Abstract
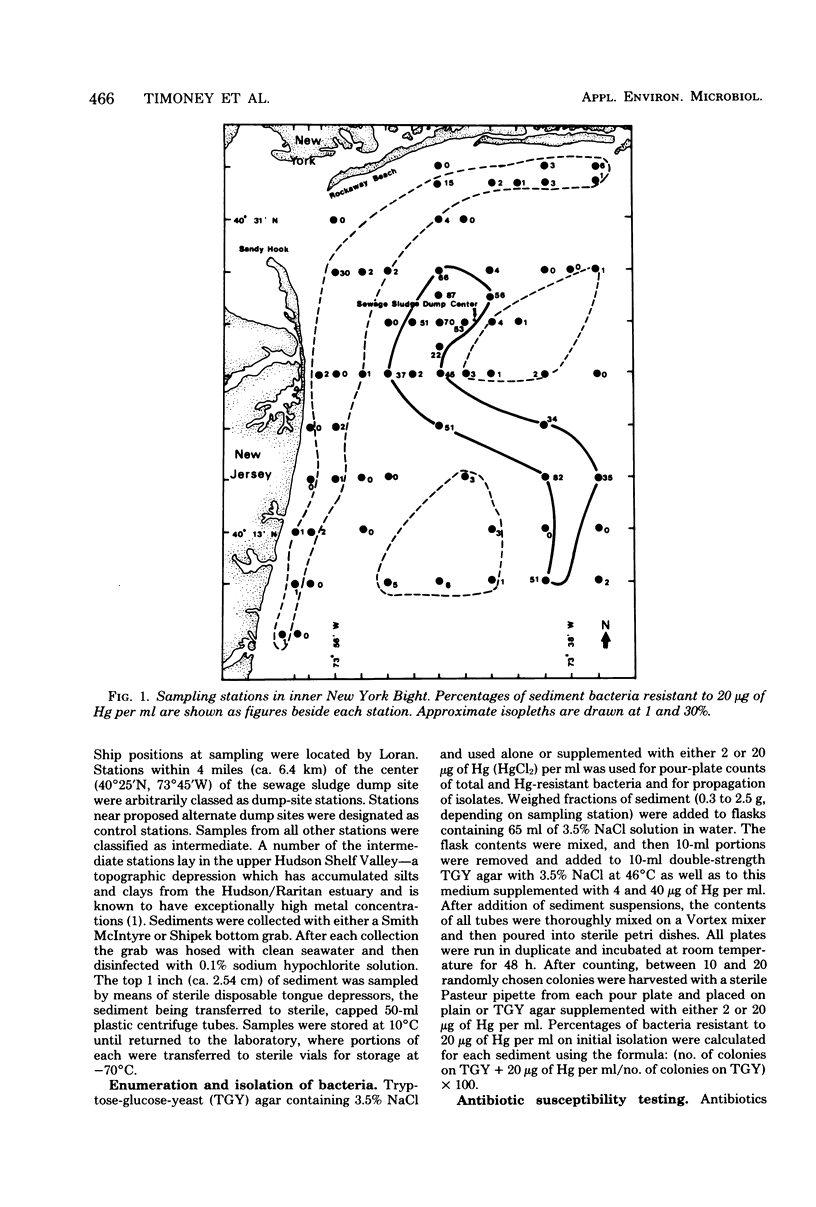
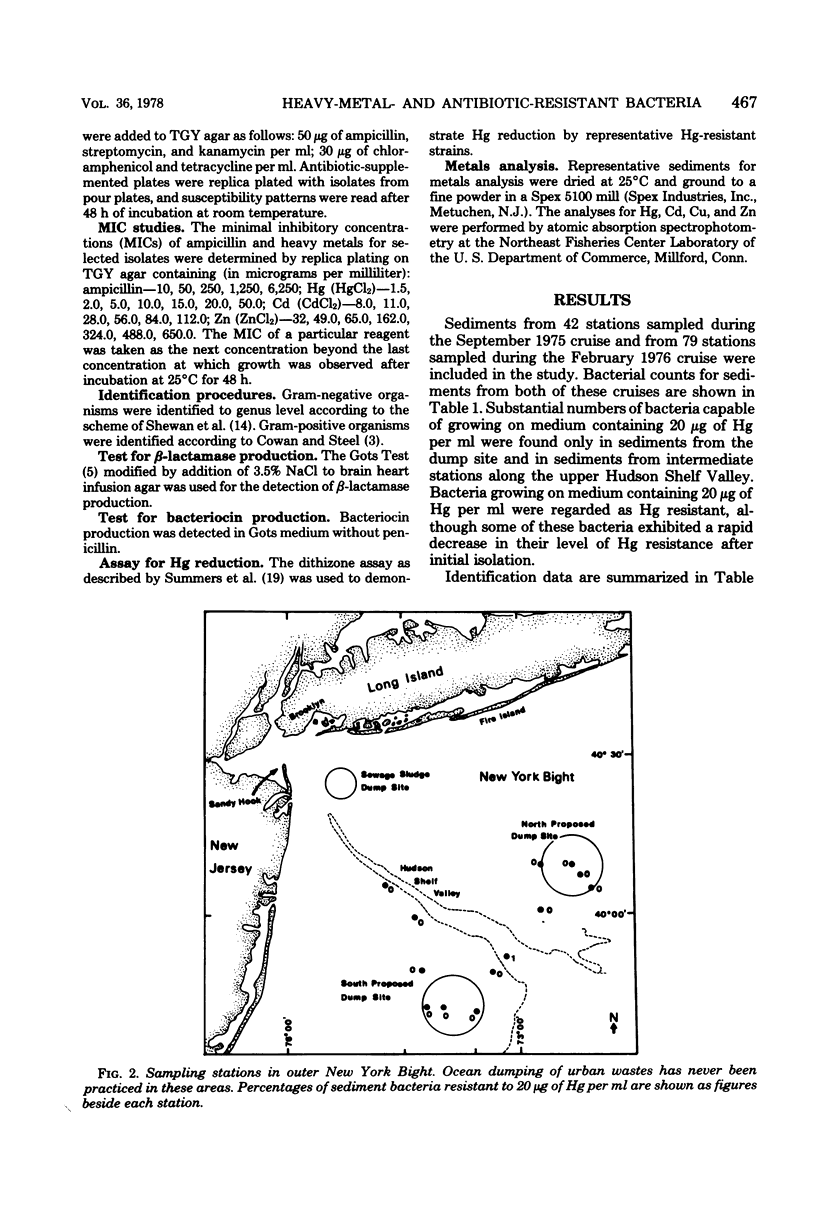
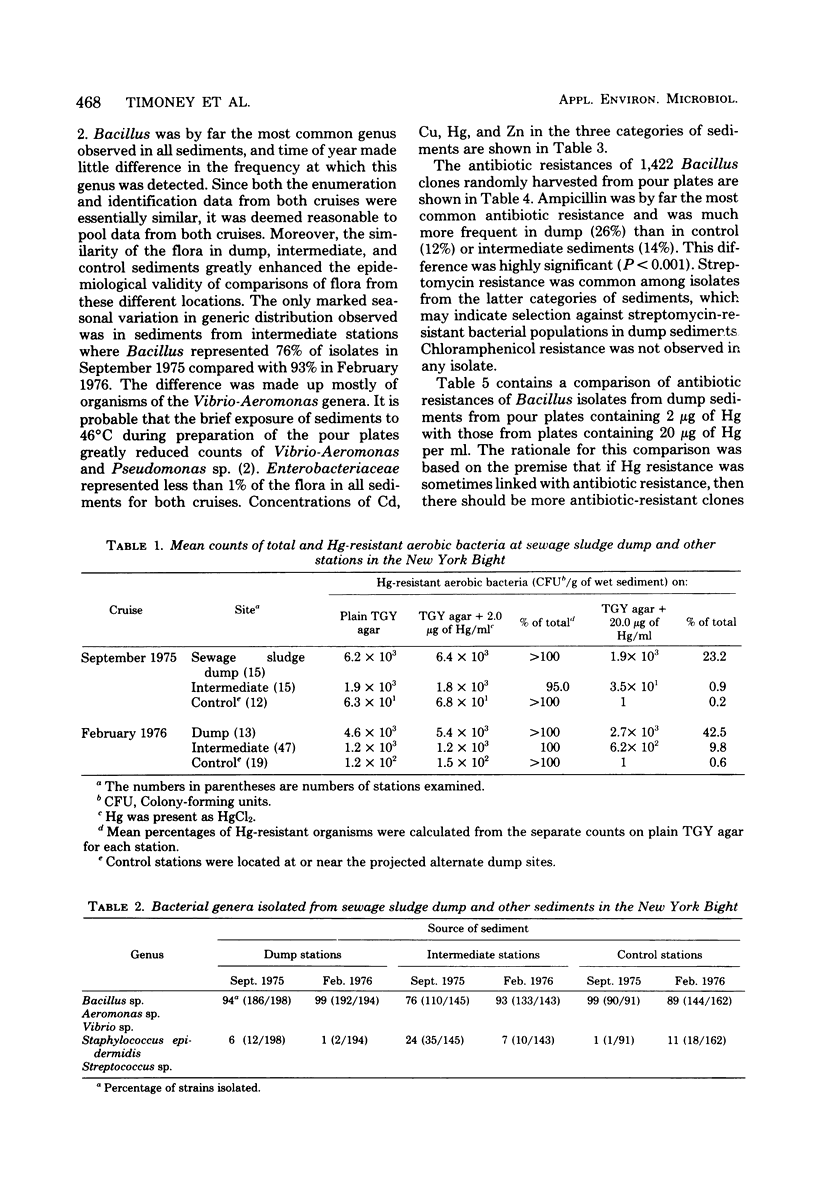
The New York Bight extends seaward some 80 to 100 miles (ca. 129 to 161 km) from the Long Island and New Jersey shorelines to the edge of the continental shelf. Over 14 × 106 m3 of sewage sludge, dredge spoils, acid wastes, and cellar dirt are discharged into this area each year. Large populations of Bacillus sp. resistant to 20 μg of mercury per ml were observed in Bight sediments contaminated by these wastes. Resistant Bacillus populations were much greater in sediments containing high concentrations of Hg and other heavy metals than in sediments from areas further offshore where dumping has never been practiced and where heavy-metal concentrations were found to be low. Ampicillin resistance due mainly to β-lactamase production was significantly (P < 0.001) more frequent in Bacillus strains from sediments near the sewage sludge dump site than in similar Bacillus populations from control sediments. Bacillus strains with combined ampicillin and Hg resistances were almost six times as frequent at the sludge dump site as in control sediments. This observation suggests that genes for Hg resistance and β-lactamase production are simultaneously selected for in Bacillus and that heavy-metal contamination of an ecosystem can result in a selection pressure for antibiotic resistance in bacteria in that system. Also, Hg resistance was frequently linked with other heavy-metal resistances and, in a substantial proportion of Bacillus strains, involved reduction to volatile metallic Hg (Hg°).
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gots J. S. THE DETECTION OF PENICILLINASE-PRODUCING PROPERTIES OF MICROORGANISMS. Science. 1945 Sep 21;102(2647):309–309. doi: 10.1126/science.102.2647.309. [DOI] [PubMed] [Google Scholar]
- McHugh G. L., Moellering R. C., Hopkins C. C., Swartz M. N. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet. 1975 Feb 1;1(7901):235–240. doi: 10.1016/s0140-6736(75)91138-1. [DOI] [PubMed] [Google Scholar]
- RICHMOND M. H., JOHN M. CO-TRANSDUCTION BY A STAPHYLOCOCCAL PHAGE OF THE GENES RESPONSIBLE FOR PENICILLINASE SYNTHESIS AND RESISTANCE TO MERCURY SALTS. Nature. 1964 Jun 27;202:1360–1361. doi: 10.1038/2021360a0. [DOI] [PubMed] [Google Scholar]
- Sizemore R. K., Colwell R. R. Plasmids carried by antibiotic-resistant marine bacteria. Antimicrob Agents Chemother. 1977 Sep;12(3):373–382. doi: 10.1128/aac.12.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. H. R factors mediate resistance to mercury, nickel, and cobalt. Science. 1967 May 26;156(3778):1114–1116. doi: 10.1126/science.156.3778.1114. [DOI] [PubMed] [Google Scholar]
- Spangler W. J., Spigarelli J. L., Rose J. M., Miller H. M. Methylmercury: bacterial degradation in lake sediments. Science. 1973 Apr 13;180(4082):192–193. doi: 10.1126/science.180.4082.192. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Mercury resistance in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1972 Dec;112(3):1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]


