rs3785883, which is associated with higher CSF tau and ptau181 levels, is associated with earlier AAO of AD. To test this hypothesis, we genotyped rs3785883 in 361 AD cases and 358 age-matched controls and compared the survival curves of the two homozygotes, using a one-tailed Log-rank test. As predicted, the minor allele homozygotes had significantly earlier AAO of AD (P = 0.031; Fig. 1) but not risk for AD (1).
Fig. 1.
Survival curves comparing age at onset of LOAD between the common (11) and rare (22) homozygotes of rs3785883.
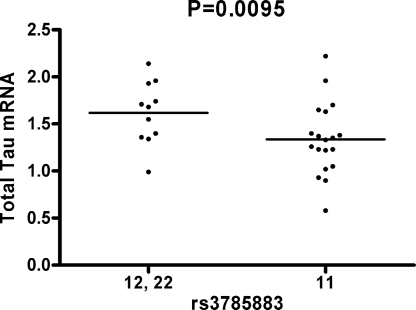
To test the hypothesis that the underlying mechanism involves regulation of gene expression we determined whether the same SNPs were also associated with total MAPT mRNA level (Ttau) or the ratio of exon 10+/exon 10− transcripts (4R/3R). We genotyped rs3785883 in 33 cases and 37 controls in whom Ttau and 4R/3R ratios in the parietal lobe were measured. Because there were only two minor allele homozygotes in the sample, we compared mRNA levels between carriers of the minor allele and noncarriers. In cases, carriers of the minor allele show increased levels of Ttau (P = 0.0095; Fig. 2) but no association with 4R/3R ratio. We failed to detect a significant effect in controls for either Ttau or 4R/3R ratio.
Fig. 2.
Expression of total tau mRNA between rs3785883 minor allele carriers (12, 22) and noncarriers (11).
Association has also been reported between the H1C haplotype of MAPT and risk for late onset Alzheimer disease (LOAD) (2) and progressive supranuclear palsy (11). Previously, we failed to detect association between the H1C haplotype and risk for LOAD (1). Here, we also failed to detect association between the H1C haplotype and CSF tau levels (Table 3). The H1U and H1H subhaplotypes showed significant association with CSF tau levels that survived the FDR = 0.1 filter. Using the same set of SNPs, we failed to detect significant association between any haplotypes and AAO or risk for LOAD in our case-control series.
Table 3.
Haplotype association with CSF tau and ptau181 levels, using UNPHASED
| Name† | Haplotype | Frequency | tau |
ptau181 |
||
|---|---|---|---|---|---|---|
| AddVal | P | AddVal | P | |||
| H2 | 2-1-1-1-1-1 | 0.22 | 0.00 | 0.36 | 0.00 | 0.56 |
| H1I | 1-2-2-1-1-2 | 0.04 | −4.95 | 0.45 | −5.90 | 0.34 |
| H1M | 1-2-2-1-1-1 | 0.03 | 3.34 | 0.21 | 3.45 | 0.42 |
| H1B | 1-2-1-1-1-2 | 0.17 | −1.44 | 0.63 | −1.12 | 0.68 |
| H1C | 1-1-2-1-2-1 | 0.07 | −3.14 | 0.99 | −3.76 | 0.70 |
| H1D | 1-1-2-1-1-2 | 0.05 | −0.76 | 0.31 | 2.53 | 0.32 |
| H1U | 1-1-2-1-1-1 | 0.03 | −8.01 | 0.0009 | −7.51 | 0.0059 |
| H1H | 1-1-1-2-1-2 | 0.06 | 2.22 | 0.023 | 3.05 | 0.017 |
| H1L | 1-1-1-2-1-1 | 0.04 | 2.10 | 0.17 | 1.07 | 0.26 |
| H1E | 1-1-1-1-1-2 | 0.08 | −3.64 | 0.11 | −4.57 | 0.095 |
Haplotypes were estimated from rs1560310, rs1467967, rs242557, rs3785883, rs2471738, and rs7521 (the common allele is represented as ″1,″ the rare allele is represented as a ″2″). For each haplotype with a frequency of >2% in this sample the frequency, additive genetic value and P value for association with CSF tau levels are shown.
†Haplotype names from Pittman et al. (27). Values in boldface indicate P values <0.05.
Discussion
Our results suggest that genetic variation in MAPT influences both CSF tau levels and tau mRNA expression. This association appears to be context-dependent; the effect is observed only in individuals with Aβ deposition. This gene by physiological environment interaction was confirmed by performing an analysis, including an interaction term between rs3785883 genotype and CSF Aβ42 levels. Our observation strongly supports the hypothesis that Aβ deposition is an early feature of AD pathogenesis that subsequently leads to tangle formation and is consistent with clinicopathological observations (12) and data suggesting that either amyloid precursor protein (APP) or Aβ influences the formation of neurofibrillary tangles (13, 14).
Association between CSF tau levels (adjusted for duration of disease but not APOE ε4 genotype) and rs242557 (P = 0.001) was reported in a series of 89 cases by Laws et al. (15). In their report, rs3785883 was marginally significant (P = 0.055). In our threefold larger sample, we failed to detect association with rs242557 but found significant association with rs3785883 and several other SNPs in MAPT. Our findings appear to be consistent with those of Laws et al. (15), suggesting that single SNPs and haplotypes in this region of MAPT are associated with CSF tau levels.
H1 homozygotes have been shown to have increased tau mRNA expression in the cortex (16). Although our sample size was not sufficient to compare expression levels in H1 and H2 homozygotes, we did perform a haplotypic association test (H1 vs. H2). We observed a P value of 0.27 in the expression controls and a P value of 0.055 in the expression cases. The lack of significance does not exclude the existence of an association as our sample size and thus power for these expression studies is very limited. However, the trend observed in the expression cases is consistent with our single SNP findings, which indicate that the association is stronger in individuals with Aβ deposition. We did not detect association between the H1/H2 haplotypes and CSF tau protein levels (see rs1560310 in Table 1). The results of our haplotype analysis (Table 3) show that, of the nine H1 subhaplotypes present in our sample, only two very rare haplotypes showed significant elevation of CSF tau relative to the H2 haplotype. Our data suggest that higher tau mRNA expression associated with H1 may be related to genetic variation within specific H1 subhaplotypes.
Although they may not directly affect whether an individual will develop AD or not, polymorphisms in MAPT are associated with the AAO of disease. This pattern of association may be part of the reason our previous study failed to replicate the associations observed by Myers et al. (17) in their neuropathologically diagnosed (ND) samples. The mean age of controls in our series is 4 years younger than the ND series. The inclusion of younger controls may result in an increase in the number of individuals with MAPT risk alleles that have not yet converted to AD thus reducing power to detect the effect. In our CSF series, 24% of the controls have CSF Aβ42 levels consistent with PIB retention in the brain indicative of Aβ deposition, which, in addition to limited sample sizes and genetic and environmental heterogeneity, may help explain the inconsistent evidence for MAPT single SNP and haplotype associations in LOAD case-control studies.
In conclusion, our data indicate that Aβ deposition directly or indirectly leads to an induction of MAPT gene expression, leading to increased CSF tau levels and increased neurodegeneration as evidenced by earlier AAO of LOAD. These findings also highlight the utility and potential of this endophenotype-based approach for understanding both the genetic etiology and pathobiology of LOAD.
Materials and Methods
Subjects.
Our initial CSF sample consists of 313 European American volunteers participating in studies of aging and dementia at the Alzheimer Disease Research Center at the Washington University School of Medicine from July of 1998 to October of 2006 (18). Approximately 63% of our sample is female, 43–95 years of age (Table 4). Approximately 72% of the sample has a clinical dementia rating (CDR) of 0 (nondemented), whereas 20% of individuals have a CDR of 0.5 (very mild dementia) and 8% of individuals have a CDR of 1.0 (mild dementia) (19). Just more than 42% of the individuals in this sample carry at least one apolipoprotein E epsilon 4 (APOE ε4) allele.
Table 4.
Summary of sample characteristics
| Sample | n | Mean age, years (range, SD) | Female, % | APOE ε4+, % | CDR |
|---|---|---|---|---|---|
| WUSM CSF | 313 | 67.5 (43–91,11.5) | 63 | 42 | 0=72%;0.5=20%; 1=8% |
| Italian CSF | 49 | 70 (55–86, 8.6) | 65 | 50 | All samples CDR > 0 |
| Expression cases | 33 | 85 (72–102, 8) | 51 | 54 | All samples CDR > 0 |
| Expression controls | 37 | 85 (60–107, 9.7) | 58 | 24 | All samples CDR = 0 |
| Cases | 361 | 83 (69–101, 6.9) | 65 | 55 | All samples CDR > 0 |
| Controls | 358 | 78 (60–102, 7.9) | 63 | 23 | All samples CDR = 0 |
Sample size (n), age, percentage of females, percentage of APOE ε4 allele carriers, and clinical dementia rating (CDR) for each sample.
The replication CSF sample is an Italian sample consisting of 49 AD cases from whom CSF has been collected. Dementia severity was assessed by using the CDR and the Mini Mental State Examination. The diagnosis of probable AD was made by exclusion according to National Institute of Neurological and Communication Disorders and Stroke-Alzheimer's Disease and Related Disorders Association criteria. Informed consent to participate in this study was given by all individuals or their caregivers.
The case-control series used in this study was collected through the ADRC patient registry. Cases in this series received a diagnosis of dementia of the Alzheimer's type (DAT), using criteria equivalent to the National Institute of Neurological and Communication Disorders and Stroke-Alzheimer's Disease and Related Disorders Association, modified slightly to include AD as a diagnosis for individuals aged >90 years (20, 21). A total of 361 unrelated DAT cases with a minimum age at onset (AAO) of 60 years were recruited for the study. DNA from 358 age- and sex-matched nondemented controls aged >60 years at assessment were obtained through the ADRC.
Endophenotypes.
CSF was collected from all individuals in the Washington University School of Medicine CSF series. CSF collection, processing, and biomarker measurements were performed as described by Fagan et al. (4). To minimize variability, all draws occurred at 8 a.m. after 12 h of fasting (22). All studies were approved by the Washington University School of Medicine Human Studies Committee, and informed consent was obtained from all subjects.
Italian CSF samples were also obtained after fasting. Samples were collected in polypropylene tubes by LP at the L4/L5 or L3/L4 interspace, centrifuged at 4°C and stored at or below −30°C until analysis. Aβ42, tau, and ptau181 CSF levels were determined with human-specific ELISA kits (Innogenetics) as reported in ref. 23.
Genotyping.
Using HapMap data, tagging SNPs for linkage disequilibrium (LD) bins (r2 > 0.8) were identified. We then added all SNPs with functional annotations in public databases and all SNPs with validated frequencies that occur in regions of conservation between humans and mice (>70% over 100 bp). SNPs were genotyped by using the Illumina Golden Gate, Taqman, and/or Sequenom genotyping technologies. SNPs with call rates of <95% and SNPs for which Hardy–Weinberg equilibrium was rejected at α = 0.05 were excluded from the analyses, which resulted in a total of 21 SNPs being genotyped in the MAPT gene.
Expression.
Total RNA was extracted from the brains (parietal lobes) of 33 autopsy confirmed AD cases and 37 nondemented controls, using the RNeasy mini kit (Qiagen) following the manufacturer's protocol. cDNAs were prepared from the total RNA, using the High-Capacity cDNA Archive kit (ABI).
Gene expression level was analyzed by real-time PCR, using an ABI-7500 real-time PCR system. Two TaqMan assays (Hs00902192_m1 (3R) and Hs00902312_m1 (4R); ABI) were used to quantify MAPT mRNA levels. Primers and TaqMan probe (sequences available on request) for the reference gene, GAPDH, were designed over exon–exon boundaries, using Primer Express software, Version 3 (ABI).
Each real-time PCR run included within-plate duplicates and each experiment was performed, at least twice for each sample. Real-time data were analyzed by using the comparative Ct method (24). The Ct values of each sample were normalized with the Ct value for the housekeeping gene, GADPH, and were corrected for the PCR efficiency of each assay (24), although the efficiency of all reactions was close to 100%. Only the samples with a standard error of <0.15 were analyzed.
Analysis.
CSF tau and ptau181 were log–log transformed to approximate a normal distribution and stepwise discriminant analysis was used to determine the significant covariates (age, gender, CDR, and APOE ε4). SNPs were analyzed for association with CSF tau and ptau181, using analysis of covariance after correction for APOE ε4 and CDR. Each SNP was tested by using an additive model with minor allele homozygotes being coded as 0, heterozygotes being coded as 1, and major allele homozygotes being coded as 2. If the observed minor allele frequency for a SNP was <0.20, we tested the dominant model. In cases where the additive model was significant at 0.05, the dominant and recessive models were tested to determine whether they were a better fit. In addition, to ensure that the log–log transform of tau and ptau181 levels was not adversely affecting our results, MAPT genotypes were tested for association with unadjusted and uncorrected tau and ptau181 levels, using both an ANOVA and a Kruskall–Wallis test. To reduce the probability of false positives, results were filtered by using an FDR of 0.1 (25).
Association between rs3785883 and age at onset (AAO) or risk for AD was evaluated in the case-control series. Allelic and genotypic association with risk for AD was tested by using Fisher's exact test. For the AAO analysis, survival fractions were calculated by using the Kaplan–Meier method and tested for significant differences, using a log-rank test. One-tailed P values were calculated for both tests, because a priori predictions concerning effects on AAO were made based on associations with CSF tau levels. MAPT mRNA levels between genotypes were compared by using ANOVA. Pairwise linkage disequilibrium was calculated by using Haploview (26). To further investigate the observed associations, we tested haplotypes for association with CSF tau levels. We used the default conditions in UNPHASED to test haplotypes from sets of SNPs that have been used for previous haplotype analyses in MAPT for association with CSF tau levels, AAO, and risk for LOAD (27).
Supplementary Material
Acknowledgments.
We thank the individuals who participated in this study; Aarti Shah for contributing to sample processing; and the Genetics, Clinical, Psychometric, and Biostatistics Cores of the Washington University Alzheimer's Disease Research Center for APOE ε4 genotyping and clinical, cognitive, and psychometric evaluation and data management. This work was supported by National Institutes of Health Grants P50-AG05681, P01-AG03991, and P01-AG026276 (to J.C.M.), R01-AG16208 (to A.M.G.), and P30-NS057105 (to Washington University); the Barnes Jewish Foundation; the Associazione “Amici del Centro Dino Ferrari,” Monzino Foundation, Ing. Cesare Cusan; a Ford Foundation Fellowship (to J.S.K.K.); and National Center for Research Resources (a component of the National Institutes of Health and National Institutes of Health Roadmap for Medical Research) Grants 1 UL1 RR024992-01, 1 TL1 RR024995-01, and 1 KL2 RR 024994-01. J.S.K.K. is a Hope Center Fellow supported by the Hope Center for Neurological Disorders and National Institutes of Health Grant T32 MH14677.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801227105/DCSupplemental.
References
- 1.Mukherjee O, Kauwe JS, Mayo K, Morris JC, Goate AM. Haplotype-based association analysis of the MAPT locus in late onset Alzheimer's disease. BMC Genet. 2007;8:3. doi: 10.1186/1471-2156-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myers AJ, et al. The H1c haplotype at the MAPT locus is associated with Alzheimer's disease. Hum Mol Genet. 2005;14:2399–2404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 3.Sunderland T, Hampel H, Takeda M, Putnam KT, Cohen RM. Biomarkers in the diagnosis of Alzheimer's disease: Are we ready? J Geriatr Psychiatry Neurol. 2006;19:172–179. doi: 10.1177/0891988706291088. [DOI] [PubMed] [Google Scholar]
- 4.Fagan AM, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 5.Fagan AM, et al. Cerebrospinal Fluid tau/beta-Amyloid42 Ratio as a Prediction of Cognitive Decline in Nondemented Older Adults. Arch Neurol. 2007;64:343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- 6.Kauwe JS, et al. Extreme cerebrospinal fluid amyloid beta levels identify family with late-onset Alzheimer's disease presenilin 1 mutation. Ann Neurol. 2007;61:446–453. doi: 10.1002/ana.21099. [DOI] [PubMed] [Google Scholar]
- 7.Mazanetz MP, Fischer PM. Untangling tau hyperphosphorylation in drug design for neurodegenerative diseases. Nat Rev Drug Discov. 2007;6:464–479. doi: 10.1038/nrd2111. [DOI] [PubMed] [Google Scholar]
- 8.Hampel H, et al. Measurement of phosphorylated tau epitopes in the differential diagnosis of Alzheimer disease: A comparative cerebrospinal fluid study. Arch Gen Psychiatry. 2004;61:95–102. doi: 10.1001/archpsyc.61.1.95. [DOI] [PubMed] [Google Scholar]
- 9.Pittman AM, et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J Med Genet. 2005;42:837–846. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rademakers R, et al. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum Mol Genet. 2005;14:3281–3292. doi: 10.1093/hmg/ddi361. [DOI] [PubMed] [Google Scholar]
- 11.Pittman AM, et al. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum Mol Genet. 2004;13:1267–1274. doi: 10.1093/hmg/ddh138. [DOI] [PubMed] [Google Scholar]
- 12.Price JL, Morris JC. Tangles and plaques in nondemented aging and “preclinical” Alzheimer's disease. Ann Neurol. 1999;45:358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 13.Gotz J, Chen F, van Dorpe J, Nitsch RM. Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Aβ42 fibrils. Science. 2001;293:1491–1495. doi: 10.1126/science.1062097. [DOI] [PubMed] [Google Scholar]
- 14.Lewis J, et al. Enhanced neurofibrillary degeneration in transgenic mice expressing mutant tau and APP. Science. 2001;293:1487–1491. doi: 10.1126/science.1058189. [DOI] [PubMed] [Google Scholar]
- 15.Laws SM, et al. Fine mapping of the MAPT locus using quantitative trait analysis identifies possible causal variants in Alzheimer's disease. Molecular Psychiatry. 2007;12:510–517. doi: 10.1038/sj.mp.4001935. [DOI] [PubMed] [Google Scholar]
- 16.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 17.Myers AJ, et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis. 2007;25:561–570. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Morris JC, McKeel DW, Jr, Fulling K, Torack RM, Berg L. Validation of clinical diagnostic criteria for Alzheimer's disease. Ann Neurol. 1988;24:17–22. doi: 10.1002/ana.410240105. [DOI] [PubMed] [Google Scholar]
- 19.Morris JC. The Clinical Dementia Rating (CDR): Current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 20.Berg L, et al. Clinicopathologic studies in cognitively healthy aging and Alzheimer's disease: Relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998;55:326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 21.McKhann G, et al. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 22.Bateman RJ, Wen G, Morris JC, Holtzman DM. Fluctuations of CSF amyloid-beta levels: Implications for a diagnostic and therapeutic biomarker. Neurology. 2007;68:666–669. doi: 10.1212/01.wnl.0000256043.50901.e3. [DOI] [PubMed] [Google Scholar]
- 23.Andreasen N, Blennow K. CSF biomarkers for mild cognitive impairment and early Alzheimer's disease. Clin Neurol Neurosurg. 2005;107:165–173. doi: 10.1016/j.clineuro.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1379. [PubMed] [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Pittman AM, et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J Med Genet. 2005;42:837–846. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.