Abstract
The mesencephalic dopaminergic (mesDA) system regulates behavior and movement control and has been implicated in psychiatric and affective disorders. We have identified a bicoid-related homeobox gene, Ptx3, a member of the Ptx-subfamily, that is uniquely expressed in these neurons. Its expression starting at E11.5 in the developing mouse midbrain correlates with the appearance of mesDA neurons. The number of Ptx3-expressing neurons is reduced in Parkinson patients, and these neurons are absent from 6-hydroxy-dopamine-lesioned rats, an animal model for this disease. Thus, Ptx3 is a unique transcription factor marking the mesDA neurons at the exclusion of other dopaminergic neurons, and it may be involved in developmental determination of this neuronal lineage.
The patterning of the developing mammalian brain is thought to involve cascades of signaling molecules and transcription factors, but the mechanisms for generation of distinct neuronal cell types during terminal differentiation are still largely speculative (1, 2). Yet, the specification of individual neuronal phenotypes underlies the assembly of neural circuits essential for brain function. The mesencephalic dopaminergic (mesDA) system consists of a limited set of neurons that are well defined anatomically and functionally (3–5). Their specific degeneration in Parkinson disease reveals their functional properties in control of behavior and movement as well as a unique vulnerability (6–10). In a search for homeobox genes associated with a unique neuronal lineage, we isolated a cDNA encoding a bicoid-related homeobox gene Ptx3, a member of the Ptx subfamily (11–14). This gene is strictly expressed in mesDA neurons.
METHODS AND MATERIALS
Cloning of Ptx3 Gene Transcripts.
Poly(A)+ RNA from hypothalamic fragments of the adult rat brain were subjected to reverse transcriptase–PCR with primers based on brain-expressed homeobox genes: upstream, 5′-GMRSCGMSAVMGSACMMBCTTYAC-3′; downstream, 5′-TGGTTYMRVAAYCGYHGMGCMARRTG-3′. The annealing temperature was 40°C. The PCR product was used to screen an adult rat hypothalamus library in λ gt11. Isolated phage DNA was cut with EcoRI, the insert of ≈1.2 kb was subcloned into pGEM7Zf(+), and both strands of the insert were sequenced.
Northern Analysis.
Total RNA extracted from tissues of the adult rat by Rnazol (Biotecx Laboratories, Houston) was fractionated on formaldehyde-agarose gels and transferred onto a nylon membrane (Hybond-N, Amersham) by downward capillary blotting. Blots were hybridized with a 32P-random-primed-labeled complete Ptx3 cDNA at 65°C overnight (15). Autoradiography was performed with a Fujix BAS1000 phosphor-imager (Fuji).
Cell Culture, Transfection, and Gel Retardation Assays.
Murine fibroblast L cells were grown in DMEM supplemented with 10% FCS. L cells were transfected by the calcium phosphate method (16). Precipitate containing 3 μg of reporter plasmid, 1 μg of effector plasmid, 1 μg of RSV-human growth hormone (hGH) internal control plasmid, and 5 μg of carrier DNA (pSP64, Promega) was applied on 1.5×105 cells in 35-mm Petri dishes. Medium was changed after 16 hr, and cells were harvested 24 hr later and assayed for luciferase activity as described previously (17). Transfections were performed in duplicate at least three times, and transfection efficiencies were corrected by measuring hGH in the medium using an RIA kit (Immunocorp, Montreal). Nuclear extracts were prepared from 300,000 L cells transfected with 20 μg of either a control vector or an expression vector for Ptx1, Ptx3, Otx1, or Otx2 as described (18). The sequence of the double-stranded oligonucleotides and binding reactions were as previously described (11).
In Situ Hybridizations.
Preparation of rat and human brain sections and in situ hybridizations were done as described (19), and sections were exposed on Betamax films (Amersham) for 3 to 7 days and then subjected to autoradiography under Hypercoat LM-1 liquid emulsion (Amersham). The cRNA probes were synthesized from an EcoRI/PstI fragment containing base pairs 1–285 and a PstI/BamHI fragment containing base pairs 799–971 of the Ptx3 cDNA. Double labeling was performed with digoxigenine-labeled BalI/EcoRI (base pairs 915-1137) fragment of the rat TH cDNA (20) and the 35S-labeled Ptx3 EcoRI/PstI fragment. CBAxC57BL6 mice were mated, and the morning when a vaginal plug was detected was considered embryonic day (E) 0.5. Pregnant mice were killed by cervical dislocation, and embryos were dissected, fixed overnight at 4°C in 4% paraformaldehyde, and embedded in paraffin. Sections (5 μm) were mounted on aminopropylethoxysilane-treated slides and used for in situ hybridization as described (11, 21). Slides were dipped in K.5 autoradiographic emulsion (Ilford), exposed for 25–30 days, developed with Kodak D-19, and counterstained with hematoxylin-eosin.
Unilateral 6-Hydroxydopamine Lesions.
Male Wistar rats (≈250 g) were injected intraperitonally (i.p.) with desipramine (DMI, 25 mg/kg). After 45 min the rats were anesthetized with Hypnorm (0.5 ml/kg body weight, intramuscular) and Dormicum (0.5 ml/kg body weight, i.p.) and placed in a stereotaxic apparatus with the incisor bar at intraaural bar level. Needles connected to a microsyringe were placed in the brain with the tips at 2.0 mm lateral of the midline, 3.8 mm posterior of bregma, and 8.6 mm below the surface of the skull aimed at the substantia nigra. During a period of 2 min, 8 μg 6-hydroxy-dopamine (6-OHDA) (4 μg/μl) was injected into the left substantia nigra region while the right was injected with vehicle (0.9% NaCl, 0.1% ascorbic acid). The animals were allowed to recover for 6 days. To test the effectiveness of the treatment, the rats were placed in a small, open field (22) and were monitored for the number of turnings over 180° during 5 min.
RESULTS AND DISCUSSION
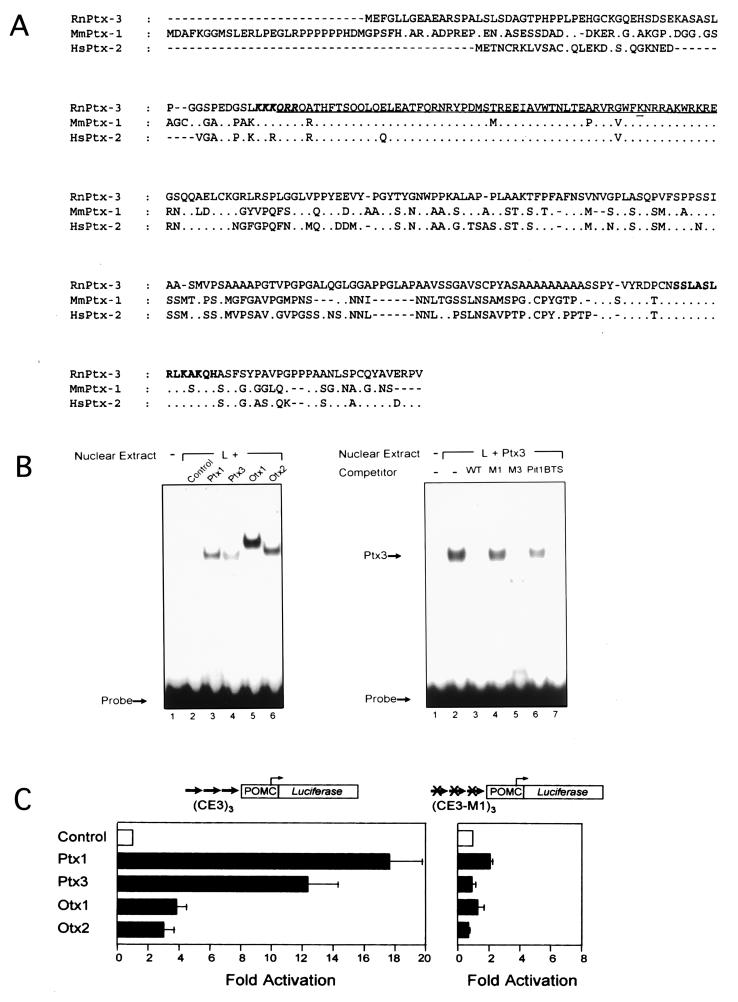
Cloning of several cDNA clones from rat brain led to the characterization of a cDNA encoding a protein of 302 aa. Comparison of its amino acid sequence to the database (Fig. 1A) revealed that it is related to Ptx1 and Rieg (hereafter named Ptx2). These two related proteins were implicated in pituitary-specific gene expression, in development of stomodeal structures (11–13), and in the Rieger syndrome, an autosomal-dominant human disorder characterized by craniofacial malformation (14), respectively. Based on homology to Ptx1 and Ptx2 (Fig. 1A), the protein was called Ptx3. The homeodomains of these three genes are highly conserved, differing only by one or two amino acids, and all display the lysine residue at position 50 of the homeodomain typical for bicoid-related proteins (Fig. 1A). The homology also extends into the proximal and extreme C terminus (≈67% homology, Ptx3 vs. Ptx1, Ptx3 vs. Ptx2). Thus, these three genes constitute a distinct and closely related subfamily within the paired-like class of homeoproteins.
Figure 1.
Structure and properties of PTX3. (A) Primary structure of Ptx3 deduced from the ORF of the Ptx3 cDNA and alignment to Ptx1 (11–13) and Ptx2 (RGS) (14). The homeodomain is underlined, with the bicoid-type-specific lysine at position 9 of the third helix double-underlined (23). A consensus nuclear localization signal is shown in boldface, italic type (24). The conserved C- terminal domain (14) is in boldface type. (B) Binding of Ptx3 to the CE3 element of the rat POMC promoter in gel shift analysis. Ptx3 binds to the CE3 element of the rat POMC promoter as was found for the related factors Ptx1, Otx1, and Otx2. Competition experiments using mutant recognition sites (11) within (M1) and outside (M3) the bicoid core of the CE3 element (200-fold molar excess) show that the ineffective mutant M3 and the wild type compete for Ptx3 binding, because the effective mutant M1 cannot. (C) Transactivational properties of ptx3 in transient transfection assays. The activity of Ptx3 on an artificial promoter construct based on three copies of the POMC-CE3 element is comparable to that of Ptx1 (11), whereas Otx1 and Otx2 are clearly less active. When using the M1 mutated binding site, the activity is lost.
Because Ptx3 is a new member of the Ptx subfamily we compared its binding and trans-activation properties to those of Ptx1 and of the related bicoid-like homeodomain proteins Otx-1 and -2 (25). In DNA-binding experiments all four homeodomain proteins bound the Ptx1-binding site of the proopiomelanocortin (POMC) gene (Fig. 1B). In contrast, both Ptx3 and Ptx1 exhibited stronger transactivating potential than Otx-1 and -2 on a reporter containing three Ptx1 binding sites (Fig. 1C). This activity was completely abolished by a mutation that prevents DNA binding (Fig. 1B), as previously shown for Ptx1 (11). Thus, Ptx3 can function as an activator of transcription on direct interaction with target genes.
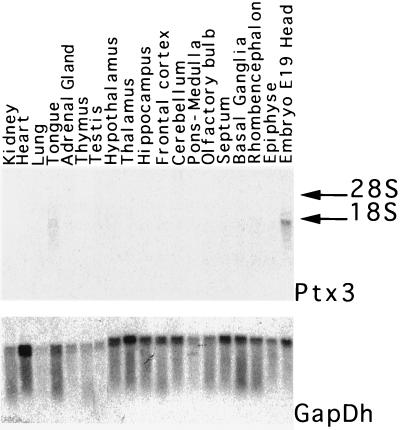
To develop a hypothesis on the function of Ptx3, its expression was analyzed in various tissues, including the pituitary gland, which is an important site of expression of Ptx1 and Ptx2 (11–14). Northern blot analysis indicated that Ptx3 is expressed in rat E19 embryo head. However, it could not be detected in dissected adult rat brain regions or in a set of peripheral organs (Fig. 2). Ptx3 is not expressed in the anterior pituitary or in a number of cell lines of pituitary origin (not shown), in contrast to Ptx1 and Ptx2. Thus, Ptx3 appears to have a very different expression pattern compared with the other subfamily members.
Figure 2.
Northern blot analysis of Ptx3 expression in the adult rat brain and peripheral organs. Blots containing 20 μg of total RNA of dissected rat brain regions and selected peripheral organs were hybridized with a full-length Ptx3 cDNA probe. GapDh was used as a control. The positions of 18S and 28S ribosomal RNAs are indicated by arrows.
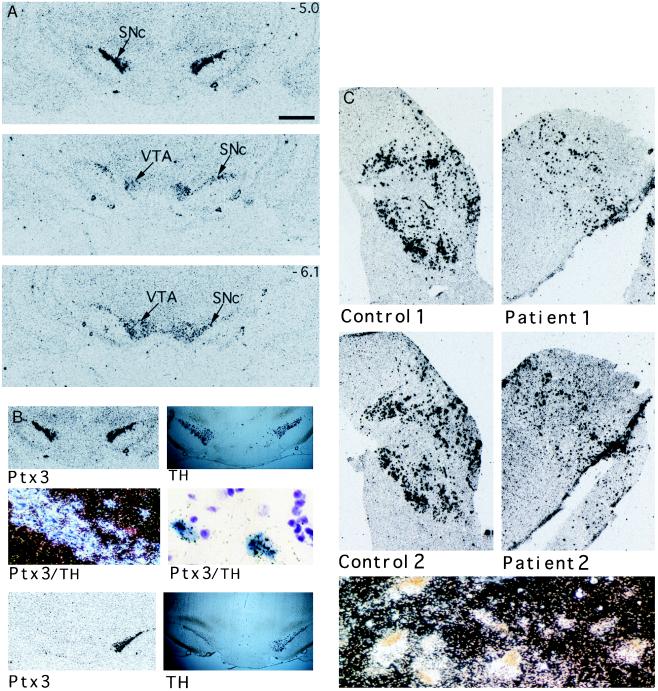
Because Ptx3 was not detected in dissected brain regions on Northern blots, despite its cloning from brain RNA, in situ hybridization was used to determine whether Ptx3 was expressed in a highly restricted manner in the brain. At the macroscopic level, hybridization of Ptx3 was confined to the substantia nigra pars compacta (SNc; A9) and the ventral tegmental area (VTA; A10) (Fig. 3A), together harboring the mesDA system (26), but was not seen in rostral regions having dopaminergic neurons nor in the adrenal gland (Fig. 2). To determine whether Ptx3 is indeed expressed in dopaminergic neurons we performed double in situ hybridization with Ptx3 and tyrosine hydroxylase (TH) cRNA probes (Fig. 3B). Ptx3 expression completely overlapped with TH-positive cells, demonstrating that Ptx3 is expressed in dopaminergic neurons of the mesDA system.
Figure 3.
Expression of Ptx3 in the mesencephalic dopaminergic system of rat and human. (A) The expression pattern of Ptx3 in rat brain starting anterior at the substantia nigra compacta (SNc) to posterior at the ventral tegmental area (VTA). The size bar (Top) represents a distance of 1 mm. The rostral–caudal positions of the Top and Bottom are indicated as millimeters relative to bregma in the upper right corner. (B) Double in situ hybridizations using a DIG-labeled tyrosine hydroxylase (TH) cRNA probe and a 35S-labeled Ptx3 cRNA probe show a similar hybridization pattern for both probes in the tegmentum of the rat brain (Top Left and Top Right). Microscopic examination (Middle) shows TH-positive cells in blue and Ptx3 labeled by silver grains (Middle Left, dark-field image; Middle Right, higher magnification, bright-field image). A complete overlap is found. In situ hybridization for TH and Ptx3 expression of animals with a unilateral lesion of dopaminergic neurons within the mesDA system. (Lower) The animals displayed increased turning behavior in the direction of the lesioned side. Both TH and Ptx3 expression was lost at the lesioned side. (C) Ptx3 expression in the human substantia nigra of two healthy controls (Control 1 and 2) and two Parkinson patients (Patient 1 and 2). The sections of the patients show a lower density of Ptx3-expressing cells, which correlates with the loss of dopaminergic neurons within this region. (Lower) A dark-field image showing Ptx3 expression (silver grains) colocalizing with typical brown-pigmented dopaminergic neurons of the human substantia nigra. Material was obtained from the Netherlands Brain Bank. Patient material was diagnosed clinically and pathologically.
Ablation of mesDA neurons by microinjection of the neurotoxin 6-OHDA has provided an animal model of motor dysfunction as in Parkinson disease (27, 28). Unilateral 6-OHDA lesions of the mesDA neurons in adult rats caused the characteristic rotational behavior and resulted in the total disappearance of TH expression on the lesioned side (Fig. 3B). Notably, Ptx3 expression was no longer detectable in the 6-OHDA-injected side but was normally expressed contralaterally. In situ hybridization experiments on the human substantia nigra showed strict colocalization of the human counterpart of Ptx3 with pigmented cells, which represent the dopaminergic neurons (Fig. 3C). Furthermore, in situ analysis of the substantia nigra of Parkinson patients revealed a reduced density of Ptx3-expressing neurons as compared with normal controls (Fig. 3C). Thus, the loss of expression correlates with loss of the mesDA neurons, both in animal models and human disease. Taken together, the results clearly indicated that mesDA neurons express Ptx3 both in rat and man.
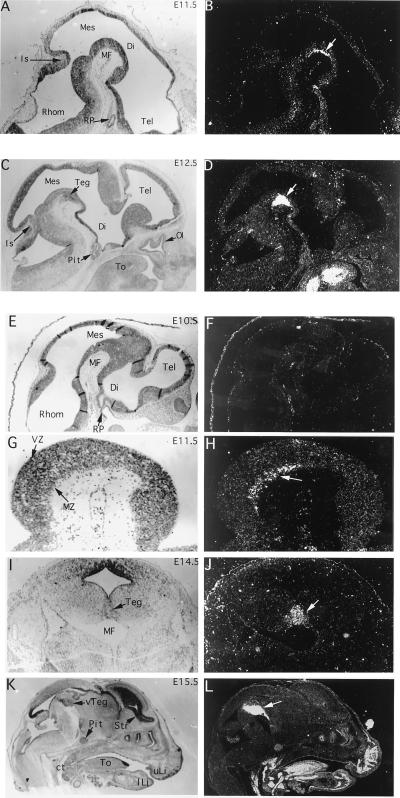
Because homeodomain proteins are usually involved in pattern formation, the close association of Ptx3 expression with an intact mesDA system suggests that Ptx3 may be involved in development and/or maintenance of this subset of dopaminergic neurons. Sectioned mouse embryos from E8.5 to E16.5 were examined by in situ hybridization to correlate Ptx3 expression with development of mesDA neurons. No signal above background was detected in the head at E8.5 (not shown) and at E10.5 (Fig. 4F). At E11.5, a small layer at the ventral surface of the mesencephalic flexure expressed Ptx3 (Fig. 4B). This group of about 50 cells corresponds to the first TH-expressing cells in the developing rodent brain (3–5, 26). At later stages, the expression remained restricted to the mesDA system (Fig. 4 D, J, and L), and this association is conserved in adult rat brain (Fig. 3). Higher magnifications show that Ptx3-positive cells are restricted to the marginal layer of the mesencephalic tegmentum (Fig. 4H); a coronal section at E14.5 reveals that the signal is restricted to the central region of the ventral tegmentum (Fig. 4J). Apart from the mesDA system, Ptx3 expression in the sclerotome and its cartilaginous derivatives as well as in the tongue was detected at this stage (Fig. 4L). The developmental pattern in brain shows that Ptx3 expression coincides spatially and temporally with the appearance of the developing mesDA system.
Figure 4.
In situ hybridization of Ptx3 in embryonic mouse brain sections between E10.5 and E15.5. Micrographs of bright-field and dark-field images of the same sections are shown. (A and B) At E11.5, expression is seen in a layer of postmitotic cells (arrow) in the mesencephalon (Mes) lining the mesencephalic flexure (MF). (C and D) At E12.5 the layer of expressing cells has thickened as part of the developing tegmentum (Teg). Extraneural expression is seen in the tongue (To) but not in the developing pituitary (Pit). (E and F) Expression of Ptx3 was not detected in the brain at E10.5. Lack of expression in Rathke’s pouch (RP), a site of Ptx1 and Ptx2 expression (11–14), confirms the specificity of the hybridization reaction. (G and H) Higher magnification of the tegmental region expressing Ptx3 at E11.5. Expression is detected ventrally in the marginal zone (MZ) of the neuroepithelium. (I and J) A coronal section of an E14.5 mouse brain shows the lateral extent of Ptx3 expression in the developing tegmentum (Teg). (K and L) A sagittal section of an E15.5 mouse head shows strong expression of Ptx3 in the ventral tegmentum (vTeg). In contrast to other hybridizations that were performed with 5′ or 3′ Ptx3 probes, the probe used for this experiment contained the homeodomain, and there is weak cross-reactivity with other members of the Ptx family outside the brain, e.g., the pituitary (Pit) (11–14). Mes, mesencephalon; Di, diencephalon; Tel, Telencephalon; Rhom, rhombencephalon; MF, mesencephalic flexure; RP, Rathke’s pouch; Is, isthmus; Pit, pituitary; To, tongue; Ol, olfactory epithelium; MZ, marginal zone; VZ, ventricular zone; Str, striatum; vTeg, ventral tegmentum; ct, cartilage of the throat; uLi, upper lip, lLi, lower lip.
Although the combinatorial action of many transcription and growth factors is likely involved in brain patterning (29, 30), the present work suggests that single homeodomain transcription factors may determine single neuronal cell identity. We have shown a highly restricted expression of Ptx3 in one neuronal lineage, the mesDA neurons. The only other example of such restricted neuronal expression is for a nematode gene, Unc-30, the closest nonmammalian homologue of the Ptx family. This gene is specifically required for the differentiation of the inhibitory GABA-ergic type D motor neurons of this nematode (31). The mesDA system is functionally heterogeneous and anatomically differentiated by different fields of axonal afferents (26, 32). Ptx3 is expressed in all these fields and, thus, its developmental association with the mesDA system suggests that Ptx3 may play a part in the development of the dopaminergic cell type rather than in axonal path finding or establishment of specific connectivity. Other factors involved in mesDA neuron development include the signaling molecule sonic hedgehog (33) and the orphan nuclear receptor Nurr1 (34). Nurr1 is expressed from E10.5 in the mouse midbrain but is not restricted to this area. Nonetheless, Nurr1 knockout mice specifically failed to develop mesDA neurons (35). These data suggest that Ptx3 and Nurr1 form a regulatory cascade for development of the mesDA system in which Nurr1 may act as an upstream activator. Because Ptx3 in contrast to Nurr1 is uniquely expressed in the mesDA system, it may activate a program for mesDA-specific gene expression and differentiation.
The degeneration of dopaminergic neurons within the mesDA system is the direct cause of Parkinson disease (7–10), and other extrapyramidal motor disorders like tardive dyskinesia (36–38) are also associated with this system. Furthermore, the mesDA system has been implicated in affective disorders like manic depression and schizophrenia (6, 39) and in behavioral reinforcement and drug addiction (40). Susceptibility to these disorders is thought to be predisposed prenatally. The identification of genes controlling developmental mechanisms of mesDA neurons can provide new insights in the etiology of these disorders. Ptx3 is a candidate gene involved in such mechanisms. Furthermore, the function of Ptx3 may be exploited for manipulation of dopaminergic cells in vitro in preparation of tissue grafting, an experimental therapy for Parkinson patients (41–43).
Acknowledgments
We thank Dr. Pieter Voorn and the Netherlands Brain Bank for preparing and providing sections of human brain material. We thank Dr. Jim Boulter for the gifts of rat cDNA libraries and Dr. A. Simeone for Otx-expression plasmids. This work was supported by grants of The Netherlands Organization for Scientific Research NWO-MW (900-546-109), the Korczak Foundation for Autism and Related Disorders, and the Medical Research Council of Canada. C.L. is a research student of the National Cancer Institute of Canada supported with funds provided by the Canadian Cancer Society.
Note Added in Proof
Recently, the mouse homologue of Ptx3 has been implicated in lens development and mapped to the aphakia region of mouse chromosome 3 (44).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbrevations: mesDA, mesencephalic dopaminergic; 6-OHDA, 6-hydroxy-dopamine; TH, tyrosine hydroxylase; POMC, proopiomelanocortin.
Data deposition: The sequence reported in this paper has been deposited in the EMBL database [accession no. P81062 (Ptx3)].
References
- 1.Lumsden A, Krumlauf R. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 2.Tanabe Y, Jessell T M. Science. 1996;274:1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]
- 3.Specht L A, Pickel V M, Joh T H, Reis D. J Comp Neurol. 1981;199:233–253. doi: 10.1002/cne.901990207. [DOI] [PubMed] [Google Scholar]
- 4.Voorn P, Kalsbeek A, Jorritsma-Byham J, Groenewegen H J. Neuroscience. 1988;25:857–887. doi: 10.1016/0306-4522(88)90041-3. [DOI] [PubMed] [Google Scholar]
- 5.Solberg Y, Silverman W, Pollack Y. Dev Brain Res. 1993;73:91–97. doi: 10.1016/0165-3806(93)90050-k. [DOI] [PubMed] [Google Scholar]
- 6.Grace A A, Bunney B S, Moore H, Todd C L. Trends Neurosci. 1997;20:31–32. doi: 10.1016/S0166-2236(96)10064-3. [DOI] [PubMed] [Google Scholar]
- 7.Jellinger K. Cesk Pathol. 1973;9:1–13. [PubMed] [Google Scholar]
- 8.Forno L S. Ann N Y Acad Sci. 1992;648:6–16. doi: 10.1111/j.1749-6632.1992.tb24519.x. [DOI] [PubMed] [Google Scholar]
- 9.Golbe L I. Rev Neurosci. 1993;4:1–16. doi: 10.1515/revneuro.1993.4.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch E C. Mol Neurobiol. 1994;9:135–142. doi: 10.1007/BF02816113. [DOI] [PubMed] [Google Scholar]
- 11.Lamonerie T, Tremblay J J, Lanctôt C, Therrien M, Gauthier Y, Drouin J. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 12.Lanctôt C, Lamolet B, Drouin J. Development. 1997;124:2807–2817. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 13.Szeto D P, Ryan A K, O’Connell S M, Rosenfeld M G. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semina E V, Reiter R, Leysens N J, Alward W L M, Small K W, Datson N A, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel B U, Carey J C, Murray J. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 15.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Therrien M, Drouin J. Mol Cell Biol. 1991;11:3492–3503. doi: 10.1128/mcb.11.7.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiber E, Matthias P, Müller M M, Schaffner W. Nucleic Acids Res. 1989;11:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopes da Silva S, Horssen A M, van Chang C, Burbach J P H. Endocrinology. 1995;136:2276–2283. doi: 10.1210/endo.136.5.7720676. [DOI] [PubMed] [Google Scholar]
- 20.Grima B, Lamouroux A, Blanot F, Biguet N F, Mallet J. Proc Natl Acad Sci USA. 1985;82:617–621. doi: 10.1073/pnas.82.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson D G. In: In Situ Hybridization: A Practical Approach. Wilkinson D G, editor. Washington, DC: IRL; 1992. [Google Scholar]
- 22.Van Ree J M, Wolterink G. Eur J Pharmacol. 1981;72:107–111. doi: 10.1016/0014-2999(81)90305-8. [DOI] [PubMed] [Google Scholar]
- 23.Bürglin T R. In: A Guidebook for Homeobox Genes. Duboule D, editor. Oxford: Oxford Univ. Press; 1993. pp. 25–71. [Google Scholar]
- 24.LaCasse E C, Lefebvre Y A. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nature (London) 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 26.Björkland L A, Lindvalt O. In: Handbook of Chemical Neuroanatomie. Björkland L A, Hökfelt T, editors. Vol. 2. London: Elsevier Science; 1984. pp. 55–122. [Google Scholar]
- 27.Ungerstedt U, Ljungberg T, Steg G. Adv Neurol. 1974;5:421–426. [PubMed] [Google Scholar]
- 28.Hudson J L, Van Horne C G, Stromberg I, Brock S, Clayton J, Masserano J, Hoffer B J, Gerhardt G A. Brain Res. 1993;626:167–174. doi: 10.1016/0006-8993(93)90576-9. [DOI] [PubMed] [Google Scholar]
- 29.Rubenstein J L R, Salvador M, Shimamura K, Puelles L. Science. 1994;266:578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- 30.Figdor M C, Stern C D. Nature (London) 1993;363:630–634. doi: 10.1038/363630a0. [DOI] [PubMed] [Google Scholar]
- 31.Jin Y, Hoskins R, Horvitz R. Nature (London) 1994;372:780–783. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- 32.Fallon J H, Loughlin S E. In: The Rat Nervous System. 2nd Ed. Paxinos G, editor. London: Academic; 1995. pp. 215–237. [Google Scholar]
- 33.Hynes M, Porter J A, Chiang C, Chang D, Tessier-Lavigne M, Beachy P A, Rosenthal A. Neuron. 1995;15:35–44. doi: 10.1016/0896-6273(95)90062-4. [DOI] [PubMed] [Google Scholar]
- 34.Zetterström R H, Williams R, Perlmann T, Olson L. Mol Brain Res. 1996;41:111–120. doi: 10.1016/0169-328x(96)00074-5. [DOI] [PubMed] [Google Scholar]
- 35.Zetterstrom R H, Solomin L, Jansson L, Hoffer B J, Olson L, Perlmann T. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 36.Klawans H L, Goetz C G, Perlik S. Am J Psychiatry. 1980;137:900–908. doi: 10.1176/ajp.137.8.900. [DOI] [PubMed] [Google Scholar]
- 37.Goetz C G, Klawans H L. Neurol Clin. 1984;2:605–614. [PubMed] [Google Scholar]
- 38.Verghese C, De Leon J, Simpson G M. Am J Psychiatry. 1994;151:1716–1717. doi: 10.1176/ajp.151.11.aj151111716. [DOI] [PubMed] [Google Scholar]
- 39.Crow T J, Johnstone E C, Longden A, Owen F. Adv Biochem Psychopharmacol. 1978;19:301–309. [PubMed] [Google Scholar]
- 40.White F J. Annu Rev Neurosci. 1996;19:405–436. doi: 10.1146/annurev.ne.19.030196.002201. [DOI] [PubMed] [Google Scholar]
- 41.Perlow M J. Neurosurgery. 1987;20:335–342. doi: 10.1227/00006123-198702000-00026. [DOI] [PubMed] [Google Scholar]
- 42.Date I, Miyoshi Y, Ono T, Imaoka T, Furuta T, Asari S, Ohmoto T, Iwata H. Cell Transplant. 1996;5:S17–S19. doi: 10.1016/0963-6897(96)00032-2. [DOI] [PubMed] [Google Scholar]
- 43.Kordower J H, Rosenstein J M, Collier T J, Burke M A, Chen E Y, Li J M, Martel L, Levey A E, Mufson E J, Freeman T B, Olanow C W. J Comp Neurol. 1996;370:203–230. doi: 10.1002/(SICI)1096-9861(19960624)370:2<203::AID-CNE6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 44.Semina, E. V., Reiter, R. S. & Murray, J. C. (1997) Hum. Mol. Genet., in press. [DOI] [PubMed]