Abstract
Studies of rare, inborn metabolic diseases establish that the phenotypes of some mutations in vitamin-dependent enzymes can be suppressed by supplementation of the cognate vitamin, which restores function of the defective enzyme. To determine whether polymorphisms exist that more subtly affect enzymes yet are augmentable in the same way, we sequenced the coding region of a prototypical vitamin-dependent enzyme, methylenetetrahydrofolate reductase (MTHFR), from 564 individuals of diverse ethnicities. All nonsynonymous changes were evaluated in functional in vivo assays in Saccharomyces cerevisiae to identify enzymatic defects and folate remediability of impaired alleles. We identified 14 nonsynonymous changes: 11 alleles with minor allele frequencies <1% and 3 common alleles (A222V, E429A, and R594Q). Four of 11 low-frequency alleles affected enzyme function, as did A222V. Of the five impaired alleles, four could be restored to normal functionality by elevating intracellular folate levels. All five impaired alleles mapped to the N-terminal catalytic domain of the enzyme, whereas changes in the C-terminal regulatory domain had little effect on activity. Impaired activity correlated with the phosphorylation state of MTHFR, with more severe mutations resulting in lower abundance of the phosphorylated protein. Significantly, diploid yeast heterozygous for mutant alleles were impaired for growth, particularly with lower folate supplementation. These results suggested that multiple less-frequent alleles, in aggregate, might significantly contribute to metabolic dysfunction. Furthermore, vitamin remediation of mutant enzymes may be a common phenomenon in certain domains of proteins.
Keywords: nutrigenetics, polymorphism, vitamin
The ability of enzyme cofactors to remedy metabolic disease-causing mutations has been well documented over the last four decades (for overviews of the field, see refs. 1 and 2). In many cases, the etiological lesion results in a missense change in a cofactor-dependent enzyme, resulting in disruption of a single metabolic step. Subsequent administration of therapeutic doses of the cognate cofactor restores some activity of the mutant enzyme, leading to clinical improvement. Cofactor-remedial mutations occur in many different enzymes that use various cofactors (2). For example, there are numerous vitamin B6 (pyridoxine)-responsive disorders that result from mutations in specific B6-dependent enzymes (2, 3). In addition, the phenylalanine hydroxylase (PAH) cofactor tetrahydrobiopterin (BH4) can be used to treat phenylketonuria caused by several different missense mutations in PAH (4). The molecular mechanisms that underlie clinical remediation are the ability of elevated levels of cofactor to either overcome binding defects of Km mutants (2) or to serve as a chemical chaperone to improve folding and/or stability of mutant enzyme variants (2–5).
Perhaps the best-studied case is the common folate-remedial polymorphism (677C→T; A222V) of 5,10-methylenetetrahydrofolate reductase (MTHFR; ref. 6). For this enzyme, folate is not a cofactor in the traditional sense but instead controls the level of the substrate 5,10-methylenetetrahydrofolate. The A222V change reduces MTHFR activity and increases its thermolability, which can lead to lower levels of serum folate, increased levels of plasma homocysteine, and genomic DNA hypomethylation, particularly in those with a diet low in folate (reviewed in ref. 7). Biochemically, the A222V variant may be less tightly bound to its flavin cofactor and more prone to dissociation into monomers but can be stabilized by reduced folates (8, 9).
Because the A222V variant can affect clinical biomarkers, such as serum homocysteine, and can be modulated by folate levels, this allele has been the subject of many epidemiological disease studies for which these biomarkers may be risk factors and for which folate is thought to be chemopreventive or therapeutic [e.g., neural tube defects (NTDs) (10), cardiovascular disease (ref. 11, but see ref. 12), and colon cancer (13)]. However, genetic association of the A222V allele to disease within all these clinical settings has not been consistent (10–14).
Among the issues that can confound association studies (15), the gene–nutrient interaction between A222V activity and folate status may affect the outcome and conclusions of such studies. Indeed, A222V is a risk factor for colon cancer when dietary folate intakes are low but may actually be protective when folate intakes are elevated (13). A second issue relevant to our work is the recent appreciation of the importance of low frequency (allele frequency <1%) nonsynonymous variants as potential genetic determinants in common diseases such as cardiovascular disease and obesity (16–18). However, the potential confounding influence of low frequency variants to disease susceptibility has not been addressed in most association studies.
We have used MTHFR as a prototype vitamin-dependent enzyme to answer several questions about the occurrence and biochemical impact of low-frequency variation in coding regions, and most importantly, the prevalence of folate-remediation of functionally impaired alleles. To this end, we sequenced the MTHFR coding region from >500 individuals of diverse ethnicities and determined the biochemical impact of all nonsynonymous changes as a function of intracellular folate status by complementation in the yeast S. cerevisiae. The results established that many nonsynonymous substitutions decreased the function of the enzyme and that folate-remediation of impaired alleles was surprisingly common. Furthermore, because cells heterozygous for MHTFR variants displayed quantitative defects, the aggregate frequencies of individually rare alleles may contribute to common phenotypes.
Results
MTHFR Variants in Humans.
The entire coding region of human MTHFR was sequenced by amplifying the coding portion in each of 11 exons from 564 individuals of diverse ethnicities. The lengths of the coding regions, the number of alleles interrogated, and all nonsynonymous substitutions are listed in Table 1. In all, we analyzed 2,081,106 bp of coding DNA and sampled every exon to a depth of >1,000 alleles. These data revealed 14 nonsynonymous changes, 11 of which show a minor allele frequency (MAF) <1%, with seven alleles seen only once. Some low-frequency alleles were seen previously (dbSNP entry in Table 1). The number of low-frequency nonsynonymous substitutions was in good agreement with other studies that sampled deeply into random populations (19–21). In addition, three well studied common substitutions were observed that displayed the expected global population frequencies (A222V, 29.3%; E429A, 23.6%; R594Q, 4.4%).
Table 1.
Spectrum of nonsynonymous MTHFR alleles observed from sampling >500 unselected individuals of diverse ethnicities
| Exon | Length, bp | Alleles sequenced | Variant | Occurrences* | dbSNP rs no. |
|---|---|---|---|---|---|
| 1 | 236† | 1,070 | None | ||
| 2 | 239 | 1,016 | M110I | 1 | Previously unrecognized |
| R134C | 1 | 45550133 | |||
| 3 | 111 | 1,068 | None | ||
| 4 | 194 | 1,050 | A222V | 308 | 1801133 |
| H213R | 1 | Previously unrecognized | |||
| D223N | 1 | Previously unrecognized | |||
| 5 | 251 | 1,056 | D291N | 1 | Previously unrecognized |
| 6 | 135 | 1,042 | None | ||
| 7 | 181 | 1,062 | E429A | 251 | 1801131 |
| G422R | 3 | 45571736 | |||
| 8 | 183 | 1,058 | None | ||
| 9 | 102 | 1,072 | R519C | 2 | 45496998 |
| R519L | 2 | Previously unrecognized | |||
| 10 | 120 | 1,072 | M581I | 1 | 45590836 |
| 11 | 219† | 1,076 | R594Q | 47 | 2274976 |
| T653M | 4 | 35737219 | |||
| Q648P | 1 | Previously unrecognized |
*Genotypes of individual Coriell samples at all nonsynonymous loci can be found in Dataset S1.
†For exons 1 and 11, only the length of the coding portion of the exon is given.
As a quality-control check on the accuracy of the base-calling, we reanalyzed eight variants (including four singletons) by TaqMan allelic-discrimination assays in 100 samples that were independently PCR amplified and saw 100% concordance of the data. Furthermore, population genotyping data from the Environmental Genome Project (www.niehs.nih.gov/envgenom/) and Perlegen, which both used Coriell Institute Cell Repository (Camden, NJ) samples that overlap some in this study (dbSNP build 128) were in concordance in 814 of 817 (99.6%) genotype calls. For two of the three discordant loci, our sequence data were unambiguous and appeared correct.
We obtained complete coding region sequence for 480 individuals. Eighteen (4%) were carriers of a low-frequency nonsynonymous variant. Significantly, the combination of the three common polymorphisms (A222V, E429A, and R594Q) with the range of the low frequency changes led to a great deal of individual heterogeneity. We observed 28 different nonsynonymous genotypes in this group whose haplotype, in most cases, could not be deduced from the data. The genotypes of individual Coriell samples at all nonsynonymous variant loci are included in supporting information (SI) Dataset S1.
MTHFR-Folate Interaction In Vivo.
Because the clinical significance of genetic variants lies in their functional consequence, we tested all nonsynonymous changes for their effect on MTHFR function, and importantly, whether impaired alleles displayed folate responsiveness. The assay is based on complementation of met13, which encodes yeast MTHFR, by human MTHFR (22) but with a modification that allowed us to study the gene–nutrient interaction between MTHFR and folate. Thus, we introduced folate auxotrophy (fol3) into a met13 strain, allowing titration of intracellular folate concentrations by varying folinic acid in the growth media. Folinic acid (5-formyl-tetrahydrofolate) can be metabolized in yeast to methenyl-tetrahydrofolate, which in turn can be converted to other folate coenzymes (23). In this way, we measured human MTHFR functionality (growth in the absence of methionine) as a function of increasingly limiting cellular folate status.
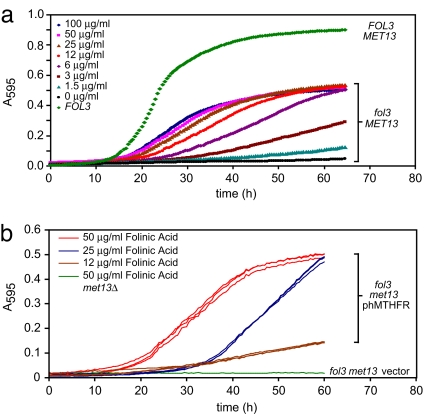
Under these conditions, folinic acid supplementation >50 μg/ml did not confer any significant growth advantage (Fig. 1a). However, at concentrations <50 μg/ml, growth clearly correlated with available folinic acid in the medium. Thus intracellular folate levels were rate limiting in this range. When compared to growth of FOL3 cells, folinic acid supplementation did not completely compensate for lack of endogenous folate biosynthesis. However, this gap was mostly reflected in the density at which cells entered stationary phase rather than growth rate, perhaps reflecting limitations in folinic acid uptake or in the utilization of folinic acid as the sole folate source.
Fig. 1.
Effects of folinic acid supplementation on growth rate of fol3Δ cells and cellular activity of human MTHFR. (a) Growth of fol3Δ MET13 haploid yeast was measured in 96-well plates as described in Materials and Methods. Media was supplemented with folinic acid at the indicated concentrations. The curve labeled FOL3 (FOL3 MET13) was from growth in medium without folinic acid. (b) Growth of fol3Δmet13Δ haploid yeast transformed with phMTHFR in media lacking methionine and supplemented with folinic acid at the indicated concentrations. Three independent transformants were tested at each folinic acid concentration to test reproducibility. The curve labeled met13Δ represented a single isolate of cells, which were transformed with empty vector and grown at 50 μg/ml folinic acid.
The ability of human MTHFR to complement fol3 met13 cells was a function of folinic acid supplementation in the media (Fig. 1b). As for folate supplementation, expression of human MTHFR from the GAL1 promoter did not completely compensate for loss of Met13p (compare Fig. 1b with fol3 MET13 cells at equivalent folate doses in Fig. 1a). Thus, when folinic acid was <50 μg/ml, both folate and MTHFR were rate limiting for growth, allowing even subtle changes in MTHFR activity to be reflected in the growth readout. Note that folinic acid supplementation of >50 μg/ml did not confer a significant growth advantage to cells expressing either the endogenous yeast MTHFR (MET13; Fig. 1a) or the major human allele (Fig. 1b) but was beneficial for impaired alleles of MTHFR (see below).
Functional Impact of MTHFR Variants.
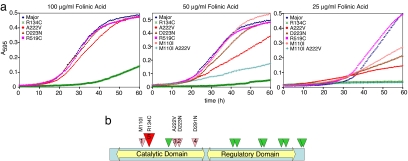
Five nonsynonymous alleles tested over a range of folate concentrations illustrated the range of functional effects observed (Fig. 2a). There was nearly complete restoration of function of the A222V variant at 100 μg/ml folinic acid and significantly less activity (relative to the major allele) at a 4-fold-lower level of supplementation (25 μg/ml). Thus, under these conditions we recapitulated the known folate remediability of the A222V defect. The exact intracellular concentrations of reduced folates in yeast under these conditions was unknown. Nevertheless, the behavior of the A222V allele effectively calibrated the intracellular concentrations in yeast and human cells. The A222V enzyme has ≈50% the intrinsic activity of common allele (19, 24), and we observed a 50% reduction in growth rate at 50 μg/ml folate supplementation. Furthermore, we observed the same 50% drop in A222V enzyme activity in cell-free assays from cells grown at 50 μg/ml folinic acid (Fig. 3). Thus, the behavior of A222V in yeast recapitulated its behavior in human cells.
Fig. 2.
Functional impact and folate remediability of nonsynonymous MTHFR population variants. (a) Six MTHFR variants were tested for the ability to rescue fol3Δ met13Δ cells in media lacking methionine at three different folinic acid concentrations. The M110I allele and the M110I A222V doubly substituted allele were tested only at 50 and 25 μg/ml folinic acid. The curve labeled Major corresponds to the most common MTHFR allele in the population. Each curve is from a pool of three to six independent transformants. (b) Schematic of the MTHFR protein (656 aa) divided into a N-terminal catalytic domain and a C-terminal regulatory domain of nearly equal size (25). Positions of all nonsynonymous changes are indicated. Benign changes are in green. Changes numbered 1–4 represent folate-remedial alleles indicated in increasing order of severity. Change 5 (R134C) was nearly loss-of-function and not designated as folate remedial (see Results) but was somewhat folate augmentable.
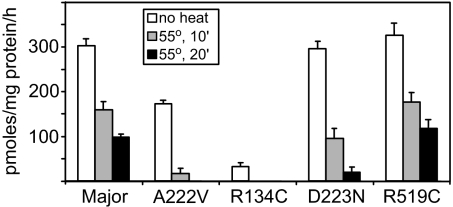
Fig. 3.
Enzyme activity of MTHFR variants. Crude yeast extract from cells transformed with the indicated MTHFR constructs was prepared and assayed for MTHFR activity as described in Materials and Methods. Heat treatment for the indicated times was done on reactions before addition of radiolabeled substrate. Measurements were averages of two independent sets of triplicate assays; error bars indicate standard deviations for the six data points.
Four low-frequency alleles were tested in the same way (Fig. 2a). R519C appeared benign because growth was unaffected at all folate concentrations. R134C was severely impaired at all folate concentrations, although activity was somewhat folate-responsive. The D223N and M110I alleles displayed folate-remedial activity similar to A222V (although less severely impaired) in that growth was similar to the major allele at ≥50 μg/ml folinic acid, but functioned poorly at <50 μg/ml folinic acid.
The MTHFR enzyme has an N-terminal catalytic domain and a C-terminal regulatory domain, which binds the allosteric inhibitor S-adenosylmethionine (AdoMet; ref. 25). Of the six alleles that fell within the catalytic domain (M110I, R134C, H213R, A222V, D223N, and D291N), only H213R was benign (Fig. 2b). M110I, A222V, D223N, and D291N displayed folate-remedial behavior in that these enzyme variants were similar to the major allele at higher concentrations of folate supplementation (50–200 μg/ml folinic acid) but were considerably weakened as folate became more rate limiting. The R134C variant never approached the capacity of the major allele to support growth at any level of folate supplementation and hence was classified as a responsive but not a remedial allele. All substitutions within the regulatory domain (from G422R through T653M) behaved similarly to the major allele (Fig. 2b).
Synergistic Interactions Between Amino Acid Substitutions.
The distribution of variants implied the existence of compound alleles containing two (or more) substitutions. Therefore we created several compound alleles (based on their occurrence in individual samples) to test whether allele combinations lead to synergistic or suppressive effects. For A222V combinations with common variants (A222V E429A and A222V R594Q), we observed minor allele homozygotes for at least one of the alleles and therefore are sure that such variants exist. However, for the low frequency variants, both the A222V variant and the novel variants always occurred as heterozygotes. Because we do not know haplotype, these individuals could harbor either the two single substitution alleles or a compound allele. Therefore we created all possible double-substitution alleles and tested their function (e.g., M110I A222V; Fig. 2a). At the two folinic acid concentrations tested, the M110I A222V variant functioned more poorly than the sum of the individual alleles, indicating synergistic defects in compound alleles. At 50 μg/ml folinic acid, the M110I variant was nearly indistinguishable from the major allele, yet it significantly enhanced the A222V defect. For all combinations tested, alleles that affected function individually (M110I and D291N) synergized when combined with A222V, whereas benign changes did not enhance the A222V defect.
Biochemical Assays Recapitulated in Vivo Function.
To evaluate the reliability of the growth assay, we performed cell-free MTHFR enzyme assays for all variants in crude yeast lysates (see Materials and Methods). In addition to measuring specific activity, variants were tested for thermolability (a measure of enzyme stability) by heat treatment at 55°C for various times. There was a good correlation between intrinsic activity and growth rate (Fig. 3; compare the activities of non-heat-treated samples for the major MTHFR allele, A222V and R134C with the growth curves in Fig. 2). Again, the A222V variant displayed ≈50% of the enzymatic activity of the major allele, as reported previously (19, 22, 24). As in the growth assay, the R519C variant exhibited similar activity to the major allele and was representative of all changes in the regulatory domain including the common E429A variant, further indicating changes in this region were benign (data not shown).
The A222V mutant enzyme is less stable and more thermolabile than the major form (8, 9), and folate remediation of this variant is thought to occur by promoting stabilization of the protein. Under the conditions used here (55°C, 20 min), A222V lost nearly all activity, whereas the major allele retained ≈30% of its original activity, in agreement with previous studies (22). The previously unrecognized D223N allele also displayed increased thermolability that may similarly explain folate remediability in this case, although the enzyme defect was not as great.
Heterozygote Phenotypes.
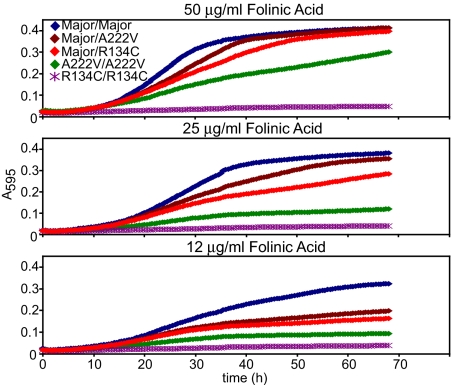
Because low-frequency alleles usually occur as heterozygotes, their significance tends to be dismissed. To understand better the functional significance of heterozygosity of MTHFR alleles, we created diploid yeast with two copies of human MTHFR by mating haploid strains that each have either the same allele expressed from an integrated expression cassette (homozygotes) or different alleles to create heterozygotes (see Materials and Methods). As above, these strains were tested for growth as a function of folate supplementation (Fig. 4). Heterozygotes displayed a growth phenotype in this assay that was exacerbated under conditions of limiting folate, indicating that the reduced-function alleles were codominant with wild type.
Fig. 4.
Heterozygote phenotypes for MTHFR variants as recapitulated in yeast. Homozygosity or heterozygosity of MTHFR alleles was recreated in diploid yeast for the major, R134C, and A222V alleles as described in Materials and Methods. Diploids were obtained from the mating of haploid strains that each expressed a single allele of MTHFR integrated in the genome. Growth as a function of folinic acid supplementation was assayed exactly as for haploids.
Cellular MTHFR activity as measured in the growth assay appeared to reflect additive effects of alleles. Furthermore, additional experiments with hemizygotes (diploids with a single integrated expressed allele; data not shown) demonstrated that the formation of heterodimers between major and minor alleles in heterozygotes offered little or no rescue of mutant alleles. For example, diploid MTHFR major allele/null cells (hemizygotes) behaved similarly to major allele/R134C heterozygotes under all conditions and similarly to major allele/A222V heterozygotes in low folate media (where A222V is inactivated). Thus, the phenotypic contribution of deleterious alleles in heterozygote cells was easily observed, raising the possibility of more widespread phenotypic consequences from heterozygosity in the human genome.
Modification of MTHFR Variants in Yeast by Phosphorylation.
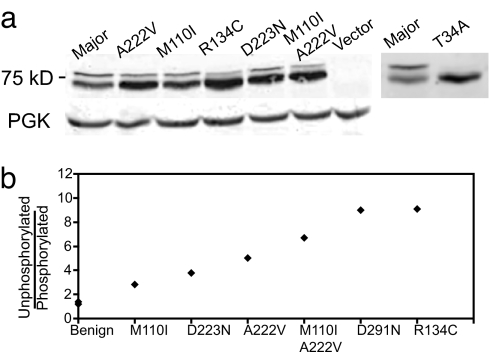
The abundance of MTHFR variant proteins was determined by immunoblotting by using antibodies directed against the N-terminal hemagglutinin A (HA) epitope tag (Fig. 5a). In all samples, the protein ran as a doublet of ≈72 kD and 78 kD. This pattern closely resembled that observed for human MTHFR expressed in insect cells (26), where the upper band represents MTHFR multiply-phosphorylated near the N terminus. Phosphorylation of MTHFR in insect cells depends on a threonine residue at position 34, and substitution of this threonine to alanine (T34A) results in an enzyme that is unable to be phosphorylated (26). This mutation had the same effect on human MTHFR expressed in S. cerevisiae and indicated that, as in insect cells, the upper band was phosphorylated MTHFR (Fig. 5a).
Fig. 5.
Immunoblot of human MTHFR variants expressed in yeast. (a) Extracts were made from yeast cells carrying different MTHFR alleles and detected with anti-HA antibody as in Materials and Methods. A222V M110I was a doubly substituted allele; Major indicates the most common MTHFR allele in the population. The two rightmost lanes were, side-by-side, the major allele and the nonphosphorylatable T34A mutant (26). (b) The ratio of signal intensities of the unphosphorylated lower band to the phosphorylated upper band for all variants of MTHFR identified in this study plotted as a function of increasing severity of functional impact. Alleles on the x axis were classified as benign or rank ordered with respect to activity. All benign alleles (including the Major allele and all regulatory domain changes) were plotted and show nearly identical ratios of the two MTHFR species, thus the symbols overlapped.
The role of phosphorylation of MTHFR is suggested to be involved in negative regulation (26). In support of this hypothesis, the phosphorylation pattern observed here directly correlated with cellular MTHFR activity. Specifically, the ratio of the abundance of the unphosphorylated/phosphorylated forms increased with decreasing activity (Fig. 5b). Interestingly, the overall abundance of all variants (phosphorylated plus unphosphorylated forms) did not appear to be strikingly different. This might not be expected if deleterious substitutions affected intrinsic enzyme stability, unless other factors are involved in determining protein levels (see Discussion).
Discussion
We investigated the prevalence of folate-remediable MTHFR enzyme variants from a large population to determine the incidence and impact of low frequency variation and explore the phenomenon of vitamin remediation. From >500 individuals, we identified 14 different nonsynonymous substitutions, five of which impaired enzyme function. Although all deleterious alleles were at least somewhat folate responsive, four of the five mutant proteins could be fully restored to normal levels by elevating intracellular folate levels. Thus vitamin-dependent rescue of enzyme variants may be a common phenomenon.
All functionally impaired alleles clustered in the N-terminal, catalytic half of MTHFR (25) which contains the folate- and FAD-binding sites. On the other hand, we identified eight nonsynonymous substitutions in the C-terminal regulatory domain of MTHFR, and all eight appeared benign in both the complementation and cell-free enzyme assays. Furthermore, we also saw no synergy between regulatory domain substitutions and A222V in compound alleles. Either these alterations were neutral, as has been reported for E429A (9, 19, 22), or the assay was insensitive to their defect. This finding however was consistent with the observation that most mutations in MTHFR that result in severe clinical phenotypes occur in the catalytic domain (www.hgmd.cf.ac.uk). The regulatory domain has been proposed to play a role in stabilization of the catalytic domain (22). If so, this role may be somewhat tolerant to amino acid substitutions and may explain how a chimeric MTHFR composed of the S. cerevisiae N-terminal domain fused to the Arabidopsis C-terminal domain (equivalent to ≈50 nonsynonymous substitutions of the yeast enzyme in the regulatory domain) does not harm enzyme activity (27). It should be noted that Martin et al. (19) reported that the common R594Q variant in the C-terminal domain affected enzyme activity when expressed in COS-1 cells. This change appeared benign, however, in cell-based and cell-free assays of the enzyme expressed in yeast. Although the reason for this discrepancy is unclear, it may be reflective of the host expression system because these authors observed only a single species of MTHFR (unknown phosphorylation status) in their immunoblot analyses.
The Phenotypes of Heterozygotes.
The behavior of diploid yeast heterozygous for functionally impaired MTHFR alleles demonstrated that heterozygote phenotypes were clearly observable, especially under conditions of limiting folate (Fig. 4). The appearance of phenotypes in heterozygotes was significant because most genetic variation occurs as heterozygosity and low frequency alleles exist primarily as heterozygotes in the population. This result is consistent with the observations that cellular MTHFR activity in lymphocyte extracts is directly correlated with genotype: individuals heterozygous for A222V (A/V) have ≈65% of the total activity seen for major allele (A/A) homozygotes, where A222V homozygotes (V/V) retain 30% of the activity of A/A homozygotes (6). In a recent study examining the full spectrum of alleles in the adipokine ANGPTL4, which affects serum triglyceride levels, heterozygosity for the nonsynonymous E40K allele was significantly associated with lower plasma triglyceride levels (17). Thus, cases in which heterozygosity is phenotypically detectable increases the significance of the contribution of low frequency variants because there can be orders of magnitude more carriers than homozygotes. Note that we observed heterozygote phenotypes under conditions in which MTHFR activity was rate limiting for cell growth. Whether enzymatic steps are rate limiting in a particular pathway in humans depends on both genetic and environmental factors.
Mutations and MTHFR Phosphorylation and Abundance.
Folate remediation of nonsynonymous changes in the catalytic domain may occur by protein stabilization (as for A222V; refs. 8 and 9) or by overcoming other aspects of molecular function such as cofactor Km (2). At least one deleterious allele, D223N, showed increased thermolability (Fig. 3) analogous to A222V, which argued for a stability defect. The hypothesis that folate-remedial alleles of MTHFR are those in which a folate species stabilizes unstable forms of the enzyme would suggest that the level of MTHFR protein be proportional to intrinsic activity of the variants, as has been suggested (19). However, our observations indicated that although phosphorylation status correlated with enzyme activity (Fig. 5), the overall abundance (phosphorylated plus unphosphorylated forms) did not appear to change strikingly (within a 2-fold range). It is unlikely that phosphorylated MTHFR is the active form of the enzyme because Yamada et al. (26) demonstrated an inhibitory effect of phosphorylation on intrinsic activity. Consistent with this, the behavior of the nonphosphorylatable T34A variant in both the growth and enzyme assays was similar to that of the major allele (data not shown). Furthermore, although low intracellular folate levels decrease MTHFR stability (as measured by abundance), this effect is not enhanced in variants that impair function. Because these results are at variance with the expected protein destabilization of deleterious changes, we deduced there must be a compensatory regulatory response that is currently under investigation. In this way the activity of variants could be strikingly different (Fig. 2), whereas the overall protein abundance may not be (Fig. 5). Although our results are consistent with feedback regulation by phosphorylation (26), the role of phosphorylation in turnover is unknown. In this vein, it will be interesting to determine the effect of the T34A change in combination with other impaired alleles.
Broader Implications.
There are ≈600 annotated cofactor-dependent enzymes in the human proteome (http://au.expasy.org/enzyme). If the number of deleterious low-frequency variants seen for MTHFR (four variants per 1,000 sequenced alleles) extends onto these genes, we would expect every individual to carry approximately five missense alleles that impair enzyme function and would be nutrient remedial. The contribution from common alleles (e.g., A222V) would further increase this incidence. Extrapolating to 30,000 proteins in the proteome, every individual would harbor ≈250 deleterious substitutions considering only the low-frequency variants. These numbers suggest that the aggregate incidence of low frequency variants could have a significant physiological impact, particularly if heterozygotes have phenotypic consequences. Furthermore, rarer alleles are predicted to be more dysfunctional (28), and the genetic heterogeneity creates the possibilities for synergies and allele–allele interactions in a individual-specific way. In addition, this study also suggests that vitamin remediation of mutant enzymes may be a common phenomenon. The ability to remedy dysfunctional enzymes in such a manner has the potential to provide corrective measures to individuals who harbor such alleles.
Materials and Methods
DNA Sample Population.
DNA samples were from the Coriell Institute Cell Repository. The make-up of the 564-member panel is included in Table S1.
MTHFR Exon Sequencing.
Eleven MTHFR coding exons were sequenced in the above samples by PCR sequencing using primer pairs commercially available from the Variant SeqR product line (Applied Biosystems) and according to the protocols supplied. The exon regions sequenced corresponded to National Center for Biotechnology Information MTHFR reference sequences for mRNA (NM_005957) and the corresponding protein (NP_005958) of 656 aa. Sequencing amplicon and probe information is available at www.ncbi.nlm.nih.gov/genome/probe for the following target amplicons: Exon 1 (RSA000045684); Exon 2 (RSA000045680); Exon 3 (RSA000577249); Exon 4 (RSA000045678); Exon 5 (RSA000045676); Exon 6 (RSA001308795); Exon 7 (RSA001253193); Exon 8 (RSA000045669); Exon 9 (RSA000580767); Exon 10 (RSA000580766); and Exon 11 (RSA000580765, RSA000027240). Only the portion of exon 11 that spanned the coding region was sequenced. To ensure high confidence in base calling, only high-quality reads were used for analysis (average QV scores >40 for the region that spanned the target exon; all exons were covered by double-strand reads). Based on these filtering criteria, success rates ranged from 89% to 95% for each exon (Table 1). All sequence information was analyzed by using the SeqScape software suite (Applied Biosystems). As a quality control measure, a subset of base calls were directly verified by TaqMan (Applied Biosystems) allelic-discrimination assays and compared with publicly available genotype data as described (see Results).
Plasmids.
Plasmid phMTHFR, which carries the 5′-end HA (hemagglutinin A) epitope-tagged human MTHFR ORF under the control of the inducible yeast GAL1 promoter and the URA3 selectable marker, was a generous gift of Warren Kruger (Fox Chase Cancer Center, Philadelphia) (22). This plasmid served as the backbone to reconstruct all MTHFR variants by site-directed mutagenesis by using the QuikChange kit (Stratagene). Integrating plasmids containing galactose-inducible MTHFR variants were created by PCR cloning the fragment containing URA3, the GAL1 promoter, and MTHFR coding region from the phMTHFR-based plasmid into pHO-polyHO (29), which enables targeted integration of this cassette at the HO locus.
Strains.
All haploid yeast strains were MATa his3 leu2 ura3 lys2 in the S288c background (30). MATa/MATα diploid strains were created by mating isogenic MATa and MATα strains. fol3Δ::KanMX and fol3Δ::KanMX met13Δ::KanMX strains were obtained by standard mating/sporulation techniques by using strains from the S. cerevisiae gene-knockout collection (Invitrogen). Diploids (homozygous or heterozygous for MTHFR variants) were created by mating fol3Δ::KanMX met13Δ::KanMX haploids that each contain an integrated version of the GAL1:MTHFR variant cassette.
Growth Conditions.
Synthetic growth media lacking folate was minimal media (31) with Yeast Nitrogen Base without Vitamins (Qbiogene), and all vitamins except folate were added back individually. All fol3Δ::KanMX cells were supplemented with 50 μg/ml folinic acid (Sigma). For kinetic growth measurements, fol3Δ::KanMX met13Δ::KanMX cells were transformed with GAL1 promoter-driven MTHFR variants and grown to log phase in synthetic galactose medium (2% galactose, 0.1% glucose) supplemented with folinic acid (50 μg/ml) and methionine (20 μg/ml). Cells were washed three times and aliquoted into 96-well plates containing fresh galactose media with various amounts of folinic acid, but lacking methionine. The volume per well was 200 μl with a starting cell density of OD595 = 0.01. Absorbance was tracked every 15–30 min for at least 60 h in a Tecan GENios plate reader at 30°C with no shaking. MET13 cells used in Fig. 1a were treated the same way except that all growth was in the absence of methionine.
MTHFR Enzyme Activity Assay.
The assay, which measures the reverse reaction of that catalyzed by MTHFR under physiological conditions, was as described (22) with the following modifications: Yeast extracts were created by bead lysis of 40 OD595 cell equivalents (fol3Δ met13Δ cells supplemented with folinic acid and methionine) in 350 μl of lysis buffer [100 mM sucrose, 50 mM KHPO4 (pH 6.3), and protease inhibitor mixture]. Extracts were clarified by a brief microcentrifugation, and 10–200 μg of extract was used to determine the linear range of activity. Radiolabeled substrate (5-[14C]MeTHF) was from GE Healthcare Life Sciences. For heat treatment, the reaction mixes without 5-[14C]MeTHF were heated to 55°C for the indicated times at which point 5-[14C]MeTHF was added back and the reaction proceeded.
MTHFR Immunoblot Analysis.
Ten OD595 cell equivalents (fol3Δ met13Δ cells supplemented with folinic acid and methionine) were extracted in 200 μl 0.1 M NaOH for 15 min. SDS sample buffer (50 μl) (0.5 M Tris 6.8, 0.4% SDS) was added to supernatants, which were then boiled, clarified, and subject to SDS/PAGE. HA-tagged MTHFR variants were detected on a LI-COR Infrared Imager. Mouse monoclonal anti-HA antibody was from Sigma. Yeast 3-phosphoglycerate kinase (Pgk1p), a loading control, was detected by mouse antibodies generously donated by Jeremy Thorner (University of California, Berkeley, CA).
Supplementary Material
Acknowledgments.
We thank Warren Kruger for human MTHFR plasmids, Jeremy Thorner for anti-yeast PGK antibody, and Dago Dimster-Denk for comments on the manuscript. This work was supported by a University of California Discovery Grant (bio03-10395), the Defense Advanced Research Projects Agency and U.S. Army Research Office Grant W911NF-06-1-0166, and National Institutes of Health Grant GM072859.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802813105/DCSupplemental.
References
- 1.Scriver CR. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw–Hill; 2001. [Google Scholar]
- 2.Ames BN, Elson-Schwab I, Silver EA. High-dose vitamin therapy stimulates variant enzymes with decreased coenzyme binding affinity (increased k(m)): Relevance to genetic disease and polymorphisms. Am J Clin Nutr. 2002;75:616–658. doi: 10.1093/ajcn/75.4.616. [DOI] [PubMed] [Google Scholar]
- 3.Clayton PT. B6-responsive disorders: A model of vitamin dependency. J Inherit Metab Dis. 2006;29:317–326. doi: 10.1007/s10545-005-0243-2. [DOI] [PubMed] [Google Scholar]
- 4.Perez B, et al. Kinetic and stability analysis of pku mutations identified in bh4-responsive patients. Mol Genet Metab. 2005;86(Suppl 1):S11–S16. doi: 10.1016/j.ymgme.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Wittung-Stafshede P. Role of cofactors in protein folding. Acc Chem Res. 2002;35:201–208. doi: 10.1021/ar010106e. [DOI] [PubMed] [Google Scholar]
- 6.Frosst P, et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y-I. 5,10-Methylenetetrahydrofolate reductase polymorphisms and pharmacogenetics: A new role of single nucleotide polymorphisms in the folate metabolic pathway in human health and disease. Nutr Rev. 2005;63:398–407. doi: 10.1111/j.1753-4887.2005.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 8.Guenther BD, et al. The structure and properties of methylenetetrahydrofolate reductase from escherichia coli suggest how folate ameliorates human hyperhomocysteinemia. Nat Struct Biol. 1999;6:359–365. doi: 10.1038/7594. [DOI] [PubMed] [Google Scholar]
- 9.Yamada K, Chen Z, Rozen R, Matthews RG. Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc Natl Acad Sci USA. 2001;98:14853–14858. doi: 10.1073/pnas.261469998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Linden IJ, Afman LA, Heil SG, Blom HJ. Genetic variation in genes of folate metabolism and neural-tube defect risk. Proc Nutr Soc. 2006;65:204–215. doi: 10.1079/pns2006495. [DOI] [PubMed] [Google Scholar]
- 11.Wald DS, Law M, Morris JK. Homocysteine and cardiovascular disease: Evidence on causality from a meta-analysis. Br Med J. 2002;325:1202. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazzano LA, Reynolds K, Holder KN, He J. Effect of folic acid supplementation on risk of cardiovascular diseases: A meta-analysis of randomized controlled trials. J Am Med Assoc. 2006;296:2720–2726. doi: 10.1001/jama.296.22.2720. [DOI] [PubMed] [Google Scholar]
- 13.Ulrich CM. Nutrigenetics in cancer research—folate metabolism and colorectal cancer. J Nutr. 2005;135:2698–2702. doi: 10.1093/jn/135.11.2698. [DOI] [PubMed] [Google Scholar]
- 14.Lewis SJ, Ebrahim S, Davey Smith G. Meta-analysis of mthfr 677c→t polymorphism and coronary heart disease: Does totality of evidence support causal role for homocysteine and preventive potential of folate? Br Med J. 2005;331:1053. doi: 10.1136/bmj.38611.658947.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton-Cheh C, Hirschhorn JN. Genetic association studies of complex traits: Design and analysis issues. Mutat Res. 2005;573:54–69. doi: 10.1016/j.mrfmmm.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 17.Romeo S, et al. Population-based resequencing of angptl4 uncovers variations that reduce triglycerides and increase hdl. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaisse C, et al. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000;106:253–262. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin YN, et al. Human methylenetetrahydrofolate reductase pharmacogenomics: Gene resequencing and functional genomics. Pharmacogenet Genomics. 2006;16:265–277. doi: 10.1097/01.fpc.0000194423.20393.08. [DOI] [PubMed] [Google Scholar]
- 20.Livingston RJ, et al. Pattern of sequence variation across 213 environmental response genes. Genome Res. 2004;14:1821–1831. doi: 10.1101/gr.2730004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glatt CE, et al. Screening a large reference sample to identify very low frequency sequence variants: Comparisons between two genes. Nat Genet. 2001;27:435–438. doi: 10.1038/86948. [DOI] [PubMed] [Google Scholar]
- 22.Shan X, Wang L, Hoffmaster R, Kruger WD. Functional characterization of human methylenetetrahydrofolate reductase in saccharomyces cerevisiae. J Biol Chem. 1999;274:32613–32618. doi: 10.1074/jbc.274.46.32613. [DOI] [PubMed] [Google Scholar]
- 23.Cherest H, Thomas D, Surdin-Kerjan Y. Polyglutamylation of folate coenzymes is necessary for methionine biosynthesis and maintenance of intact mitochondrial genome in saccharomyces cerevisiae. J Biol Chem. 2000;275:14056–14063. doi: 10.1074/jbc.275.19.14056. [DOI] [PubMed] [Google Scholar]
- 24.Rozen R. Genetic predisposition to hyperhomocysteinemia: Deficiency of methylenetetrahydrofolate reductase (MTHFR) Thromb Haemost. 1997;78:523–526. [PubMed] [Google Scholar]
- 25.Sumner J, Jencks DA, Khani S, Matthews RG. Photoaffinity labeling of methylenetetrahydrofolate reductase with 8-azido-s-adenosylmethionine. J Biol Chem. 1986;261:7697–7700. [PubMed] [Google Scholar]
- 26.Yamada K, Strahler JR, Andrews PC, Matthews RG. Regulation of human methylenetetrahydrofolate reductase by phosphorylation. Proc Natl Acad Sci USA. 2005;102:10454–10459. doi: 10.1073/pnas.0504786102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roje S, et al. Isolation, characterization, and functional expression of cdnas encoding nadh-dependent methylenetetrahydrofolate reductase from higher plants. J Biol Chem. 1999;274:36089–36096. doi: 10.1074/jbc.274.51.36089. [DOI] [PubMed] [Google Scholar]
- 28.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voth WP, Richards JD, Shaw JM, Stillman DJ. Yeast vectors for integration at the HO locus. Nucleic Acids Res. 2001;29:e59. doi: 10.1093/nar/29.12.e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Sherman F. Getting started with yeast. In: Guthrie C, Fink G, editors. Guide to Yeast Genetics and Molecular Biology. New York: Academic; 2002. pp. 3–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.