Abstract
Epigenetic changes in multiple genes are emerging as an important mechanism for tumour cells to acquire resistance to chemotherapy. In the present work, we test the hypothesis that epigenetic organisation in cancer cells can be affected by cytostatic drugs. Colorectal cancer cells were cultured for several weeks in the presence of 6-thioguanine. Bisulphite sequencing of the CpG-rich promoter regions of two expressed genes showed a significantly increased frequency of methylated CpG sites in drug-treated cells, as compared with controls: 4.7% and 1.7%, respectively, for the HPRT gene; and 11.1% and 8.2% for CDX1. Essentially, all of the increase for the CDX1 gene was in a four CpG sub-region previously found to correlate with gene activity (P = 0.006). This pattern of sparse promoter methylation fits with a recently proposed ‘seeding’ two-step mechanism leading up to gene inactivation in cancer cells. Taken together, our findings suggest activation in cancer cells of an epigenetic process enabling a tumour to generate drug-resistant variant cells.
Keywords: DNA methylation, Drug resistance, 6-Thioguanine, Colorectal cancer, HPRT
1. Introduction
Resistance to cancer chemotherapy can be mediated by mutations in specific genes such as MDR1 (P-glykoprotein).1 This has been well documented in cells in vitro and in transgenic mice. Nevertheless in a clinical setting neither the status of MDR1 or any other so far identified individual gene has served as a useful predictor of therapy response (reviewed in Ref. 2). The contribution of epigenetic changes to treatment failure has, however, been reported recently, including CpG methylation of the hMLH1 promoter in ovarian cancer occurring during development of drug resistance. This has even been reported to become detectable as free tumour DNA circulating in patient sera.3 At very high drug levels (typically giving >95% cell kill and DNA synthesis inhibition) the changes in overall DNA methylation have been documented, with some drug types causing hypermethylation, for example carboplatin. When a lung cancer cell line was exposed to 6-thioguanine (6TG) at 100 μM there was a decrease in the total DNA content of methylated bases, but there was no effect at 10 μM.4 There are also data suggesting that an interplay between epigenetic changes in multiple genes is needed for a tumour to acquire resistance.5,6 However, an unresolved issue is whether resistant cells are present as rare epigenetic variants already at the start of treatment, eventually to become enriched by selection, or whether in sensitive cells epigenetic regulatory mechanisms can be affected by a cytostatic drug stress. It has been known for some time that environmental stress factors are associated with epigenetic changes. In plants, chemical mutagens and cold can lead to epigenetic modification.7,8 Such observations have also been made in mammals. Tumours induced in rodents by exposure to chemical carcinogens have been found to contain not only oncogene mutations, but also epigenetically silenced tumour suppressor genes including p169; moreover, cells or mice treated with nickel or DNA alkylating agents develop cancer-related epigenetic changes.10–12
In the present work, we attempt to answer the question as to whether tumour cells respond to clinically attained chemotherapy levels with epigenetic modifications. If that is the case then this may help in designing strategies to combat an important source of therapy failure. Colorectal cancer (CRC) cell lines were exposed to relatively low levels of 6-thioguanine (6TG), a drug in clinical use, allowing some growth to continue for several weeks. Bisulphite sequencing of the promoter regions of two genes was then carried out. The first gene, HPRT, is involved in cellular resistance to this drug, as it is essential for 6TG to become incorporated into DNA in order to exert its cytostatic effect, and the second gene, CDX1, has no known functional relationship to the effects of 6TG.
2. Materials and methods
2.1. Cells and 6TG treatment
Mononuclear cells were separated from freshly collected venous blood from healthy donors by Lymphoprep (Axis-Shield, Oslo, Norway) gradients and used directly. The T84 and SW620 CRC cell lines both originate from male patients. They have both been shown to be hMLH1-positive, suggesting normal DNA mismatch repair function. The CRC lines were cultured under standard conditions with 10% (v/v) bovine serum in the medium, and split as appropriate before confluence was reached. 6TG was from Sigma (A4882), and a stock solution of 10 mM in 0.2 M NaOH was prepared and kept in aliquots at −20 °C. The cultures were washed and fresh medium with 6TG added at least once per week, in order to maintain the desired 6TG concentration. The CFDA (carboxyfluorescein diacetate) fluorescent dye exclusion test was used to determine cell viability, as described previously.13
2.2. Bisulphite sequencing
DNA was extracted from the cells using the DNeasy kit (Qiagen, Crawley, UK). DNA (0.5 μg) was bisulphite-modified using the EZ DNA Methylation-Gold Kit, as recommended by the manufacturer (Zymo Research, Orange, CA). A promoter region was amplified by PCR as described below, and the PCR product was purified from a solubilised agarose gel slice using a silica column (Qiagen), cloned (TopoTA cloning kit, Invitrogen, Purchase, NY), and finally submitted to the local sequencing core facility for ABI sequencing (Applied Biosystems).
2.3. PCR
PCR primers were designed based on the HPRT and CDX1 5′-flanking regions given on the University of California at Santa Cruz website http://genome.ucsc.edu. For the sequencing PCR reaction the primers bind to non-CpG sequences in order to yield products irrespective of the template’s CpG methylation status (positions indicated in Fig. 1). The forward primer was 5′-ATTGAGTTGGGAGGGAAAGG, and the reverse primer was 5′-CCATTTCCACCTTCTCTTCCCA. Cycling conditions were 95 °C for 12 min followed by 40 cycles of 95 °C 1 min, 58 °C 1 min 30 s, 72 °C 2 min 30 s, and then finally 72 °C for 10 min. The 25 μl reaction solution contained 0.2 μM each primer, 0.1 U Hotstar Taq polymerase (Qiagen), 1.5 mM MgCl2, 200 μM each dNTP and 2 μl of the bisulphite-modified DNA. The PCR products were visualised after gel electrophoresis through 1.5% (w/v) agarose containing ethidium bromide. The primer positions for the HPRT methylation-specific PCR are also shown in Fig. 1. These primers were designed to bind to DNA methylated at several CpG sites: the forward primer was 5′-AATTGGTAGGCGTCGGCGTAGGCGCGC, and the reverse primer was 5′-CCGACAAACCGAACTACTCACCACGAC. The cycling conditions were similar to those for the HPRT sequencing PCR, with the following differences: annealing at 63 °C, elongation for 1 min 30 s. For CDX1 sequencing PCR was performed as describer earlier.14 For all PCR reactions, only the bisulphite-modified sense strand was amplified, starting with the reverse primer binding to this sense strand, and the complementary molecule thus produced serving as a template for the forward primer.
Fig. 1.
Sequences of the 5′-flanking regions for human and mouse HPRT. The bisulphite-modified sequence is human DNA, assuming all CpG sites are unmethylated. CpG’s are underlined. Mouse portions homologous to the human sequence are in bold. Positions relative to the human exon 1 start are shown above the bisulphite line. The arrows indicate the PCR primer positions.
2.4. Statistical analysis
The significance of all the 2 × 2 comparisons was assessed using Fisher’s exact test.
3. Results
3.1. Definition of 6TG concentrations used for long-term treatment of CRC cell lines
The T84 and SW620 CRC cell lines were kept in medium containing up to 16 μM 6TG, and split as needed to keep the cultures proliferating at 20–80% confluence. In patients, plasma concentrations of up to 10 μM 6TG have been documented for the Lanvis pharmaceutical preparation (RxMed website http://www.rxmed.com). Expression of the HPRT gene in both lines was confirmed by RT-PCR, and there was no apparent change in the level of expression following 6TG treatment (results not shown). Total cell number and the fraction of dead cells, as determined by the fluorescent dye exclusion test, were analysed after 1 week of culture (Table 1). No growth was seen at the highest concentrations of 6TG (8–16 μM), whereas sustained strong growth was seen at 1 μM 6TG, with both T84 and SW620 reaching 25–60% of the total cell number achieved by the untreated controls. T84 had a slightly higher growth rate than SW620, and there was no apparent effect of 6TG on cell death for either of the two cell lines. Paradoxically, the more marked growth retardation of SW620 in 6TG was coupled with less death. A similar sensitivity to 6TG was also seen with other CRC lines (COLO201, COLO678 and SW1116; results not shown). Based on these findings, 1 μM 6TG was considered to provide an optimal level of cytostatic drug stress on the CRC lines during long-term culture. T84 was still proliferating after one month in 1 μM 6TG (see T84 data at 17 days in Table 1). A minor reduction of the growth rate was also seen at 0.25 μM 6TG for SW620 (results not shown). For all long-term experiments, a 16 μM 6TG culture was set up, in addition to cultures at 0 and 1 μM. Only a few cells were occasionally detectable after one month of culture. This suggests that cells harvested from cultures maintained for several weeks in low concentrations of 6TG do not significantly represent outgrowth of rare variants with a complete lack of HPRT expression likely to be present at the start of the culture. Such variants are expected to occur at a frequency of approximately 10−6 in CRC lines,15 while for the cloning of HPRT-mutants much higher 6TG concentrations, namely around 10–15 μM, are usually used.16
Table 1.
Sensitivity of CRC cell lines to 6TG
| CRC | 6TG (μM) | Total cell number, relative to absence of 6TG (% dead cells) |
|
|---|---|---|---|
| 7 days 6TG | 17 days 6TG | ||
| T84 | 0 | 100 (7) | 100 (3) |
| 1 | 60 (10) | 67 (4) | |
| 2 | 22 (11) | 13 (3) | |
| 4 | 4 (11) | 8 (7) | |
| 8 | 2 (9) | 1 (5) | |
| 16 | 1 (14) | <1 (6) | |
| SW620 | 0 | 100 (1) | n.d. |
| 1 | 25 (2) | n.d. | |
| 2 | 14 (1) | n.d. | |
| 4 | 2 (2) | n.d. | |
| 8 | 1 (1) | n.d. | |
| 16 | <1 (2) | n.d. | |
3.2. Analysis of CpG methylation in the HPRT promoter region
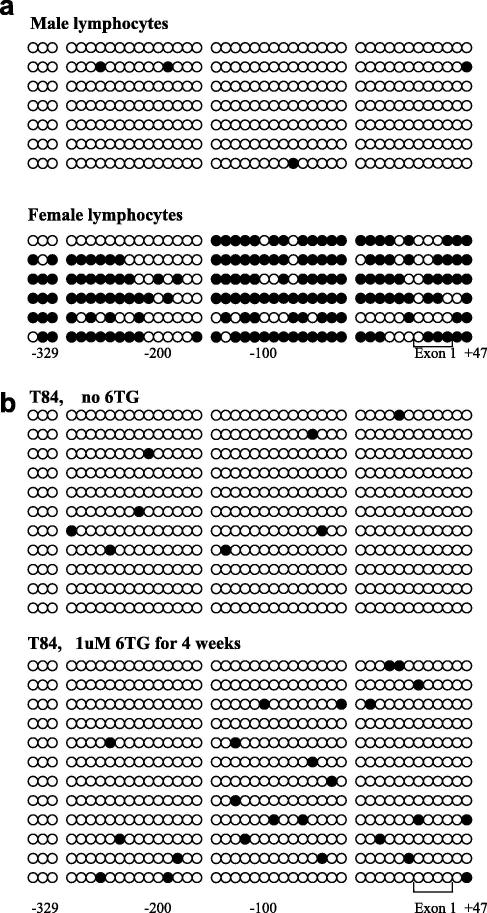
The HPRT gene has been extensively studied.17–19 There is a very good evidence indicating that its expression is regulated by a CpG-rich approximately 500 bp upstream region. Due to the X chromosome location of HPRT, and its house-keeping function in the nucleotide salvage pathway, human cells in general carry only a single active allele. However, which CpGs are most important for the control of HPRT gene expression has not been clearly established. In the present work, we have chosen to study the 357 bp region 5′ to exon 1 because of its relatively high homology with the corresponding mouse sequence (61%, as compared to 34% in the further upstream 250 bp) (Fig. 1). This 357 bp segment contains a total of 43 CpG sites, and several transcription factor-binding motifs including, for example, four GGGCGG Sp1 sites. After DNA extraction and bisulphite-modification, the promoter region was PCR-amplified, cloned and sequenced. In accordance with published data,19 the promoter in male lymphocytes was virtually free from methylated CpGs, whereas clones with dense methylation were found in female lymphocytes (Fig. 2a). The over-representation of the inactive female allele (all six clones were hyper-methylated) indicates that bisulphite-modified molecules containing relatively few long thymine-rich stretches (due to the conversion by bisulphite of cytosine to thymine, while methylated cytosines remain as cytosine) are preferentially amplified by the current PCR protocol. T84 cells, originating from a tumour of a male patient, showed a very low level of methylation (Fig. 2b, upper panel), similar to that in male lymphocytes. T84 cells exposed for 28 days to 6TG showed no signs of the heavy methylation found in female lymphocytes (Fig. 2b, lower panel). However, the total number of methylated CpGs among the total of 12 clones analysed from the 6TG-treated cultures (4.7%) was significantly higher than that in the 11 clones from the untreated cells (1.7%). These results are given in Table 2, together with an estimate of the possible artefactual contribution to the scoring of methylated CpGs arising from bisulphite-modification errors. In the analysed 376 bp HPRT promoter region there is a total of 88 cytosines at non-CpG positions, all of which are assumed to be unmethylated, and therefore should be converted to thymines by bisulphite. The number of cytosines remaining after bisulphite-modification at these 88 positions can be used as a measure of the bisulphite failure rate assuming that this is the same for C at CpG and non-CpG sites. Thus, for example, the 1.5% of errors for 6TG untreated T84 suggests that the observed figure of 1.7% of C’s at CpG sites mainly reflects bisulphite modification failures, whereas for the 6TG-treated T84 cells subtraction of the 2.1% errors from the 4.7% of observed cytosines at CpG sites indicates a true presence of 2.6% methylated CpG sites, the difference presumably being due to the effects of culturing in the presence of low concentrations of 6TG. There was no significant change in the numbers of methylated C sites at the non-CpG positions. Line SW620 showed a slightly lower increase in the number of methylated C sites at CpG positions, but this was not significant because fewer clones were analysed.
Fig. 2.
HPRT promoter bisulphite DNA sequencing results for individual CpG sites: (a) seven clones from male lymphocytes and six from female lymphocytes and (b) 11 clones from untreated T84 cells and 12 from 6TG-treated T84. Each of the 43 CpG sites is shown as a circle, and methylated CpGs are highlighted in black. The base positions relative to the exon 1 start site are shown. Each line represents one clone.
Table 2.
HPRT promoter: bisulphite DNA sequencing results
| Cells | Number of sequenced clones | 6TG (μM/days) | C at CpG sites (=methylated CpG) | C at non-CpG site (=bisulphite failure) |
|---|---|---|---|---|
| Male lymphocytes | 7 | 0 | 0 4/301 (1.3%) | 6/616 (1.0%) |
| Female lymphocytes | 6 | 0 | 168/258 (65.1%) | 21/528 (4.0%) |
| T84 | 11 | 0 | 8/473 ( 1.7%) | 15/968 (1.5%) |
| T84 | 12 | 1/28 d | 24/516 (4.7%)a | 22/1056 (2.1%)b |
| SW620 | 2 | 0 | 0/86 (<1.2%) | 0/176 (<0.6%) |
| SW620 | 2 | 0.25/7 d | 0/86 (<1.2%) | 0/176 (<0.6%) |
| SW620 | 5 | 1/7 d | 4/215 (1.9%)c | 1/440 (0.2%) |
All significance tested by Fisher’s exact test, two sided (except where indicated), results as P-values.
Control versus 6TG, P = 0.0108.
NS.
Control and 0.25 6TG versus 1 6TG, P = 0.1323 (one sided p = 0.0941).
3.3. Analysis of CpG methylation in the CDX1 5′-flanking region
CDX1 is involved in colonic epithelium differentiation and was chosen as a representative gene with no known specific role in 6TG toxicity. In a previous report on a panel of 37 CRC lines, we found evidence for a role for CDX1 as tumour suppressor in colorectal cancer,14 with 12 of the analysed lines showing either CDX1 silencing due to promoter methylation or a pattern of partial methylation. We now find lymphocytes to have heavy CDX1 promoter methylation, in accordance with our previous result for a lymphoblastoid cell line, suggesting, as expected, that CDX1 is not expressed in these cells (Fig. 3a). In our earlier study, T84 was positive for CDX1 mRNA and showed no signs of heavy promoter methylation as judged by methylation-specific PCR. In line with these published data, we now see a sparse CDX1 promoter methylation among the 15 analysed T84 clones grown in the absence of 6TG. There is, in particular, very little methylation at the four CpG sites shown to be strongly correlated with gene expression14 (Fig. 3b, upper panel). Interestingly, however, the region containing the 29 CpGs before the four CpG sites differs significantly in the proportion of CpG sites with methylated C versus that containing the five CpG sites after, and neither show any significant change with growth in 6TG (including control and 6TG data, 7.2% versus 18%, P = 0.001). T84 cells exposed to 6TG for 28 days showed a slightly higher overall proportion of methylated CpGs, similar to the results for HPRT, with an increase from a relatively high basal level of 8.2–11.1% in the 6TG-treated culture (Fig. 3b, lower panel, and Table 3). However, essentially all of the increase was in the four CpG sub-region previously found to correlate with gene expression (marked with a box in Fig. 3), where the change from 7% to 27% was highly significant with a P-value of 0.006, which remains significant even allowing for the selection made of the four CpG box sequence for which the comparisons were done. The bisulphite modification errors were at a clearly lower level, similar to that seen in the HPRT experiments (2.5% and 3.0%, respectively).
Fig. 3.
CDX1 promoter bisulphite DNA sequencing results for individual CpG sites: (a) six clones from lymphocytes and (b) 15 clones from untreated and 6TG-treated T84 cells, respectively. Each of the 38 CpG sites is shown as a circle, and methylated CpGs are highlighted in black. The base positions relative to the transcription start site are shown. The box denotes the four CpG sites correlating strongly with gene expression.14 Each line represents one clone.
Table 3.
CDX1 promoter: bisulphite DNA sequencing results
| Cells | Number of sequenced clones | 6TG (μM/days) | C at CpG sites (=methylated CpG) | C at non-CpG site (=bisulphite failure) |
|---|---|---|---|---|
| Female lymph | 6 | 0 | 162/228 (77.1%) | 14/702 (2.0%) |
| T84 | 15 | 0 | 47/570 (8.2%) | 44/1755 (2.5%) |
| T84 | 15 | 1/28 d | 63/570 (11.1%)a | 53/1755 (3.0%)b |
All significance by Fisher’s exact test, two sided except where indicated, P-values given.
P = 0.1321 (one sided 0.0661).
NS.
3.4. Methylation-specific PCR in search of rare heavy promoter methylation events
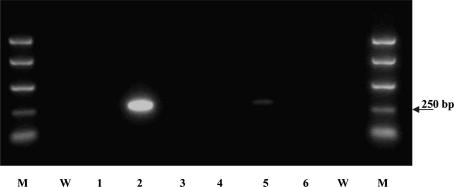
Although sequencing revealed no signs of cells with a densely methylated HPRT promoter suggestive of gene silencing, a low frequency of such variants could not be excluded. Recently two groups have, for example, presented evidence for a ‘seeding’ two-step mechanism operating during tumour suppressor gene silencing.20–23 The first step is suggested to involve a low-density methylation occurring in a high fraction of the tumour population, similar to that we have observed for both HPRT and CDX1, while the second step involving high-density promoter methylation leads to gene inactivation seen only in a low fraction of the ‘seeded’ cells, presumably as a result of the selection for reduced gene expression. Stimulated by these reports, we performed methylation-specific PCR, using primers binding only to methylated CpG stretches. This assay was sensitive enough to detect as little as one methylated copy among 100 unmethylated molecules (Fig. 4, lane 5). However, no PCR band was seen with the same 6TG-treated T84 cells that were used in Figs. 2 and 3, suggesting that the frequency of T84 cells becoming heavily methylated during 6TG treatment was less than 1% (Fig. 4).
Fig. 4.
No evidence for clones with high-density HPRT promoter methylation in 6TG-treated T84. Methylation-specific PCR was performed with primers binding only to the methylated CpG stretches (as shown in Fig. 1). Bisulphite-modified DNA was from female lymphocytes (lane 2), T84 without 6TG treatment (lane 3), T84 after 4 weeks with 1 μM 6TG (lane 4), male lymphocytes spiked with a 100× dilution of the female lymphocyte DNA solution used in lane 2 (lane 5) (a faint band could be seen in the illuminated gel), male lymphocytes as in lane 5 with no spiking (lane 6). Lane 1: female lymphocyte native DNA not modified with bisulphite. W, water control; M, size marker.
4. Discussion
We report here a significant, though low-level, increase in promoter CpG methylation in colorectal cancer cells treated with a cytostatic drug, 6TG, at a clinically relevant concentration. The data, which show variability in methylation within the promoter regions of two genes both in controls and following 6TG treatment, indicate the potential complexity of factors controlling DNA methylation at CpG sites. In addition to the fact that the observed sparse methylation was statistically significant for both of the studied genes, a case can be made in support of the relevance of our observations. The determination of cytosine methylation at CpG sites is based on the conversion of non-methylated cytosines to thymine, whereas a 5′-methylated cytosine is resistant, resulting in a cytosine rather than thymine on sequencing analysis of bisulphite-modified DNA. Since cytosines located at non-CpG positions are considered not to become methylated, it is possible to estimate the error rate of the bisulphite process by scoring the number of cytosines remaining as a cytosine at non-CpG sites. For the HPRT promoter in untreated T84 cells, the frequency of such methodological errors was no different from the frequency of methylation observed at CpG sites (1.5% and 1.7%, respectively). Assuming a uniform error rate at all cytosine positions, the true frequencies of CpG methylation in T84 cells are therefore 1.7–1.5 = 0.2% for no 6TG and 4.7–2.1 = 2.6% for 6TG-treated cells. These figures suggest that during 6TG treatment CpG methylation went from a virtually methylation-free state (0.2%) to a sparsely methylated level of 2.6%. Superficially, the corresponding picture for the CDX1 gene was not as clear-cut, with a true basal CpG methylation frequency of 8.2–2.5 = 5.7%, going up to 11.1–3.0 = 8.1% in 6TG-treated T84 cells. However, this overall analysis hides the fact that essentially all the change in the frequency of methylation occurred in the region of four CpGs (from 7% to 27% in that region), which has been shown to be critical for the control of expression of the CDX1 gene.14 Thus, among the total excess of 16 methylated CpG’s in 6TG-treated cells, 12 (75%) occurred in a region representing only 11% (4/38) of the total analysed CpG content.
If, as we believe our data show, there is a real, though low-level drug-associated increase in promoter methylation, does this represent a toxic and non-specific effect, or does it in any way relate to the inactivation of tumour suppressor genes in cancer? As already discussed, it has been suggested that ‘seeds of methylation’ may be an important precursor to dense methylation.23 Thus, CpG silencing of a glutathione-S-transferase (GSTP1) construct in prostate cancer cells has been shown to require treatment with HpaII methylase, mediating sparse methylation.20 Similar data come from gastric cancer cell lines where low-density methylation in most cells, as we have found in 6TG-treated cells, was associated with dense methylation in a small fraction of the cells for all the five analysed genes including E-cadherin.22 These reports support that our findings of sparse methylation may reflect a mechanism which provides the basis for an increased frequency of gene silencing in cancer cells during anti-cancer treatment, presumably following the selection for reduced gene expression. Our findings are also in agreement with previous reports on the cumulative influence of epigenetic changes in multiple genes for emergence of drug resistance, on the assumption that drugs other than 6TG may act in a similar way to increase methylation at CpG sites.5,6 It may be speculated that besides HPRT and CDX1, a number of other CpG island-containing loci in tumour cells exposed to cytostatic therapy stress may acquire a concordant low-level methylation.
Although our findings cannot exclude that variants with a subtle reduction in HPRT expression or with some other resistance mechanism were enriched for, it seems unlikely for at least three reasons that the CpG methylation seen in our HPRT promoter clones represents the outgrowth of a rare subpopulation of cells already present before 6TG treatment. Firstly, there is no reason to assume that such a sparse promoter methylation would in any way affect gene expression or yield a selectable phenotype. This agrees with the cited report on gastric cancer cell lines, showing no correlation between the fraction of methylation-seeded cells and the level of gene expression,22 and also with our results on the level of HPRT expression after growth in 6TG. Secondly, there was a continuous increase in cell number at the 6TG concentration (1 μM) used by us, which argues against significant enrichment of preformed variants. Thirdly, a 16 μM 6TG culture (a growth-inhibitory concentration used for the selection of HPRT-negative cell clones) was always run in parallel with the 1 μM culture. A steady decrease in cell number was then observed, with outgrowth of clones occurring in only a small minority of all culture flasks.
In conclusion, our data show that treatment with low levels of the cytotoxic drug 6TG leads to low-level increases of methylation at CpG sites, which suggests that this may also be seen with other drug treatments. In that case, such drug treatments may be analogous to treatments which increase the mutation rate, in that increasing the level of non-specific CpG methylation may increase the probability of functionally relevant methylation changes occurring, which are then selected for. Future work is needed to document promoter methylation in association with other treatment modalities such as ionising radiation and alkylating agents, as well as to test whether inhibition of CpG methylation may interfere with the development of drug resistance.
Conflict of interest statement
None declared.
Acknowledgements
Supported by a core programme grant from Cancer Research UK (to WB); ICRETT Fellowship, UICC, Geneva and the Malmö Hospital and Gunnar Nilsson Cancer Funds (to AB).
References
- 1.Gottesman M.M. How cancer cells evade chemotherapy. Cancer Res. 1993;53:747–754. [PubMed] [Google Scholar]
- 2.Glasspool R.M., Theodoridis J.M., Brown R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br J Cancer. 2006;94:1087–1092. doi: 10.1038/sj.bjc.6603024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gifford G., Paul J., Vasey P.A., Kaye S.B., Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10:4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 4.Nyce J.W. Drug-induced DNA hypermethylation: a potential mediator of acquired drug resistance during cancer chemotherapy. Mutation Res. 1997;386:153–161. doi: 10.1016/s1383-5742(96)00051-8. [DOI] [PubMed] [Google Scholar]
- 5.Theodoridis J.M., Strathdee G., Brown R. Epigenetic silencing by CpG island methylation: potential as a therapeutic target and as a biomarker. Drug Res Update. 2004;7:267–278. doi: 10.1016/j.drup.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Wei S.H., Brown R., Huang T.H. Aberrant DNA methylation in ovarian cancer: is there an epigenetic predisposition to drug response? Ann NY Acad Sci. 2003;983:243–250. doi: 10.1111/j.1749-6632.2003.tb05979.x. [DOI] [PubMed] [Google Scholar]
- 7.Stokes T.L., Kunkel B.N., Richards E.J. Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 2002;16:171–182. doi: 10.1101/gad.952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steward N., Ito M., Yamaguchi Y., Koizumi N., Sano H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J Biol Chem. 2002;277:37741–37746. doi: 10.1074/jbc.M204050200. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu K., Kato A., Shigemura M. Aberrant methylation patterns of the Rassf1a gene in rat lung adenocarcinomas induced by N-nitrosobis(2-hydroxypropyl)amine. Mol Carcinogen. 2005;45:112–117. doi: 10.1002/mc.20173. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland J.E., Costa M. Epigenetics and the environment. Ann NY Acad Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 11.Murphy S.P., Holtz R., Lewandowski N., Tomasi T.B., Fuji H. DNA alkylating agents alleviate silencing of class II transactivator gene expression in L1210 lymphoma cells. J Immunol. 2002;169:3085–3093. doi: 10.4049/jimmunol.169.6.3085. [DOI] [PubMed] [Google Scholar]
- 12.Barton T.S., Robaire B., Hales B.F. Epigenetic programming in the preimplantaion rat embryo is disrupted by chronic paternal cyclophosphamide exposure. Proc Natl Acad Sci USA. 2005;102:7865–7870. doi: 10.1073/pnas.0501200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodmer W.F., Tripp M., Bodmer J. Application of a fluorochromatic cytotoxicity assay to human leukocyte typing. In: Curtoni E.S., editor. Histocompatibility testing 1967. Munksgaard; Copenhagen: 1967. pp. 341–350. [Google Scholar]
- 14.Wong N.A.C.S., Britton M.P., Choi G.S. Loss of CDX1 expression in colorectal carcinoma: promoter methylation, mutation, and loss of heterozygosity analyses of 37 cell lines. Proc Natl Acad Sci USA. 2004;101:574–579. doi: 10.1073/pnas.0307190101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eshleman J.R., Markowitz S.D., Donover P.S. Increased mutation rate at the hprt locus accompanies microsatellite instability in colon cancer. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 16.Hou S.M., van Dam F.J., de Zwart F. Validation of the human T-lymphocyte cloning assay – ring test report from the EU concerted action on HPRT mutation (EUCAHM) Mutation Res. 1999;431:211–221. doi: 10.1016/s0027-5107(99)00164-5. [DOI] [PubMed] [Google Scholar]
- 17.Stout J.T., Caskey C.T. HPRT: gene structure, expression, and mutation. Ann Rev Genet. 1985;19:127–148. doi: 10.1146/annurev.ge.19.120185.001015. [DOI] [PubMed] [Google Scholar]
- 18.Holliday R., Ho T. DNA methylation and epigenetic inheritance. Methods. 2002;27:179–183. doi: 10.1016/s1046-2023(02)00072-5. [DOI] [PubMed] [Google Scholar]
- 19.Kang S.L., Kiefer C.M., Yang T.P. Role of the promoter in maintaining transcriptionally active chromatin structure and DNA methylation patterns in vivo. Mol Cell Biol. 2003;23:4150–4161. doi: 10.1128/MCB.23.12.4150-4161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J.Z., Stirzaker C., Harrison J., Melki J.R., Clark S.J. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–1061. doi: 10.1038/sj.onc.1205153. [DOI] [PubMed] [Google Scholar]
- 21.Stirzaker C., Song J.Z., Davidson B., Clark S.J. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004;64:3871–3877. doi: 10.1158/0008-5472.CAN-03-3690. [DOI] [PubMed] [Google Scholar]
- 22.Ushijima T., Watanabe N., Shimizu K. Decreased fidelity in replicating CpG methylation patterns in cancer cells. Cancer Res. 2005;65:11–17. [PubMed] [Google Scholar]
- 23.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]