Abstract
Yersinia pestis, the agent of bubonic plague, evolved from the enteric pathogen Yersinia pseudotuberculosis within the past 20,000 years. Because ancestor and descendant both exist, it is possible to infer steps in molecular evolution by direct experimental approaches. The Y. pestis life cycle includes establishment of a biofilm within its vector, the flea. Although Y. pseudotuberculosis makes biofilms in other environments, it fails to do so in the insect. We show that rcsA, a negative regulator of biofilms that is functional in Y. pseudotuberculosis, is a pseudogene in Y. pestis. Replacement of the pseudogene with the functional Y. pseudotuberculosis rcsA allele strongly represses biofilm formation and essentially abolishes flea biofilms. The conversion of rcsA to a pseudogene during Y. pestis evolution, therefore, was a case of negative selection rather than neutral genetic drift.
Keywords: biofilm, flea, bubonic plague, phosphorelay, Caenorhabditis elegans
Evolution leaves pseudogenes in its wake. Pseudogenes are DNA sequences that have been inactivated by one or more mutations, but retain enough structure to be recognizable as the remnants of functional genes. Pseudogenes can be the result of neutral genetic drift: If a gene becomes unnecessary, random mutations accumulate in the absence of selection (1). A second possibility is that a pseudogene arises by negative selection when a functional gene becomes deleterious in a new environment.
Distinguishing between drift and negative selection experimentally is impossible in most cases because ancestral organisms are extinct. However, laboratory analysis is feasible for bacteria whose ancestors remain extant. A study of the enteric pathogen Shigella flexneri found that it lacks a large locus present in its ancestor, Escherichia coli. Heterologous expression in S. flexneri of a single E. coli gene from the locus reduced the pathogen's virulence, suggesting that negative selection was involved in the S. flexneri deletion (2).
Similar analysis, examining individual genes rather than large deletions, is possible using the bubonic plague bacterium Yersinia pestis and its ancestor, Yersinia pseudotuberculosis. Molecular evidence indicates that Y. pestis evolved from Y. pseudotuberculosis within the past 20,000 years (3), and DNA sequences of most of their common genes are at least 97% identical (4). Recently evolved pathogens are predicted on theoretical grounds to contain many pseudogenes (1). This was confirmed for Y. pestis by a comparison to Y. pseudotuberculosis that identified 208 putative Y. pestis pseudogenes, or ≈5% of the genome (4).
Plague is spread by flea bites. After feeding on a septicemic mammal, the insects can become “blocked” by a chronic Y. pestis infection in the digestive tract that prevents food from reaching the midgut. This starves the flea and prompts it to bite mammals repeatedly in futile attempts to feed, inoculating new hosts in the process. Not all infected fleas become blocked, and unblocked fleas are also capable of spreading disease, but it is nevertheless believed that blockage is crucial for long-term enzootic persistence of Y. pestis (5–7). Blockage is due to a biofilm that binds tightly in the proventriculus, an organ in the flea digestive tract; this makes the infection less susceptible to clearance by peristalsis and defecation and thus enhances high-titer colonization for extended periods (8, 9). The in vivo biofilm, consisting of bacteria in a self-synthesized, exopolysaccharide-rich matrix, requires the hmsHFRS operon of polysaccharide biosynthetic genes (8, 9). Y. pseudotuberculosis contains a functional hmsHFRS operon (10, 11), but it is unable to make biofilms in fleas (11). This suggests that the different biofilm capabilities in fleas are due to functions other than exopolysaccharide (EPS) biosynthesis.
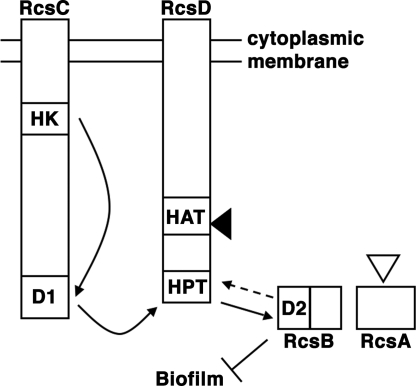
Bacterial phosphorelays are signal transduction pathways that are similar biochemically to two-component systems but more complex (12–14). In two-component systems, a sensor histidine kinase autophosphorylates, then transfers the phosphate to an aspartate in the receiver domain of a response regulator. In phosphorelays, there are multiple signaling steps between these amino acids, e.g., His-Asp-His-Asp (12). The Rcs phosphorelay is conserved in Enterobacteriaceae, including E. coli and Y. pestis (15). The current understanding of Rcs, based on work in E. coli, is summarized in Fig. 1. At the end of the relay is RcsB, a DNA-binding protein activated by phosphorylation. RcsB regulates transcription of some targets on its own, whereas other targets require RcsA, an accessory protein that has no known RcsB-independent activity.
Fig. 1.
Model of Rcs phosphorelay signaling. The histidine kinase domain (HK) of RcsC autophosphorylates, after which phosphate is transferred intramolecularly to aspartate in a receiver domain (D1). Next, phosphate is transferred intermolecularly from D1 to the histidine phosphotransfer domain (HPT) of RcsD. Last, phosphate is transferred from HPT to the RcsB receiver domain (D2). Biofilms are repressed by RcsB, and repression is increased by functional RcsA, as described in Results. Solid arrows show phosphate transfers that are well established experimentally (13, 14); dashed arrow indicates hypothesized phosphatase activity by RcsC, RcsD, or both (13, 16, 17). The histidine kinase-like ATPase (HAT) domain of RcsD does not participate in phosphotransfer. Filled arrowhead, site of frameshift in Y. pestis RcsD. Open arrowhead, site of inactivating insertion in Y. pestis RcsA.
In this report, we show that Y. pestis rcsA is a pseudogene. Substitution with the functional homologue from Y. pseudotuberculosis abolishes biofilms in fleas, evidence that the Y. pestis pseudogene arose by negative selection. We also show that rcsD, despite an apparent frameshift that gives it the appearance of a pseudogene, is functional.
Results
rcsB Negatively Regulates Biofilms.
Y. pestis biofilm production is robust at 26°C but almost absent at 37°C (18). When grown at 26°C on agar media containing Congo red (CR), colonies adsorb the dye and become red, whereas at 37°C they remain white (19); the red phenotype correlates well, although not perfectly, with biofilm formation (20). By screening transposon mutants for aberrant red colonies at 37°C, Kirillina et al. identified a negative regulator of biofilms, hmsP (18). To identify additional negative regulators, we screened for mutants that made darker-than-normal red colonies at 25°C. From 4,000 transposon insertions examined, we obtained seven mutations: one in hmsP, five that will not be described here, and one in rcsB. We constructed a deletion strain, ΔrcsB, and it had the identical dark-red CR phenotype as the transposon mutant. Plasmid complementation produced colonies that were pink rather than red, presumably because of slight overexpression. These findings indicate that rcsB is a negative regulator. Y. pestis RcsB is 91% identical to the well characterized E. coli protein and therefore is presumptively a transcriptional regulator.
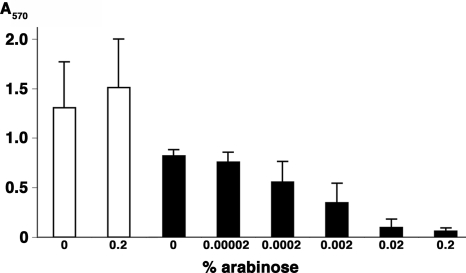
Colonies of ΔrcsB were smaller than those of the wild type and had a rugose (wrinkled) morphology (Fig. 2). Studies in Vibrio cholerae indicate that the rugose phenotype requires EPS production (21); the ΔrcsB phenotype therefore suggests that EPS is overproduced. A strain defective for EPS, ΔhmsS, made smooth colonies slightly larger than those of the wild type (Fig. 2). In vitro biofilm assays confirmed that rcsB is a negative regulator: Biofilms made by ΔrcsB on polystyrene surfaces were substantially denser than those of the wild type (Fig. 3). Conversely, when rcsB expression was put under the control of an arabinose-inducible promoter, biofilms diminished with increasing concentration of inducer (Fig. 4).
Fig. 2.
Colony size and morphology of mutants. ΔrcsB overproduces biofilms and makes small, rugose (wrinkled) colonies. ΔhmsS is defective for biofilm formation and makes colonies slightly larger than those of the wild type. Typical colonies of each strain were photographed at identical magnifications.
Fig. 3.
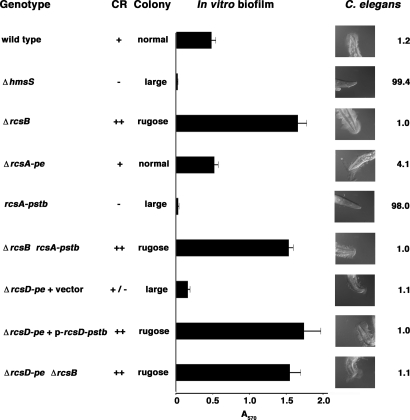
Phenotypes of rcs mutants and substitutions. “Δ” indicates deletion; “-pe” and “-pstb” denote Y. pestis and Y. pseudotuberculosis alleles. ΔrcsD-pe experiments used the empty plasmid vector pET-32a(+); p-rcsD-pstb denotes the Y. pseudotuberculosis allele cloned into this plasmid under its native promoter. Congo red (CR) colony color phenotypes: −, white; +/−, pink; +, red; ++, dark red. Colony size and morphology are described in the Fig. 2 legend. In vitro biofilms grown on polystyrene culture dishes were quantified by staining with crystal violet (Methods); data are mean ± SD of two to four trials. Biofilms on C. elegans were photographed after overnight incubation on Y. pestis lawns; rare animals on rcsA-pstb acquired traces of biofilm (data not shown). Numbers are percentage of worms that grew to L4 stage in 2 d; data are aggregate of two to four trials in which a minimum of 1,100 worms were scored for each bacterial genotype.
Fig. 4.
rcsB represses biofilms. In vitro biofilms were grown in the ΔrcsB strain transformed with vector control (white) or rcsB (black) under an arabinose-inducible promoter. Inducer was added at indicated concentrations. Data are mean ± SD of two (vector) or three (rcsB) trials. The difference between control and rcsB samples in the absence of arabinose is apparently due to basal rcsB expression.
Y. pestis rcsA Is a Pseudogene.
Y. pestis YPO2449 has 25% amino acid identity to E. coli RcsA. Comparison with ancestral Y. pseudotuberculosis shows that YPO2449 contains a 30-bp internal duplication, resulting in a 10-aa insertion in an otherwise identical protein, and on this basis, YPO2449 was predicted to be a pseudogene (4). We refer to YPO2449 hereafter as rcsA-pe and to the Y. pseudotuberculosis allele as rcsA-pstb.
Deletion of a pseudogene should produce no phenotype, and when we deleted rcsA-pe from Y. pestis, there were indeed no changes in CR adsorption, colony size, colony morphology, and in vitro biofilm growth (Fig. 3). Another assay for Y. pestis biofilms uses the nematode Caenorhabditis elegans (10, 22). Wild-type Y. pestis made biofilms on the worms' heads, which blocked feeding and inhibited growth, such that only 1.2% of eggs laid synchronously on bacterial lawns developed to the fourth larval stage (L4) in 2 days (Fig. 3). Slightly more nematodes (4.1%) developed normally on the ΔrcsA-pe strain. However, the growth inhibition by ΔrcsA-pe was still strong compared with that of the EPS-defective ΔhmsS, which made no detectable biofilms and permitted >99% of worms to reach L4. (Because wild-type Y. pestis inhibits growth almost completely, overproduction of biofilms by ΔrcsB or other mutants is not detectable with the C. elegans assay.)
In contrast to the deletion results, replacement of rcsA-pe with rcsA-pstb produced dramatic differences. Y. pestis with rcsA-pstb was negative for CR binding and made colonies resembling those of the EPS-defective mutant ΔhmsS, i.e., they were smooth and larger than wild-type colonies (Fig. 3). In vitro biofilms were absent, also like ΔhmsS. In the C. elegans assay, there was a slight inhibition of nematode growth: 98% of animals reached L4 with the rcsA-pstb substitution, whereas 99.4% did so on the EPS-defective ΔhmsS strain. Consistent with this weak growth inhibition, traces of extracellular matrix were observed on some worms exposed to rcsA-pstb, whereas none was ever present with ΔhmsS. The rcsB deletion was epistatic to rcsA-pstb for all phenotypes, indicating that in Y. pestis, as in E. coli, RcsA activity requires RcsB.
Functional RcsA Prevents Flea Blockage.
To investigate the importance of the rcsA-pe pseudogene in its native environment, we experimentally infected the Oriental rat flea, Xenopsylla cheopis. With wild-type Y. pestis, 21% of fleas were blocked by biofilms (Fig. 5A). This is consistent with a previous study using the same Y. pestis background, in which 24–38% of fleas were blocked by the wild type, whereas none were blocked by an EPS-defective hmsHFRS mutant (8). The rcsA-pstb strain was severely defective for flea blockage (Fig. 4A), i.e., it resembles an EPS-defective strain (8, 9). The ability of the rcsA-pstb strain to establish a chronic infection was not compromised (Fig. 4B), and it grew in fleas to levels only slightly less than the wild type (Fig. 4C), indicating that the defect is specific for blockage.
Fig. 5.
Flea blockage and colonization by rcsA-pstb. Data are mean ± SEM of three trials. (A) Cumulative blockage frequency 4 weeks after infection with Y. pestis. At least 80 fleas were analyzed for each sample. (B) Percentage of fleas infected initially and after 4 weeks. (C) Colony-forming units (CFU) in fleas sampled immediately after infection (n = 20 per strain per trial) and in fleas that remained colonized after 4 weeks (n = 4–19).
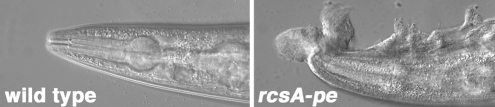
To further establish the importance of rcsA-pe in Y. pestis evolution, we examined rcsA in Y. pseudotuberculosis biofilms. Y. pseudotuberculosis strain IP32953 (4) failed to produce detectable biofilms when adult C. elegans were incubated overnight on lawns of the bacteria. When rcsA-pstb was replaced with the nonfunctional rcsA-pe allele, robust biofilms were formed (Fig. 6). This indicates that RcsA negatively regulates biofilms in the ancestral organism and is additional evidence that conversion to a pseudogene was necessary in Y. pestis evolution.
Fig. 6.
RcsA negatively regulates Y. pseudotuberculosis biofilms. No biofilm formed on adult C. elegans placed on lawns of wild-type strain IP32953 and incubated overnight. Replacement of the wild-type allele rcsA-pstb with nonfunctional rcsA-pe resulted in the production of substantial biofilms.
rcsD Is Not a Pseudogene.
The rcsD-pstb ORF is 2,691 bp long, encoding a predicted 897-aa protein. The sequence includes a run of eight consecutive thymines (bases 1921–1928). The Y. pestis sequence is identical except that one thymine is deleted, causing a frameshift after codon 642. The frameshift is predicted to eliminate the HPT domain that directly participates in phosphorelay signaling (Fig. 1); it also disrupts the HAT domain, whose function is not known. Thus, the Y. pestis rcsD-pe allele has the appearance of a pseudogene.
We deleted most of rcsD-pe 5′ to the frameshift and found that biofilms were decreased, although less so than with ΔhmsS (Fig. 3). The existence of a mutant phenotype indicates that rcsD-pe is not a pseudogene, and the phenotype is consistent with a positive regulatory function. Substitution of rcsD-pstb for rcsD-pe slightly increased biofilms (data not shown), and plasmid-based expression of rcsD-pstb in the ΔrcsD-pe background greatly enhanced biofilms, also suggestive of positive regulation by RcsD (Fig. 3). A hypothesis that RcsD can have phosphatase activity on RcsB (13, 17) is consistent with this regulatory polarity. A ΔrcsD-pe ΔrcsB double mutant was fully derepressed, making biofilms equal to those of the ΔrcsB strain, indicating that RcsD regulation acts through RcsB.
Discussion
Because some bacteria coexist with their evolutionary ancestors, direct experimental comparisons are possible that can elucidate steps in molecular evolution. An early study found evidence for negative selection during S. flexneri evolution from E. coli. The cadA biosynthetic gene, part of a large E. coli locus that is missing in S. flexneri, was sufficient to reduce virulence when heterologously expressed in S. flexneri (2). Independent cadA losses in other Shigella sp. provided additional evidence for negative selection (23), and similar findings were made for other Shigella genes (24).
We used a similar approach to analyze a regulatory pathway in Y. pestis and its living ancestor, Y. pseudotuberculosis. We replaced the predicted Y. pestis rcsA pseudogene with the functional Y. pseudotuberculosis allele and also performed the reciprocal experiment, placing the pseudogene in Y. pseudotuberculosis. These experiments provided strong evidence that loss of functional rcsA was necessary for Y. pestis to occupy its unique niche in the flea vector.
Despite their recent divergence, the two Yersinia spp. differ markedly in disease severity, mode of transmission, and epidemiology. Y. pseudotuberculosis is not vector-borne, and it is not among the most virulent of pathogens. It is transmitted by the oral–fecal route and most often causes self-limiting gastroenteritis and lymphadenitis (25). Y. pestis is among the most lethal bacterial pathogens. It is spread by fleas, and this mode of transmission helps explain its extraordinary virulence: Only if a mammal is septicemic will a naïve flea that feeds on it become a vector (26).
The differences that allow Y. pestis but not Y. pseudotuberculosis to make biofilms in fleas are not completely known. Ymt, a Y. pestis factor encoded on a plasmid absent from Y. pseudotuberculosis, promotes survival in fleas, but it is not involved in biofilm formation per se (27). Ymt expression in Y. pseudotuberculosis was not sufficient to produce biofilms in the insect (11). Y. pseudotuberculosis produces robust hmsHFRS-dependent biofilms in the C. elegans model (10) and in vitro (11), indicating that the difference is not one of EPS production. Y. pseudotuberculosis is acutely toxic to fleas, whereas Y. pestis is not; although the genetic basis and mechanism of the toxicity are unknown, presumably it was lost in Y. pestis evolution (28).
The results presented here indicate that Y. pestis biofilm formation in fleas is due in part to an evolutionary change in the Rcs regulatory pathway. Heterologous expression of functional RcsA strongly represses Y. pestis biofilms in fleas and prevents blocking the insect digestive tract (Fig. 5). RcsA is functional in Y. pseudotuberculosis and represses biofilms in the ancestral organism (Fig. 6). Conversion of rcsA to a pseudogene thus appears to have been required for Y. pestis to occupy the flea niche in a manner that promotes high transmissibility and enzootic persistence. We conclude that the pseudogene is a product of negative selection. Although several recent reports have shown that fleas can transmit plague without becoming blocked (5, 6, 29), the importance of blocked fleas in plague transmission is long established (7) and not refuted by those studies.
Y. pestis RcsB is functional and negatively regulates biofilms. Its deletion from the wild type results in small rugose colonies (Fig. 2) and aberrantly large biofilms in vitro (Fig. 3), whereas its overexpression produces large, smooth colonies (data not shown) and reduced biofilms in vitro (Fig. 4). RcsB is required for biofilm repression by functional RcsA (Fig. 3). The phenotypes of the rcsA and rcsB variants analyzed here are consistent with a model in which RcsB represses biofilms to some degree, but permits biofilm formation sufficiently to block fleas. When functional RcsA is present, RcsB-dependent repression is increased, such that essentially no biofilms are formed. This regulation appears to be indirect: We fused a lacZ reporter to chromosomal hmsH in the rcsA-pe and rcsA-pstb backgrounds and found no significant difference in expression of the EPS biosynthetic locus (data not shown).
rcsA-pe was predicted on bioinformatic grounds alone to be a pseudogene (4). Consistent with this hypothesis, there was no difference in colony phenotypes or in vitro biofilm formation in the ΔrcsA-pe strain. A slight defect was observed for C. elegans biofilms (Fig. 3), which could be interpreted to mean that wild-type rcsA-pe retains weak function. Alternatively, the result may indicate that rcsA-pe has a dominant-negative effect, i.e., the aberrant protein could be synthesized in the wild type and interfere with RcsB, lessening RcsB-mediated biofilm repression. This second interpretation is supported by the observation that the CR colony-color phenotype of the rcsA-pstb strain was partially rescued when rcsA-pe was overexpressed from a plasmid (data not shown).
Fully functional rcsA-pstb strongly repressed Y. pestis biofilms in vitro (Fig. 3), on C. elegans (Fig. 3) and in the natural niche, the flea (Fig. 5). rcsA-pstb bacteria colonized as many fleas as the wild type and grew in the insects to almost the same titers but failed to block the proventriculus. These phenotypes are virtually identical to those of an EPS-defective hmsHFRS mutant (8). We conclude that the failure to block fleas is specifically due to biofilm repression in vivo.
By inspection alone, rcsD-pe also appears to be a pseudogene: A single base pair is deleted, causing an apparent frame shift. However, rcsD-pe has some function, because biofilms are substantially reduced in the ΔrcsD-pe strain (Fig. 3). One possible explanation for RcsD function is suggested by the frameshift context, which occurs in a run of thymines; transcriptional slippage that changes reading frame is known to occur in polythymine tracts (30). Programmed translational frameshifting has also been observed in bacteria (31). Thus, it is possible that full-length RcsD is synthesized, perhaps at low levels, despite the putative frameshift. A second possibility is that the N-terminal portion of RcsD confers some function, perhaps as part of an interaction with RcsC. A hypothesis that RcsD has phosphatase activity on RcsB (13, 15, 17) is consistent with the regulatory polarity we observe; however, inhibition of RcsC signaling would also account for the Y. pestis effect. Regardless of the explanation, our findings indicate that caution is warranted in assigning pseudogene status on the basis of sequence data alone: rcsA-pe is a single ORF but is a pseudogene, whereas rcsD-pe is frameshifted yet functional.
The experimental approach described here is not limited to analysis of biofilms. The great differences between Y. pestis and Y. pseudotuberculosis in mammalian infection route and disease severity could also be due in part to conversion of genes to pseudogenes. Systematic substitution of Y. pseudotuberculosis functional genes for predicted Y. pestis pseudogenes, with tests in both flea and rodent models, could yield significant insights into the molecular evolution of an extraordinarily virulent pathogen.
Methods
Y. pestis Strains.
All strains used in this study are in the KIM6+ background, which is derived from the sequenced strain KIM (32) but cured of the pCD1/pYV plasmid required for mammalian virulence. It is competent for flea blockage (8) and C. elegans biofilms (10) and for convenience is referred to as wild type.
The ΔhmsS mutant and the substitution of rcsA-pstb for rcsA-pe were made by two-step allelic replacement (22, 33). hmsS was replaced with a chloramphenicol resistance gene. An unmarked substitution of rcsA-pstb for rcsA-pe was made by using a 2.2-kb PCR product amplified from Y. pseudotuberculosis YPIII that contained rcsA-pstb and flanking DNA; the sequence was verified to be identical to the rcsA gene YPTB2486 of the sequenced Y. pseudotuberculosis strain IP32953. The rcsA-pe substitution in Y. pseudotuberculosis was made in the same manner, by using a PCR product amplified from Y. pestis. Complete deletions of rcsA-pe (gene y1741 in KIM; YPO2449 in Y. pestis strain CO92) and rcsB (y2970; YPO1218) and a deletion of nucleotides 1–1846 of rcsD (y2968; YPO1219) were made by using a one-step method to recombine PCR products into the chromosome (34). Strains made with this procedure were cured of the temperature-sensitive recombinase-encoding plasmid by overnight incubation at 37°C. Double mutants were made by sequential application of the procedures described. Oligonucleotide primers used are shown in supporting information (SI) Table S1. All strains were verified by PCR, Southern blot hybridization, DNA sequencing, and plasmid complementation, as appropriate. For inducible rcsB expression, the gene was cloned downstream of the arabinose-inducible promoter of plasmid pBADMycHis (Invitrogen). For plasmid expression of rcsD-pstb, the native promoter and ORF were cloned into vector pET-32a(+) (Novagen).
Screen for CR Phenotypes and Colony Morphology Assays.
Transposon mutagenesis with Tn5-RL27 and identification of insertion sites was essentially as described (35). Mutants were plated on LBCR agar (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.01% Congo red, 1.5% agar) with 30 μg/ml of kanamycin and screened for color phenotypes after growth for 2 d at 25°C. To analyze colony size and morphology, strains were grown on the same medium, but with Congo red omitted, for 4 d at 25°C.
In Vitro Biofilm Assays.
Bacteria were grown in Luria–Bertani broth for 24 h and diluted to OD600 0.3 in 40% brain-heart infusion broth. Four 750-μl aliquots per strain were added to wells of 24-well polystyrene dishes, which were incubated with shaking at 200 rpm for 16–18 h at 26°C. Media and planktonic cells were removed, the wells were washed with 2.5 ml of water, and the adherent biofilm was stained with 2.5 ml of 0.01% crystal violet for 12 min. The wells were washed three times with water, bound dye was solubilized with 1.5 ml of 80% ethanol-20% acetone, and the A570 was measured. Background absorbance for uninoculated control wells was subtracted.
C. elegans Biofilm Assays.
A previous method (22) was modified slightly to increase growth inhibition by Y. pestis biofilms. Gravid adult C. elegans were washed to remove E. coli on which they are fed, placed on lawns of Y. pestis for several hours to lay eggs, then removed. After incubation for 2 d at 20°C, growth of the broods to the fourth larval stage (L4) was scored. Micrographs of representative worms after overnight incubation on lawns were obtained by using differential interference contrast optics.
Nematode growth could not be used to assay Y. pseudotuberculosis IP32953 biofilms, because this strain has an uncharacterized activity that is lethal to unhatched C. elegans eggs. Therefore, Y. pseudotuberculosis biofilms were analyzed solely by placing adult C. elegans on bacterial lawns and incubating overnight.
Flea Blockage.
Experiments were performed as described (8). X. cheopis fleas were fed a single blood meal containing Y. pestis and scored for blockage at twice-weekly intervals for 4 weeks. Additional samples of 20 fleas were collected immediately after the infectious blood meal, and at 4 weeks after infection, to determine the infection rate and bacterial load. Colony-forming unit (CFU) counts were made of individually triturated fleas.
Supplementary Material
Acknowledgments.
We thank Elizabeth Carniel (Institut Pasteur, Paris, France) and Emilio Garcia (Lawrence Livermore National Laboratory, Livermore, CA) for Y. pseudotuberculosis strains. This work was supported by National Institutes of Health Grant AI057512 and the Sandler Family Supporting Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803525105/DCSupplemental.
References
- 1.Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 2.Maurelli AT, Fernandez RE, Bloch CA, Rode CK, Fasano A. “Black holes” and bacterial pathogenicity: A large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achtman M, et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chain PS, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisen RJ, et al. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci USA. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen RJ, Lowell JL, Montenieri JA, Bearden SW, Gage KL. Temporal dynamics of early-phase transmission of Yersinia pestis by unblocked fleas: Secondary infectious feeds prolong efficient transmission by Oropsylla montana (Siphonaptera: Ceratophyllidae) J Med Entomol. 2007;44:672–677. doi: 10.1603/0022-2585(2007)44[672:tdoeto]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Pollitzer R. Plague. Geneva: World Health Organization; 1954. [Google Scholar]
- 8.Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 9.Jarrett CO, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 10.Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: Plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- 11.Erickson DL, Jarrett CO, Wren BW, Hinnebusch BJ. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J Bacteriol. 2006;188:1113–1119. doi: 10.1128/JB.188.3.1113-1119.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Appleby JL, Parkinson JS, Bourret RB. Signal transduction via the multi-step phosphorelay: Not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 13.Majdalani N, Gottesman S. The Rcs phosphorelay: A complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 14.Takeda S, Fujisawa Y, Matsubara M, Aiba H, Mizuno T. A novel feature of the multistep phosphorelay in Escherichia coli: A revised model of the RcsC → YojN → RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol Microbiol. 2001;40:440–450. doi: 10.1046/j.1365-2958.2001.02393.x. [DOI] [PubMed] [Google Scholar]
- 15.Huang YH, Ferrieres L, Clarke DJ. The role of the Rcs phosphorelay in Enterobacteriaceae. Res Microbiol. 2006;157:206–212. doi: 10.1016/j.resmic.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Fredericks CE, Shibata S, Aizawa S, Reimann SA, Wolfe AJ. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol Microbiol. 2006;61:734–747. doi: 10.1111/j.1365-2958.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 17.Majdalani N, Heck M, Stout V, Gottesman S. Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J Bacteriol. 2005;187:6770–6778. doi: 10.1128/JB.187.19.6770-6778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- 19.Lillard JW, Jr, Fetherston JD, Pedersen L, Pendrak ML, Perry RD. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- 20.Forman S, et al. Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology. 2006;152:3399–3410. doi: 10.1099/mic.0.29224-0. [DOI] [PubMed] [Google Scholar]
- 21.Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darby C, Ananth SL, Tan L, Hinnebusch BJ. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect Immun. 2005;73:7236–7242. doi: 10.1128/IAI.73.11.7236-7242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Day WA, Jr, Fernandez RE, Maurelli AT. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect Immun. 2001;69:7471–7480. doi: 10.1128/IAI.69.12.7471-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prunier AL, Schuch R, Fernandez RE, Maurelli AT. Genetic structure of the nadA and nadB antivirulence loci in Shigella spp. J Bacteriol. 2007;189:6482–6486. doi: 10.1128/JB.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koornhof HJ, Smego RA, Jr, Nicol M. Yersiniosis. II: The pathogenesis of Yersinia infections. Eur J Clin Microbiol Infect Dis. 1999;18:87–112. doi: 10.1007/s100960050237. [DOI] [PubMed] [Google Scholar]
- 26.Prentice MB, Rahalison L. Plague Lancet. 2007;369:1196–1207. doi: 10.1016/S0140-6736(07)60566-2. [DOI] [PubMed] [Google Scholar]
- 27.Hinnebusch BJ, et al. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- 28.Erickson DL, et al. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: Implications for the evolution of vector-borne transmission of plague. Cell Microbiol. 2007;9:2658–2666. doi: 10.1111/j.1462-5822.2007.00986.x. [DOI] [PubMed] [Google Scholar]
- 29.Eisen RJ, Wilder AP, Bearden SW, Montenieri JA, Gage KL. Early-phase transmission of Yersinia pestis by unblocked Xenopsylla cheopis (Siphonaptera: Pulicidae) is as efficient as transmission by blocked fleas. J Med Entomol. 2007;44:678–682. doi: 10.1603/0022-2585(2007)44[678:etoypb]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 30.Wagner LA, Weiss RB, Driscoll R, Dunn DS, Gesteland RF. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 1990;18:3529–3535. doi: 10.1093/nar/18.12.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engelberg-Kulka H, Schoulaker-Schwarz R. Regulatory implications of translational frameshifting in cellular gene expression. Mol Microbiol. 1994;11:3–8. doi: 10.1111/j.1365-2958.1994.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 32.Deng W, et al. Genome sequence of Yersinia pestis KIM. J Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen RA, Wilson MM, Guss AM, Metcalf WW. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol. 2002;178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.