Abstract
Human Ape1 is a multifunctional protein with a major role in initiating repair of apurinic/apyrimidinic (AP) sites in DNA by catalyzing hydrolytic incision of the phosphodiester backbone immediately adjacent to the damage. Besides in double-stranded DNA, Ape1 has been shown to cleave at AP sites in single-stranded (ss) regions of a number of biologically-relevant DNA conformations and in structured ssDNA. Extension of these studies has revealed a more expansive repertoire of model substrates on which Ape1 exerts AP endonuclease activity. In particular, Ape1 possesses the ability to cleave at AP sites located in (i) the DNA strand of a DNA/RNA hybrid, (ii) “pseudo-triplex” bubble substrates designed to mimic stalled replication or transcription intermediates, and (iii) configurations that emulate R-loop structures that arise during class switch recombination (CSR). Moreover, Ape1 was found to cleave AP site-containing ssRNA, suggesting a novel “cleansing” function that may contribute to the elimination of detrimental cellular AP-RNA molecules. Finally, sequence context immediately surrounding an abasic site in duplex DNA was found to have a < 3-fold effect on the incision efficiency of Ape1, and ATP was found to exert complex effects on the endonuclease capacity of Ape1 on double-stranded substrates. The results suggest that in addition to abasic sites in conventional duplex genomic DNA, Ape1 has the ability to incise at AP sites in DNA conformations formed during DNA replication, transcription and CSR, and that Ape1 can endonucleolyticly destroy damaged RNA.
Keywords: Ape1, DNA repair, polymerase stalling, class switch recombination, RNA cleavage, nucleotide sequence context
Introduction
Apurinic/Apyrimidinic (AP) endonuclease 1 (Ape1) is a multifunctional enzyme of 318 amino acids (35.5 kDa) with redox-dependent regulation of transcription factors1, 3′ to 5′ exonuclease2, 3′ phosphodiesterase3, RNaseH4, and class II type AP endonuclease activities5. The endonuclease function entails incision of the phosphodiester bond immediately 5′ to an AP site, generating a single-strand break with 5′ deoxyribose phosphate (dRP) and 3′ hydroxyl ends6. This action creates a substrate on which subsequent enzymes of the base excision repair (BER) pathway can act to fill the nucleotide gap and seal the nick in the DNA backbone to complete AP site repair (i.e. DNA polymerase β and XRCC1-DNA ligase IIIα). Ape1 is the major mammalian protein for initiating removal of abasic sites in DNA7, although a second Ape1-like protein has been identified in mammalian cells that acts as a 3′ to 5′ exonuclease and possesses a weak AP endonuclease activity8, 9.
Abasic sites are estimated to arise spontaneously ~10,000 times per mammalian genome equivalent per day10. In addition to spontaneous base loss, abasic intermediates are formed via enzyme (DNA glycosylase)-catalyzed base release during the process of BER11. Unrepaired, non-instructional abasic sites have mutagenic and cytotoxic consequences to the cell12. For instance, elongating replicative DNA polymerases and RNA polymerases will pause or arrest at abasic products, leading to collapse of the synthetic process and possibly activation of cell death responses13. Abasic sites encountered during DNA replication can also lead to error-prone bypass synthesis by translesion DNA polymerases14. In Saccharomyces cerevisiae, enhanced mutagenesis and severe inhibition of transcription occurs in strains defective in the removal of AP sites and that lack RAD26, a SWI/SNF family ATPase and a homolog of the human Cockayne syndrome B (ERCC6/CSB) gene15. In all, current experimental evidence supports the idea that (i) AP sites lead to enhanced DNA mutagenesis, (ii) natural and oxidized abasic forms are blocks to RNA polymerase progression, and (iii) CSB functions to promote AP site processing, presumably by facilitating efficient transcription and/or preventing the production of mutant RNA templates.
The presence of damaged or mutant RNA molecules can have many consequences to the cell. For instance, damaged or miscoding ribosomal RNAs have the potential to poison an entire ribosome and lead to collapse of the translational machinery16. Damaged or miscoding tRNAs can lead to mutagenic protein synthesis17. Aberrant mRNAs have the capacity to do both, as a ribosome may stall upon encountering the damage or incorporate improper amino acids to produce mutant polypeptides18. Pathways for dealing with altered mRNAs exist in mammalian cells, such as nonsense-mediated decay and non-stop decay19. In addition to pathways that degrade improper mRNAs, enzymes have evolved to repair damage to RNA molecules. The AlkB family of oxidative demethylases removes 1-methyladenine, 3-methylcytosine, and 1-methylguanine from both DNA and RNA20. It is not currently known whether other types of RNA damage are subject to “repair”, although oxidative RNA modifications have been shown to be increased in both Alzheimer’s21 and atherosclerosis22.
Complex nucleic acid structures form not only during the processes of DNA replication and RNA transcription, but also during other cellular events. Antibody class switch recombination (CSR) occurs at immunoglobulin (Ig) switch regions via the creation and joining of DNA double-strand breaks (DSBs)23. The process of CSR is presumably mediated by transcription through the GC rich switch regions, which leads to the formation of DNA/RNA hybrids within so-called R-loops24. The establishment of DSBs is thought to be initiated by the action of activation-induced cytidine deaminase (AID), which deaminates cytosine to uracil within these loop structures. The uracil bases are then processed to abasic sites and single-strand breaks by as yet undefined repair components. One model proposes that the creation of juxtaposed nicks in DNA by processing of opposed uracil residues gives rise to the DSBs necessary for CSR and antibody isotype diversity25. Recent evidence has implicated Ape1, and a paralogous protein Ape2, in AP site cleavage during CSR26, yet a precise biochemical activity on R-loop substrates has not been examined.
Here, we explored the activities of human Ape1 on a variety of alternative substrates. We have designed abasic site-containing substrates that mimic a stalled DNA replication or RNA transcription “pseudo-triplex” bubble to determine if Ape1 can cleave in these contexts. We have also designed “pseudo-triplex” R-loop type substrates to examine the influence of this configuration on Ape1 AP site incision during simulated CSR. In addition, we analyzed whether Ape1 exhibits endonuclease activity on single-strand AP-RNA molecules, as well as various lesion-containing DNA/RNA arrangements. Lastly, we determined the effect of the DNA sequence immediately surrounding an abasic site on Ape1 incision activity. Our findings demonstrate that Ape1 can endonucleolytically cleave abasic sites in complex biologically-relevant contexts related to dysfunction during the essential processes of DNA replication, transcription and translation, and can cleave at AP sites in ssDNA regions of R-loops related to CSR.
Results
Incision by Ape1 of AP-DNA/RNA duplexes
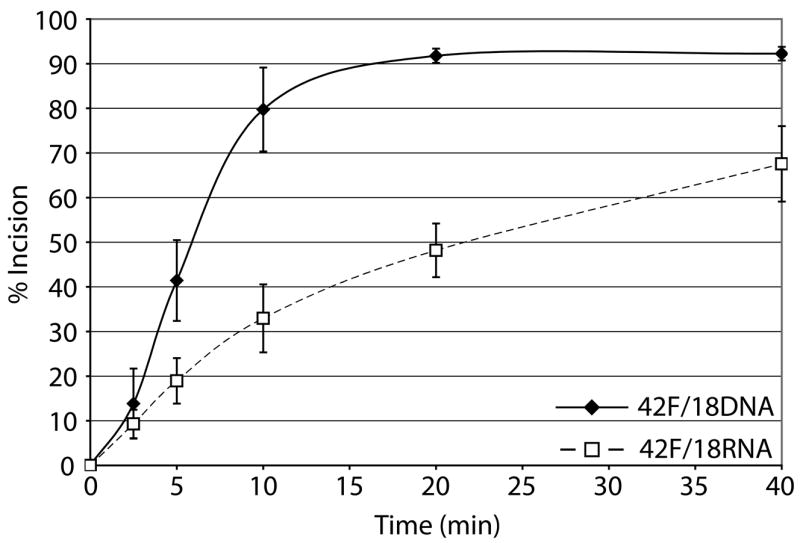
Given the prior evidence that Ape1 can cleave at AP sites in transcription-like bubble structures27, 28, we evaluated, in vitro, the AP endonuclease activity of human Ape1 on synthetic DNA/RNA hybrids that would simulate a transcription intermediate. Substrates analyzed were 42mer DNA strands containing a centrally located abasic site analog (42F) that were annealed to either a 18mer DNA (control) or a 18mer RNA (Table 1). We found that Ape1 had only slightly more robust activity on the partial DNA/DNA duplex than on the comparable DNA/RNA duplex (Figure 1). Kinetic parameters for Ape1 endonuclease activity on the AP-DNA/DNA and AP-DNA/RNA are as follows: Km values for DNA/DNA and DNA/RNA were 23.8 nM and 5.7 nM; Vmax values were 4.2 nM product min−1 and 1.3 nM product min−1; kcat values were 2.9 min−1 and 0.9 min−1; and catalytic efficiencies (kcat/Km) were 0.12 and 0.16, respectively.
Table 1. Oligonucleotides used in this study.
F denotes synthetic abasic site analog tetrahydrofuran.
| DNA/DNA, DNA/RNA, ssDNA, ssRNA
| |
|---|---|
| Name | Sequence (5′-3′) |
| 42F | CCGCTGAATTGCACCCTCGAFCTAGGTCGATGATCCTAAGCA |
| 42bubbleComp | TGCTTAGGATCATCGAGGATCGAGCTCGGTGCAATTCAGCGG |
| 42comp | TGCTTAGGATCATCGACCTAGGTCGAGGGTGCAATTCAGCGG |
| 18DNA | TCGACCTAGATCGAGGGT |
| 18RNA | rUrCrGrArCrCrUrArGrArUrCrGrArGrGrGrU |
| 18RNAfor54comp | rUrGrGrGrArGrCrUrUrGrArUrCrCrArGrCrU |
| 54Fendbubble18 | CCGCTGCCGCTGAATTGCFCCCTCGATCTAGGTCGATGATCCTAAGCATAAGCA |
| 54Fcenterbubble18 | CCGCTGCCGCTGAATTGCACCCTCGAFCTAGGTCGATGATCCTAAGCATAAGCA |
| 54bubble18comp | TGCTTATGCTTAGGATCAAGCTGGATCAAGCTCCCAGCAATTCAGCGGCAGCGG |
| 26FDNA | AATTCACCGGTACGFACTAGAATTCG |
| 26FRNA | rArArUrUrCrArCrCrGrGrUrArCrGFrArCrUrArGrArArUrUrCrG |
| 34FDNA | CTGCAGCTGATGCGCFGTACGGATCCCCGGGTAC |
| 34FRNA | rCrUrGrCrArGrCrUrGrArUrGrCrGrCFrGrUrArCrGrGrArUrCrCrCrCrGrGrGrUrArC |
| Sequence Context
| |
| Name | Sequence (5′-3′) |
|
| |
| GFA | AATTCACCGGTACGFACTAGAATTCG |
| GFA-C | CGAATTCTAGTCCGTACCGGTGAATT |
| GFA-G | CGAATTCTAGTGCGTACCGGTGAATT |
| CFT | AATTCACCGGTACCFTCTAGAATTCG |
| CFT-C | CGAATTCTAGACGGTACCGGTGAATT |
| CFT-G | CGAATTCTAGAGGGTACCGGTGAATT |
| GFT | AATTCACCGGTACGFTCTAGAATTCG |
| GFT-C | CGAATTCTAGACCGTACCGGTGAATT |
| GFT-G | CGAATTCTAGAGCGTACCGGTGAATT |
| CFA | AATTCACCGGTACCFACTAGAATTCG |
| CFA-C | CGAATTCTAGTCGGTACCGGTGAATT |
| CFA-G | CGAATTCTAGTGGGTACCGGTGAATT |
Figure 1. Ape1 can cleave AP sites in DNA/RNA duplex substrates.
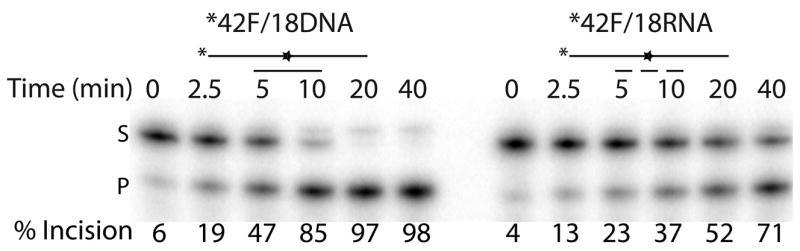
a. Representative denaturing polyacrylamide gel of AP site incision by Ape1 on partially duplex DNA/DNA (left) and DNA/RNA (right). DNA is represented by solid lines, RNA is represented by dashed lines, and abasic site position is represented as a star. Asterisk indicates site of radiolabel. S denotes substrate position and P denotes product position. Time points are shown on top and percent of substrate converted to product is shown below for each time point analyzed.
b. Graph depicting time course kinetics of Ape1 incision of DNA/DNA and DNA/RNA substrates. Average values are plotted with standard deviations of at least 3 independent experiments.
AP site position affects Ape1 incision efficiency
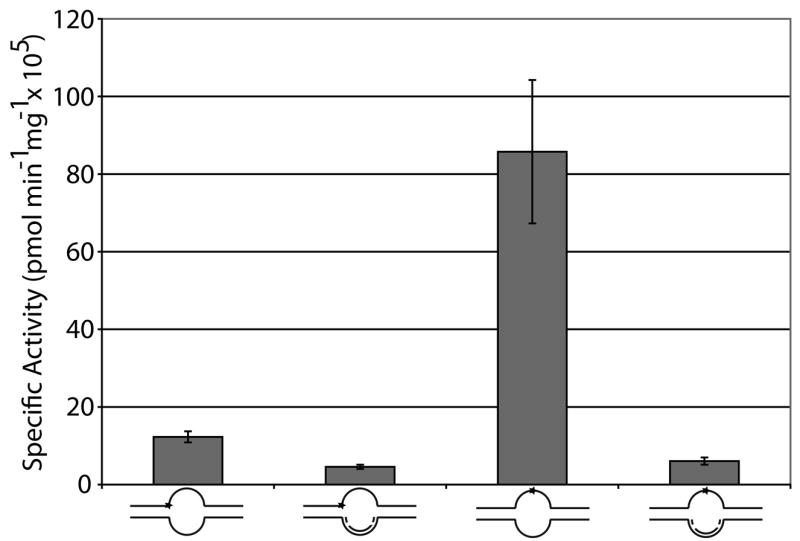
We next sought to determine the effect of the location of an abasic site in partial double-stranded DNA and in bubble duplex DNA. Oligonucleotides used were 54mers annealed to a complementary 18mer DNA to form the partial duplex substrates (54Fendbubble or 54Fcenterbubble with 18DNA, Table 1) and two 54mers annealed to create an 18 nucleotide (nt) bubble structure (54Fendbubble or 54Fcenterbubble with 54bubble18comp, Table 1). The abasic site was either centrally located (center bubble DNAs) or located at the ssDNA/dsDNA junction (end bubble DNAs) (Figure 2a, top). We found that Ape1 most efficiently incised at an abasic site located centrally in the 18 nt bubble structure (54Fcenterbubble18/54bubblecomp), followed by the centrally located abasic site in the partial duplex DNA (54Fcenterbubble18/18DNA), the abasic site at a ssDNA/dsDNA junction in the bubble conformation (54Fendbubble18/54bubblecomp), and the abasic site at a ssDNA/dsDNA junction in partial duplex DNA (54Fendbubble18/18DNA) (Figure 2a and b).
Figure 2. Ape1 incises at AP sites in complex nucleic acid structures formed during replication or transcription.

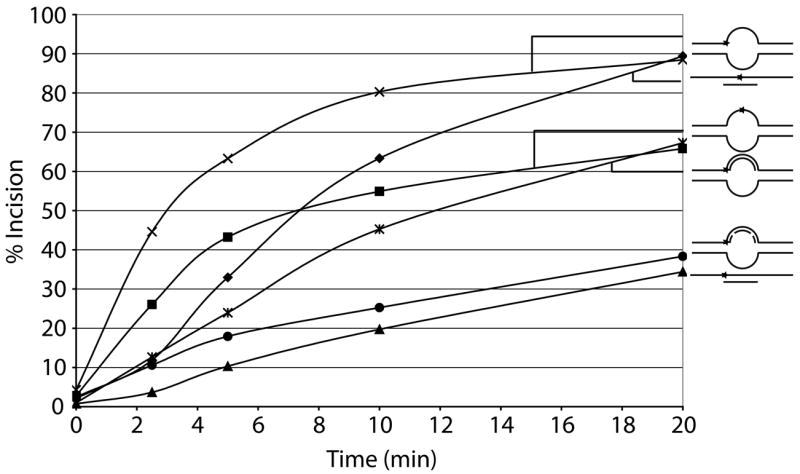
a. Substrate composition, representation of substrate configuration, and specific activities of Ape1 with standard deviations from at least 3 independent experiments are shown, along with relative incision capacity compared to standard partial duplex substrate (54Fcenterbubble18/18DNA). DNA is represented by solid lines, RNA is represented by dashed lines, and abasic site position is represented as a star. Asterisk denotes site of radiolabel.
b. Graph depicting time course kinetics of Ape1 incision of partial duplex, bubble duplex, replication bubble mimic, and transcription bubble mimic substrates. Average values of at least 3 independent experiments are shown; standard deviations were omitted to simplify figure (see panel “a” for variability). Substrates are depicted next to the graph with DNA represented by solid lines, RNA represented by dashed lines, and abasic site position represented as a star. Reactions with center-positioned abasic site contained 10 pg (0.3 fmol, 30 fM) Ape1 and reactions with junction-positioned abasic site contained 100 pg (3 fmol, 300 fM) Ape1.
Ape1 incises at AP sites in replication and transcription intermediates
As we observed that Ape1 could incise at AP sites in the DNA strand of a DNA/RNA duplex (Figure 1) and within a bubble structure (Figure 2 and previous results 27, 28), we next sought to determine the effect of a DNA strand or a RNA strand positioned within the bubble. These “pseudo-triplex” nucleic acid substrates contained an abasic damage at the ssDNA/dsDNA junction and reflect potential intermediates formed when either a DNA or RNA polymerase stalls at an abasic site in the leading strand during DNA replication (54Fendbubble18/54bubblecomp/18DNA) or the transcribed strand during transcription (54Fendbubble18/54bubblecomp/18RNA), respectively (Figure 2a, bottom). Strikingly, creation of these three strand substrates provided a 2–2.5 fold increase in the specific activity of Ape1 over the partial duplex with the abasic lesion positioned at the ss/dsDNA junction (54Fendbubble18/18DNA), indicating that Ape1 can efficiently incise complex substrates that resemble both arrested replication and transcription intermediates (Figure 2b). Pseudo-triplexes containing a centrally located abasic site were not analyzed as annealing of the third 18mer DNA or RNA strand was inefficient, likely due to the position of the AP lesion (data not shown).
ATP has complex effects on Ape1 incision activity
Previously, it had been demonstrated that the SWI/SNF DNA-dependent ATPase CSB and Ape1 physically interact in vitro and exist in a common protein complex in vivo28. CSB was also found to stimulate AP site incision by Ape1 (in the absence of ATP) on both fully-paired duplex DNA (42F/42Comp, Table 1) and an 11 nt bubble duplex substrate containing a centrally located abasic site (42F/42bubbleComp, Table 1). Here, we evaluated the effect of CSB on Ape1 incision of our partial duplex DNA substrate (42F/18DNA, Table 1), the previously employed fully-paired duplex DNA (42F/42Comp) and 11nt bubble substrates (42F/42bubbleComp), and our more complex 18 nt bubble DNA replication (54Fendbubble18/54bubblecomp/18DNA) and transcription (54Fendbubble18/54bubblecomp/18RNA) “pseudo-triplex” intermediates. CSB stimulated Ape1 incision on the 42F/18DNA, 42F/42Comp, and 42F/42bubbleComp substrates, recapitulating the previous observations (data not shown). However, with both “pseudo-triplex” substrates, no significant effect of CSB was observed on Ape1 AP site incision in the absence of ATP (data not shown), suggesting, as previously proposed, that activation by CSB is substrate influenced dependent28. Notably, when 2.5 mM ATP was included in the reactions with 1 mM MgCl2, we detected an inhibition of Ape1 incision activity (data not shown). We therefore sought to further investigate the effect of ATP on Ape1 AP site cleavage.
Using 42mer fully-paired abasic site-containing duplex DNA (42F/42comp, Table 1), we examined the effects of different concentrations of ATP on Ape1 incision at either 1 mM or 10 mM MgCl2. We found that ATP had complex effects on Ape1 cleavage activity (Figure 3a). At 1 mM MgCl2, Ape1 abasic endonuclease activity was stimulated ~1.75- and ~1.25-fold at 0.5 mM and 1 mM ATP concentrations, respectively and Ape1 incision activity was inhibited at higher ATP concentrations (2–5 mM). At 10 mM MgCl2, the higher concentrations of ATP had stimulatory effects on AP site incision by Ape1, whereas the lower concentrations had no effect (Figure 3a). Interestingly, when the ratio of Mg2+ to ATP was 2:1 (see 1 mM MgCl2 and 0.5 mM ATP vs. 10 mM MgCl2 and 5 mM ATP), Ape1 AP site incision activity was activated (1.7 to 3.3-fold) most significantly (Figure 3b). Steady-state kinetic analysis of Ape1 AP endonuclease activity at 10 mM MgCl2 with and without 5 mM ATP indicated a less then 2-fold difference in Km, but a ~20-fold enhancement in kcat in the presence of ATP, suggesting an enhancement of the catalytic reaction specifically. We further examined the effect of other nucleotides (TTP, CTP, GTP, and dATP) on Ape1 incision activity at a Mg2+:(d)NTP ratio of 2:1 (10 mM MgCl2 and 5 mM (d)NTP). We found that ATP, dATP, and CTP had a similar degree of stimulation, and that TTP and GTP activated incision, but to a lesser extent (data not shown).
Figure 3. Relative ATP and Mg++ concentration has complex effect on Ape1 incision activity of AP dsDNA.
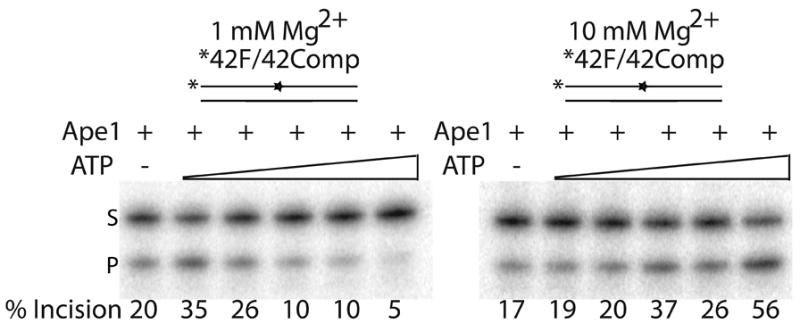
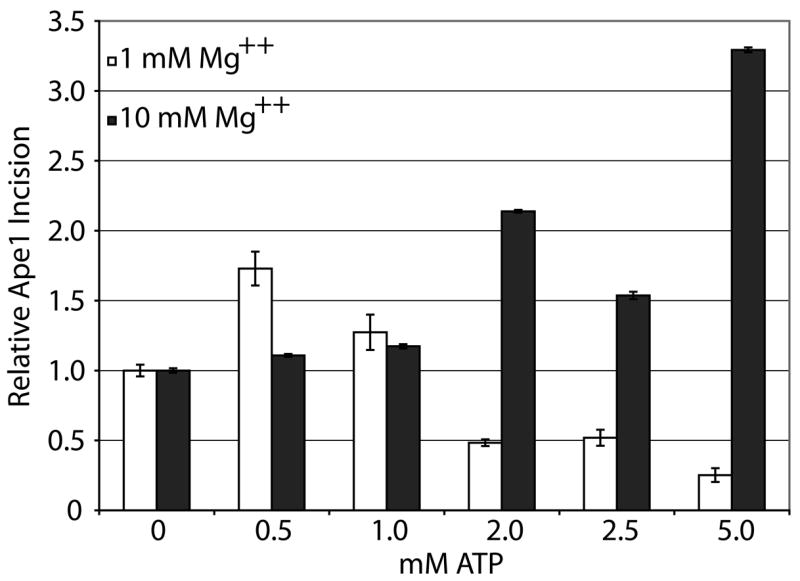
a. Representative denaturing polyacrylamide gel displaying the effects of ATP and Mg++ concentration on Ape1 incision activity. DNA (42F/42comp) is represented by solid lines and abasic site position is represented as a star. Asterisk denotes site of radiolabel. Ape1 concentration was constant with increasing concentrations of ATP (0, 0.5, 1, 2, 2.5, 5 mM) for 1 mM Mg++ (left) or 10 mM Mg++ (right). S denotes substrate position and P denotes product position. Percentage of substrate incised is indicated below each lane.
b. Graph of relative Ape1 incision activity vs. ATP concentration on DNA/DNA substrates. Average values with standard deviations of at least 3 independent experiments are plotted (White bars = 1 mM Mg++, Black bars = 10 mM Mg++).
Ape1 incises at AP sites within ssDNA regions of R-loops
As Ape1 has been implicated in incising at abasic sites during CSR26, we sought to examine the effect of a model R-loop structure on Ape1 incision efficiency. We employed the two AP site-containing 54mer oligos, i.e. 54Fendbubble and 54Fcenterbubble (Table 1), annealed to both 54bubblecomp and 18RNAfor54comp to create two R-loop intermediates (54Fendbubble/54bubblecomp/18RNAfor54comp and 54Fcenterbubble/54bubblecomp/18RNAfor54comp), which differed only in the location of the AP site within the single-stranded portion of the “pseudo-triplex” (Figure 4a). Ape1 was able to incise both substrates with similar efficiency (Figure 4b), indicating that Ape1 can cleave within structures that reflect R-loop intermediates formed during CSR. We note that addition of the RNA strand to the undamaged DNA strand of the “pseudo-triplex” lowered the specific activity of the enzyme ~3 to 14-fold, depending on the location of the abasic site.
Figure 4. Ape1 incises at AP sites in ssDNA region of R-loops.
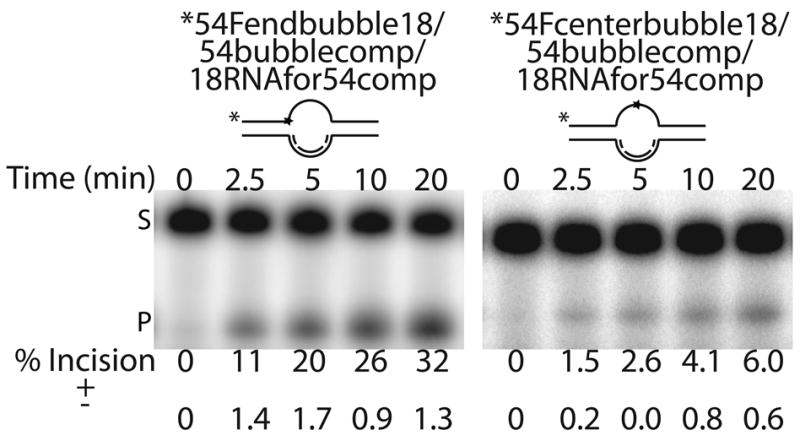
a. Representative denaturing polyacrylamide gels of AP site incision by Ape1 on R-loop substrates. DNA is represented by solid lines, RNA is represented by dashed lines, and abasic site position is represented as a star. Asterisk denotes site of radiolabel. Time points are shown on top and average percent of substrate converted to product is shown below for each time point analyzed along with standard deviations calculated from triplicate experiments. S denotes substrate position and P denotes product position.
b. Graph of Ape1 AP endonuclease specific activity, calculated from the 2.5 minute timepoint for the “pseudo-triplex” R-loop substrates. The specific activities for the two-stranded bubble substrates were derived from the data presented in Figure 2. Substrates are shown below with DNA represented by solid lines, RNA represented by dashed lines, and abasic site position represented as a star. Asterisk denotes site of radiolabel. Reactions with center-positioned abasic site contained 10 pg (0.3 fmol, 30 fM) Ape1 and reactions with junction-positioned abasic site contained 100 pg (3 fmol, 300 fM) Ape1.
Ape1 incises AP site-containing ssRNA
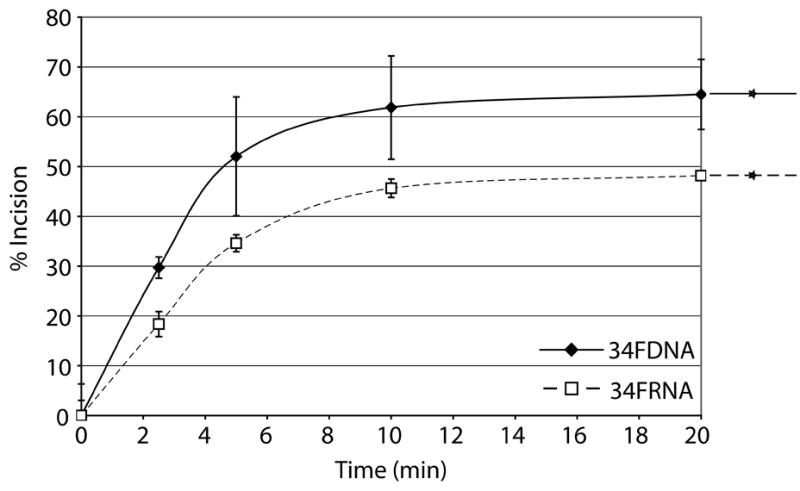
AlkB homologs have been shown to repair methyl lesions in RNA via oxidative demethylation in order to prevent damaged RNA from causing ribosome stalling or being translated20. As a single unrepaired MMS-induced lesion has the potential to block translation29, we sought to determine whether Ape1 had AP endonuclease activity on RNA containing a centrally located abasic site analog, as abasic sites are common alkylation products due to the increased rate of hydrolysis of the N-glycosidic bond12. We used two AP-ssRNAs, a 26mer and a 34mer, and two AP-ssDNAs of the same sequence (Table 1). Surprisingly, Ape1 incised the 26FRNA more efficiently then the 26FDNA (Figure 5A). With the 34mers, Ape1 incised the ssDNA much more efficiently then the ssRNA (Figure 5B; note the difference in Ape1 concentration used with the two substrates). Steady-state kinetic parameters for Ape1 incision of 34FDNA and 34FRNA are as follows: Km values were 179.2 nM and 1561.0 nM; Vmax values were 12.4 nM product min−1 and 61.4 nM product min−1; kcat values were 441.6 min−1 and 7.3 min−1; and catalytic efficiencies were 2.5 and 0.005, respectively.
Figure 5. Ape1 possesses the ability to cleave AP ssRNA.
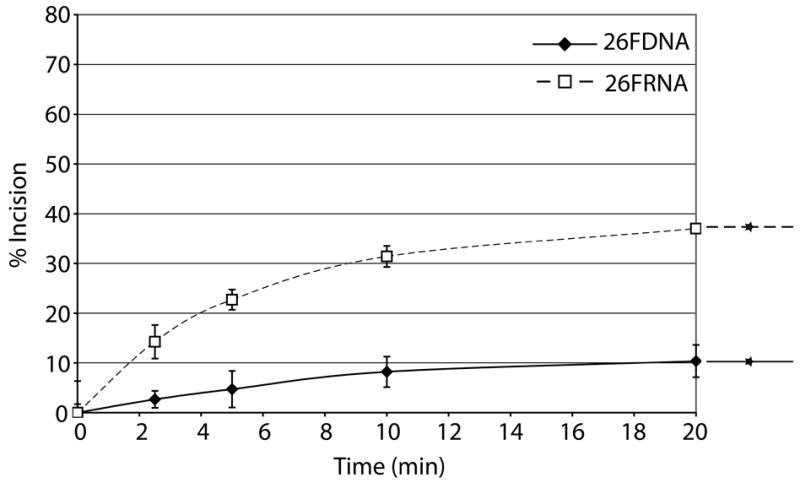
a. Graph depicting Ape1 time course kinetics on 26mer ssDNA and 26mer ssRNA. Average values with standard deviations of at least 3 independent experiments are plotted. Substrates are depicted next to the graph with DNA represented by solid lines, RNA represented by dashed lines, and abasic site position represented as a star.
b. Graph depicting Ape1 time course kinetics on 34mer ssDNA and 34mer ssRNA. Note that the reactions with 34FRNA contained 3 ng (90 fmol, 9 nM) Ape1, whereas 10 pg (0.3 fmol, 30 fM) was used with 34FDNA. See panel “a” for further details.
Neighboring sequence context has subtle effects on Ape1 incision efficiency
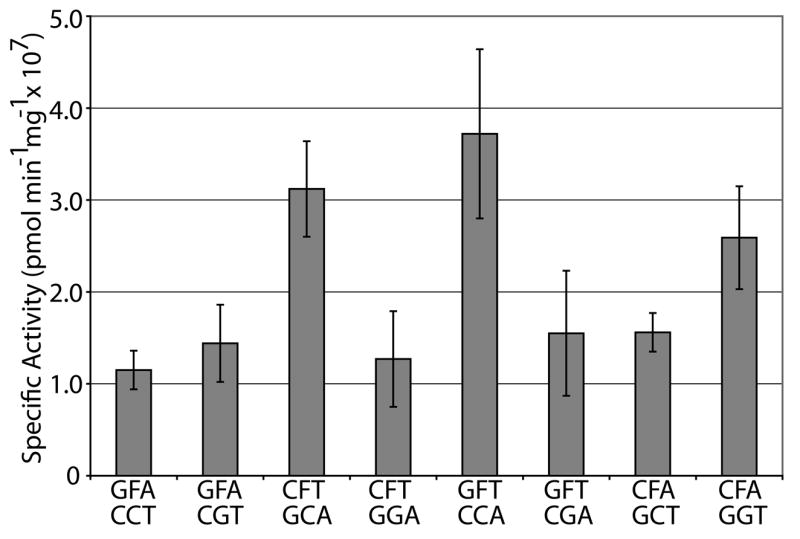
Sequence context surrounding an abasic site can affect the local structure of the DNA duplex, with the bases flanking and the base opposite the AP site having the most significant effect30. Generally speaking, AP sites opposite a purine base maintain B-form DNA with the purine base located within the helix, whereas AP sites opposed by a pyrimidine base can take on localized non-B-form DNA, with the pyrimidine base capable of extrahelical positioning depending on the stacking energies of the bases flanking the abasic site31. To determine the combined effect of the bases flanking and opposite an AP site on Ape1 incision efficiency, we utilized a set of AP site analog-containing 26mer oligonucleotides of the same sequence except for the five nucleotides surrounding the lesion. In particular, we examined substrates with either a purine (G) or a pyrimidine (C) positioned opposite the AP site and: 2 purines flanking the abasic site (GFA paired with CCT or CGT); 2 pyrimidines flanking (CFT with GCA or GGA); a 5′ purine and a 3′ pyrimidine flanking (GFT with CCA or CGA); or a 5′ pyrimidine and a 3′ purine flanking (CFA with GCT or GGT) (Figure 6a). Our results revealed no obvious unified pattern of incision efficiency with respect to the bases flanking or the base opposite the AP site. In short, Ape1 incision activity was 2 to 3-fold higher with the GFT/CCA, CFA/GCT and CFT/GGA arrangements relative to GFA/CCT, while Ape1 showed similar activity for the other sequence contexts (Figure 6b).
Figure 6. Sequence context immediately surrounding an AP site has little effect on Ape1 endonuclease activity.
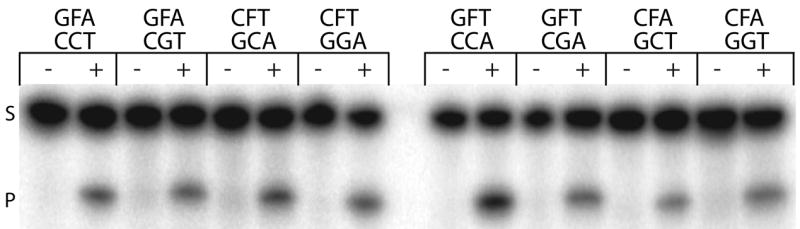
a. Representative denaturing polyacrylamide gel of AP site incision by Ape1 on duplex DNA/DNA containing different sequence contexts immediately surrounding the abasic site. Sequence of nucleotides immediately surrounding the abasic site (denoted as F) shown above lanes either not containing Ape1 (−) or containing Ape1 (+). See Table 1 for full sequence information of oligonucleotides. S denotes substrate position and P denotes product position.
b. Graph of specific activities of Ape1 on duplex DNA/DNA containing different sequence contexts immediately surrounding the abasic site. Average values with standard deviations of at least 3 independent experiments are plotted.
Discussion
We have demonstrated herein that human Ape1 can incise at AP sites in (i) the DNA strand of DNA/RNA hybrids, (ii) complex substrates that mimic a stalled replication or transcription intermediate, (iii) ssDNA regions of R-loop structures and (iv) single-stranded RNA molecules. In addition, we found that the nucleotide sequence surrounding an AP site, i.e. both immediately flanking and directly opposite, had a less than three-fold effect on Ape1 cleavage activity. This latter observation suggests that nucleotide sequence context is likely to have a minimal role in determining the biological impact of abasic sites within naked DNA, specifically in terms of repair-ability. The contribution of sequence context on AP site bypass synthesis (i.e. mutagenesis) is less clear.
Previous biochemical work had demonstrated that salt conditions can have a profound effect on the efficiency of Ape1 as an AP endonuclease or 3′ to 5′ exonuclease in vitro. For instance, both MgCl2 and KCl strongly influence the efficiency of Ape1 as a ssDNA or dsDNA AP endonuclease27. Furthermore, the endo- and exonuclease activity of Ape1 was found to be influenced by the MgCl2 concentration, with optimal exonuclease activity at low MgCl2 concentration (0.1–2 mM) and optimal endonuclease activity at high MgCl2 concentration (10–15 mM)32. We report here that depending on the relative concentration of MgCl2, ATP can have both inhibitory and stimulatory consequences on Ape1 incision capacity. Most notably, when the MgCl2:ATP ratio was 2:1, Ape1 AP endonuclease activity was highest. Under the same reaction conditions, other (d)NTPs were also found to stimulate Ape1 AP site incision activity.
Previous limited proteolysis and peptide mapping of native Ape1 revealed that chymotrypsin is able to cleave within the tight globular domain of the protein, unveiling 19.8 kDa (aa 1–179), 19.5 kDa (aa 174–318), and 12.8 kDa (aa 206–318) major digestion fragments33. When abasic-site containing DNA was incubated with Ape1 and chymotrypsin, inhibition of cleavage at the three major protease sites was observed, delaying Ape1 degradation. We found that when Ape1 was digested with chymotrypsin in the presence of ATP a similar delay was seen (Supplementary Figure 1), suggesting that DNA and ATP bind Ape1 in a similar manner. In line with this conclusion, dCTP has been detected in the crystal structure of the Ape1 ortholog, Escherichia coli ExoIII34. The dCTP moiety, as well as a Mn2+ ion, were specifically complexed within the enzyme active site, which is conserved in AP endonucleases from both pro- and eukaryotes. This observation may suggest that nucleotides, such as ATP, and Mg2+ can bind simultaneously to Ape1 and modulate its AP endonuclease function via a mechanism not fully understood.
The spectrum of AP site-containing DNA substrates for Ape1 has expanded in recent years. In particular, we had previously reported that Ape1 can cleave AP sites positioned within the single-stranded regions of fork and bubble configurations that mimic replication and transcription intermediates27, as well as at AP sites within single-stranded DNA molecules in a manner that is influenced by secondary structure35. Significantly, we have found that the ability to cleave at AP sites within single-stranded regions of bubble duplexes is evolutionarily conserved, as members of both families of AP endonucleases, i.e. E. coli ExoIII and EndoIV, incise efficiently at AP sites within the ssDNA region of the 11 nt bubble 42mer duplex DNA (42F/42bubbleComp, Table 1; Supplementary Figure 2). Indeed, both ExoIII (1.8 to 2.6-fold) and EndoIV (4.3 to 29.5-fold) exert greater AP endonuclease activity on an abasic site located in the single-stranded region of the bubble duplex relative to an abasic site located in a classical duplex substrate (42F/42comp). A similar preference for an abasic site located in the single-stranded region of a bubble duplex is seen with Ape1 (Figure 2a, 54Fcenterbubble/54comp vs. 54Fcenterbubble/18DNA).
We have demonstrated herein that Ape1 can incise at AP sites in more complex DNA and/or RNA intermediates. In particular, Ape1 can cleave AP sites in substrates that reflect a stalled DNA replication fork (54Fendbubble18/54bubblecomp/18DNA, Figure 2a). Creation of a three-strand containing “pseudo-triplex” intermediate, in fact, increased the specific activity of Ape1 ~3-fold over the partial double-stranded counterpart (54Fendbubble18/18DNA). This action of Ape1 may be promoted to avert error-prone bypass synthesis and instead create a one-ended DSB (following incision) that can be faithfully resolved by homologous recombination (HR)36. A role for HR in AP site repair is supported by genetic evidence in bacteria and yeast37–39. Alternatively, in the absence of AP site cleavage, or bypass synthesis, the leading DNA strand could be displaced (or degraded) to permit re-annealing of the downstream DNA, ultimately permitting classic template-driven BER prior to the resumption of replicative synthesis. Ape1, as well as other BER enzymes, has been found to be physically associated with components of the DNA replication machinery, including DNA polymerases α, δ, ε, DNA ligase I, MCM7, and cyclin A40, indicating that BER factors are intimately linked to actively elongating replication forks.
In addition to DNA polymerases stalling at a blocking lesion, the progression of RNA polymerases is also affected by the presence of DNA damage. Of particular interest is the fact that an abasic site in the transcribed strand has been identified as a potent block to RNA polymerase II progression41. We find that Ape1 exhibits significant activity on “pseudo-triplex” substrates designed to replicate a stalled transcription bubble. This finding suggests that a stalled transcription intermediate containing partially transcribed RNA has the potential to be acted upon by a BER enzyme. Whether BER substrates are processed via transcription-coupled repair (TCR) is controversial42, 43. That said, Ape1 has been shown to interact with the human TCR factor CSB, and this interaction enhances Ape1 AP site incision activity on 11 nt bubble substrates, suggesting that certain forms of endogenous DNA damage may be repaired in a gene-specific manner28. Even without invoking TCR, it seems reasonable that immediate processing of lesions that both stall RNA polymerase progression and potentially generate mutagenic RNA templates would be advantageous.
CSR involves transcription through Ig switch regions, which ultimately generates R-loop DNA/RNA hybrid structures between the transcribed strand and the nascent transcript, and leaves the untranscribed strand as ssDNA24. ssDNA is a preferred substrate on which AID acts to deaminate cytosine to uracil. While it is unclear how the uracil generated in DNA by AID is converted to DSBs, evidence suggests that the aberrant base is acted upon by uracil DNA glycosylase (UNG), leaving behind an abasic site that can be cleaved by Ape126 and/or the Mre11/Rad50/NBS1 (MRN) complex44 to create a single-strand break. A set of closely opposed nicks would create a DSB25. We demonstrate within that Ape1 possesses the ability to efficiently incise at AP sites in either the center or at the ssDNA/dsDNA junction of the single-stranded portion of an R-loop intermediate. Coupled with the fact that Ape1 can incise at AP sites within a DNA/RNA hybrid (Figure 1), the results herein unveil a plausible mechanism for generating DSBs during CSR.
We have shown for the first time that Ape1 possesses the ability to cleave AP sites within RNA molecules. For the 26mer substrates, Ape1 cleaved the AP-ssRNA over 5-fold more efficiently than the cognate AP-ssDNA. For the 34mer substrates, Ape1 cleaved the AP-ssDNA (which has been shown previously to be a very potent Ape1 substrate, even more so than double-stranded AP-DNA27, 35, 45), over 500-fold more efficiently than the AP-ssRNA. Prediction of substrate secondary structures revealed that although possessing the same nucleotide sequence (26mer ssDNA and 26mer ssRNA; 34mer ssDNA and 34mer ssRNA, Table 1), the RNA and DNA for each set has the potential to form different conformations (Figure 7). Previous work has shown that secondary structure significantly influences Ape1 ssDNA incision activity35, likely explaining the variability seen in incision activity for the disparate single-stranded oligonucleotides. RNA substrates are likely, in general, recognized with relatively low affinity, as suggested by the high Km value of Ape1 for 34FRNA (~1.5 μM).
Figure 7. Predicted secondary structure(s) of 26 and 34-mer ssDNA and ssRNA substrates.
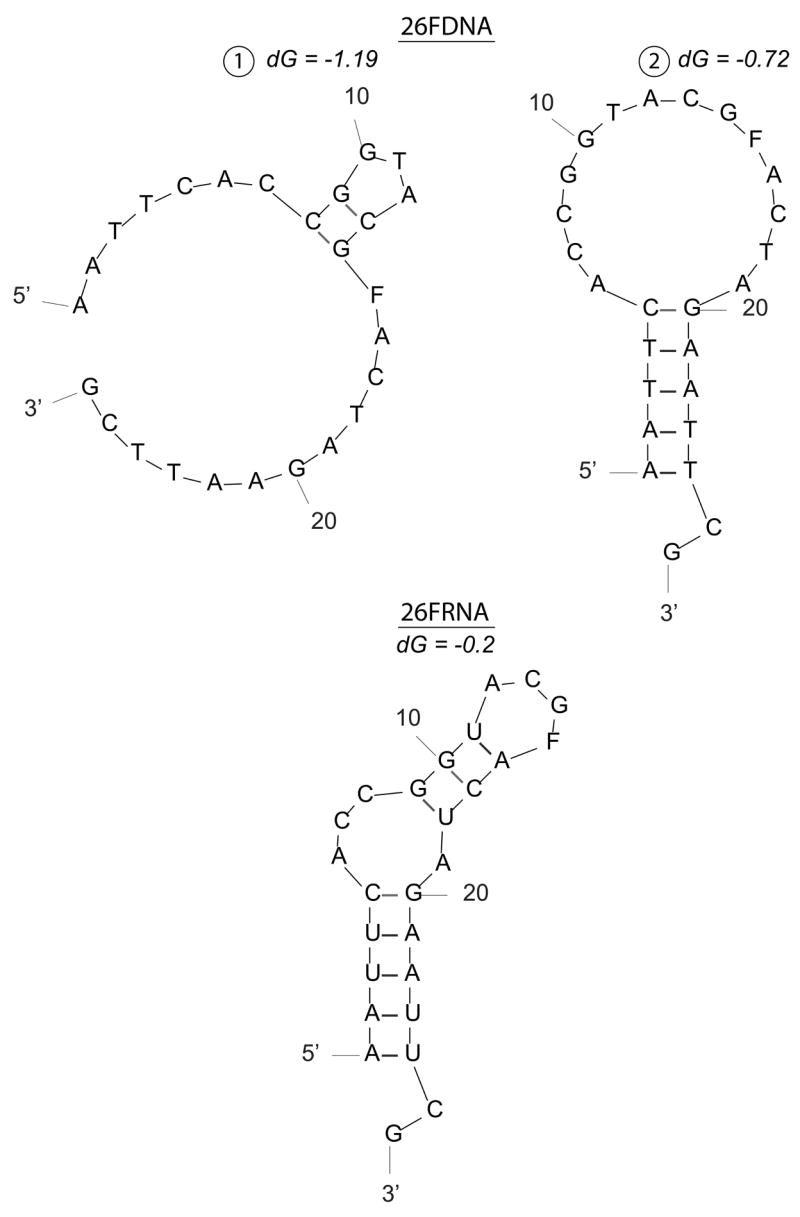
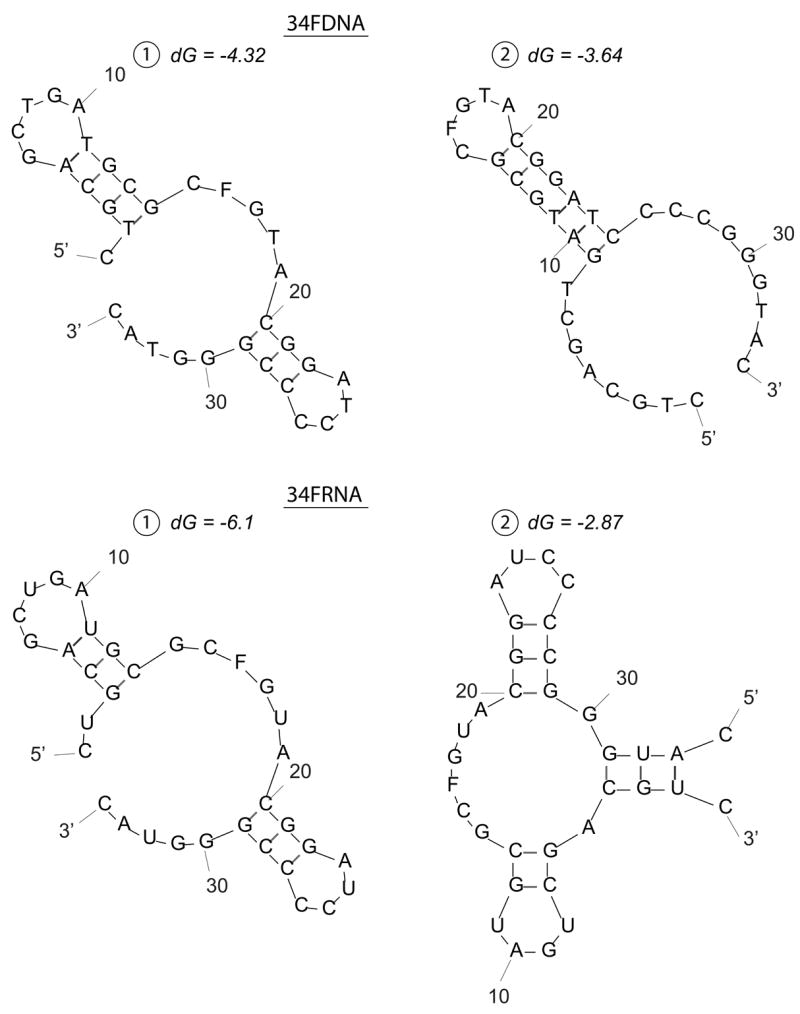
a. 26mer ssDNA and ssRNA predicted structures.
b. 34mer ssDNA and ssRNA predicted structures.
F denotes position of abasic site analog tetrahydrofuran. Secondary structures were predicted using mfold (http://frontend.bioinfo.rpi.edu/applications/mfold/) with standard prediction parameters. dG represents the minimum free energy for folding.
The ability to cleave AP site-containing RNA implies that Ape1 can remove damaged templates from the endogenous pool. It has previously been shown that RNA is oxidized to a greater extent than cellular DNA by treatment with H2O246, that abasic RNA intermediates can arise via the action of RNA N-ribohydrolases, including the toxin ricin47, and that oxidatively damaged mRNAs are present in high quantity in brains from Alzheimer’s patients48. Damaged mRNA either cannot be translated or is mistranslated to produce small peptides due to premature termination of translation or by proteolytic degradation of proteins containing destabilizing amino acid errors18. The ability of Ape1 to cleave abasic site-containing ssRNA provides a possible mechanism for the cell to eliminate damaged mRNA prior to association with the ribosome and execution of non-productive translation. Damage to rRNA can completely shut down protein synthesis16, and damage to tRNA can abolish the ability to read codons17. Taking into consideration the organized secondary structure of rRNA and tRNA molecules, Ape1 is a candidate to cleave abasic site-containing rRNA and tRNA molecules as well. Ape1 has been found in the nucleus and cytoplasm of a number of cell types, subcellular locations where RNA “cleansing” would presumably take place49.
Materials and Methods
Proteins and oligonucleotide substrates
Recombinant human Ape1 protein was purified as previously described50. Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) or Midland Certified Reagent Company, Inc. (Midland, TX). Oligonucleotide sequences are listed in Table 1. For AP site incision assays, oligonucleotides containing an abasic site analog (tetrahydrofuran) were radiolabeled at the 5′ end using [γ-32P] ATP and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). Substrates were annealed by heating at 95°C for 3 minutes and then cooling 2°C every 2 minutes to 4°C in a thermocycler. Annealing of the DNA or RNA “pseudo-triplex” strand was verified by native polyacrylamide electrophoresis, as shown in Supplementary Figure 3.
AP site incision assays
Unless otherwise specified, Ape1 was incubated with 5′ 32P labeled abasic site analog-containing oligonucleotide substrates at 37°C under physiologically-relevant reaction conditions (25 mM MOPS, pH 7.2;100 mM KCl; 1 mM MgCl2). Reactions were inhibited by the addition of stop buffer (95% formamide, 20 mM EDTA, 0.5% bromphenol blue, and 0.5% xylene cyanol), and then heated at 95°C for 5 minutes. Reaction products were resolved by 15% polyacrylamide urea denaturing gel electrophoresis, imaged, and quantified using a Typhoon phosphoimager and ImageQuant TL software (GE Healthcare, Piscataway, NJ).
Time course reactions with the 42F partial duplex substrates contained 500 pg (i.e. 15 fmol, 1.5 nM) Ape1 protein. Steady-state kinetic parameters were determined by incubating 500 pg of Ape1 with 1, 3, 10, 30, 100 or 300 nM DNA/DNA or DNA/RNA substrate for 2.5 minutes at 37°C. Reaction velocities were calculated following denaturing gel electrophoresis by phosphoimager analysis. Km and Vmax values were determined using Lineweaver-Burk plots. Time course reactions with the 54mer oligonucleotides contained 10 pg (0.3 fmol, 30 fM) (center positioned AP site) or 100 pg (3 fmol, 300 fM) (junction positioned AP site) Ape1. Reactions with the fully-paired 42mer dsDNA substrates contained either 1 mM or 10 mM MgCl2 and 30 pg (0.9 fmol, 90 pM) Ape1, respectively. Time course reactions with 26FDNA, 26FRNA, and 34FRNA contained 3 ng (90 fmol, 9 nM) Ape1, whereas reactions with 34FDNA contained 10 pg (0.3 fmol, 30 fM). Kinetic parameters for the 34mer ssDNA and ssRNA molecules were measured by incubating either 10 pg or 3 ng of Ape1 (see above) with 10, 30, 100, 300 or 600 nM substrate for 5 minutes at 37°C. Km and Vmax values were determined as above.
For sequence context reactions, buffer conditions used were 50 mM HEPES-KOH (pH 7.5), 50 mM KCl, 10% glycerol, 10 mM MgCl2, 0.1 mg/ml BSA, and 0.5% TritonX-100. Reactions contained 20 pg (0.6 fmol, 60 fM) Ape1, 1 pmol of substrate, and were incubated at 37°C for 10 min. The reactions were stopped by the addition of stop buffer and then heated at 95°C for 5 minutes. Reaction products were resolved by 18% polyacrylamide urea denaturing gel electrophoresis, imaged, and quantified as above.
Supplementary Material
Acknowledgments
We thank Dr. Heng-Kuan Wong (CNRS, Illkirch, France) for executing the incision studies involving the bacterial AP endonucleases. We thank Drs. P.J. Brooks, Lior Weissman, Tom Kulikowicz, and Melissa Hefferin for reading and critical input on the manuscript. This work was supported by the Intramural Research Program of the NIH, NIA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DM, III, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 3.Chen DS, Herman T, Demple B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barzilay G, Walker LJ, Robson CN, Hickson ID. Site-directed mutagenesis of the human DNA repair enzyme HAP1: identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res. 1995;23:1544–1550. doi: 10.1093/nar/23.9.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kane CM, Linn S. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J Biol Chem. 1981;256:3405–3414. [PubMed] [Google Scholar]
- 6.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 7.Wilson DM, III, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 8.Hadi MZ, Wilson DM., III Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutagen. 2000;36:312–324. [PubMed] [Google Scholar]
- 9.Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34:2508–2515. doi: 10.1093/nar/gkl259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 11.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 12.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 13.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Yu SL, Lee SK, Johnson RE, Prakash L, Prakash S. The stalling of transcription at abasic sites is highly mutagenic. Mol Cell Biol. 2003;23:382–388. doi: 10.1128/MCB.23.1.382-388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korennykh AV, Correll CC, Piccirilli JA. Evidence for the importance of electrostatics in the function of two distinct families of ribosome inactivating toxins. RNA. 2007;13:1391–1396. doi: 10.1261/rna.619707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc Natl Acad Sci U S A. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 20.Falnes PO, Klungland A, Alseth I. Repair of methyl lesions in DNA and RNA by oxidative demethylation. Neuroscience. 2007;145:1222–1232. doi: 10.1016/j.neuroscience.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 21.Shan X, Tashiro H, Lin CL. The identification and characterization of oxidized RNAs in Alzheimer’s disease. J Neurosci. 2003;23:4913–4921. doi: 10.1523/JNEUROSCI.23-12-04913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinet W, De Meyer GR, Herman AG, Kockx MM. Reactive oxygen species induce RNA damage in human atherosclerosis. Eur J Clin Invest. 2004;34:323–327. doi: 10.1111/j.1365-2362.2004.01343.x. [DOI] [PubMed] [Google Scholar]
- 23.Maizels N. Immunoglobulin gene diversification. Annu Rev Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 24.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 26.Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. J ExpMed. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson DM., III Ape1 abasic endonuclease activity is regulated by magnesium and potassium concentrations and is robust on alternative DNA structures. J Mol Biol. 2005;345:1003–1014. doi: 10.1016/j.jmb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 28.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., III Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Stivers JT. 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res. 1998;26:3837–3844. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barsky D, Foloppe N, Ahmadia S, Wilson DM, III, MacKerell AD., Jr New insights into the structure of abasic DNA from molecular dynamics simulations. Nucleic Acids Res. 2000;28:2613–2626. doi: 10.1093/nar/28.13.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou KM, Cheng YC. The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1) Biochemical properties and inhibition by the natural dinucleotide Gp4G J Biol Chem. 2003;278:18289–18296. doi: 10.1074/jbc.M212143200. [DOI] [PubMed] [Google Scholar]
- 33.Strauss PR, Holt CM. Domain mapping of human apurinic/apyrimidinic endonuclease. Structural and functional evidence for a disordered amino terminus and a tight globular carboxyl domain J Biol Chem. 1998;273:14435–14441. doi: 10.1074/jbc.273.23.14435. [DOI] [PubMed] [Google Scholar]
- 34.Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 35.Fan J, Matsumoto Y, Wilson DM., III Nucleotide sequence and DNA secondary structure, as well as replication protein A, modulate the single-stranded abasic endonuclease activity of APE1. J Biol Chem. 2006;281:3889–3898. doi: 10.1074/jbc.M511004200. [DOI] [PubMed] [Google Scholar]
- 36.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otterlei M, Kavli B, Standal R, Skjelbred C, Bharati S, Krokan HE. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 2000;19:5542–5551. doi: 10.1093/emboj/19.20.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendricks CA, Razlog M, Matsuguchi T, Goyal A, Brock AL, Engelward BP. The S. cerevisiae Mag1 3-methyladenine DNA glycosylase modulates susceptibility to homologous recombination. DNA Repair (Amst) 2002;1:645–659. doi: 10.1016/s1568-7864(02)00072-1. [DOI] [PubMed] [Google Scholar]
- 39.Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parlanti E, Locatelli G, Maga G, Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tornaletti S, Maeda LS, Hanawalt PC. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem Res Toxicol. 2006;19:1215–1220. doi: 10.1021/tx060103g. [DOI] [PubMed] [Google Scholar]
- 42.Hanawalt PC. DNA repair. The bases for Cockayne syndrome Nature. 2000;405:415–416. doi: 10.1038/35013197. [DOI] [PubMed] [Google Scholar]
- 43.Cozzarelli NR. Editorial expression of concern. Proc Natl Acad Sci U S A. 2003;100:11816. doi: 10.1073/pnas.2034938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larson ED, Cummings WJ, Bednarski DW, Maizels N. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Mol Cell. 2005;20:367–375. doi: 10.1016/j.molcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 45.Marenstein DR, Wilson DM, III, Teebor GW. Human AP endonuclease (APE1) demonstrates endonucleolytic activity against AP sites in single-stranded DNA. DNA Repair (Amst) 2004;3:527–533. doi: 10.1016/j.dnarep.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Hofer T, Badouard C, Bajak E, Ravanat JL, Mattsson A, Cotgreave IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol Chem. 2005;386:333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 47.Schramm VL. Enzymatic N-riboside scission in RNA and RNA precursors. Curr Opin Chem Biol. 1997;1:323–331. doi: 10.1016/s1367-5931(97)80069-5. [DOI] [PubMed] [Google Scholar]
- 48.Shan X, Chang Y, Lin CL. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007;21:2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 49.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 50.Erzberger JP, Barsky D, Scharer OD, Colvin ME, Wilson DM., III Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 1998;26:2771–2778. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


